Long Noncoding RNA in Myeloid and Lymphoid Cell Differentiation, Polarization and Function
Abstract
:1. Introduction
2. Myeloid Cells
2.1. A. Macrophages
2.2. Dendritic Cells
3. Lymphoid Cells
3.1. T Cells
3.1.1. T helper (Th) Cells
3.1.2. CD8+ T Cells
3.1.3. Regulatory T cells (Tregs)
3.2. B Cells
3.3. Natural Killer Cells
4. LncRNAs in Immune-Related Diseases
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.M.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Salzberg, S.L. Open questions: How many genes do we have? BMC Boil. 2018, 16. [Google Scholar] [CrossRef]
- Dempsey, J.L.; Cui, J.Y. Long noncoding RNAs: A novel paradigm for toxicology. Toxicol. Sci. 2017, 155, 3–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Losko, M.; Kotlinowski, J.; Jura, J. Long Noncoding RNAs in Metabolic Syndrome Related Disorders. Mediat. Inflamm. 2016, 2016, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Böcker, W.; Brosius, J.; Tiedge, H. Expression of neural BC200 RNA in human tumours. J. Pathol. 1997, 183, 345–351. [Google Scholar] [CrossRef]
- Hawkins, P.G.; Morris, K.V. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription 2010, 1, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Perry, R.B.-T.; Ulitsky, I. The functions of long noncoding RNAs in development and stem cells. Development 2016, 143, 3882–3894. [Google Scholar] [CrossRef] [Green Version]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef]
- Gibb, E.A.; Brown, C.J.; Lam, W.L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer 2011, 10, 38. [Google Scholar] [CrossRef] [Green Version]
- Sigdel, K.R.; Cheng, A.; Wang, Y.; Duan, L.; Zhang, Y. The Emerging Functions of Long Noncoding RNA in Immune Cells: Autoimmune Diseases. J. Immunol. Res. 2015, 2015, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Rossi, M.N.; Antonangeli, F. LncRNAs: New Players in Apoptosis Control. Int. J. Cell Boil. 2014, 2014, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206. [Google Scholar] [CrossRef] [Green Version]
- Dhanoa, J.K.; Sethi, R.S.; Verma, R.; Arora, J.S.; Mukhopadhyay, C.S. Long non-coding RNA: Its evolutionary relics and biological implications in mammals: A review. J. Anim. Sci. Technol. 2018, 60, 25. [Google Scholar] [CrossRef] [Green Version]
- Salviano-Silva, A.; Lobo-Alves, S.C.; De Almeida, R.C.; Malheiros, D.; Petzl-Erler, M.L. Besides Pathology: Long Non-Coding RNA in Cell and Tissue Homeostasis. Non-Coding RNA 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, M. Lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. Immunol. Rev. 2010, 238, 37–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atianand, M.K.; Fitzgerald, K.A. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol. Med. 2014, 20, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Kotzin, J.J.; Spencer, S.P.; McCright, S.J.; Kumar, D.B.U.; Collet, M.A.; Mowel, W.K.; Elliott, E.N.; Uyar, A.; Makiya, M.A.; Dunagin, M.C.; et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature 2016, 537, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Tian, J.; Tang, X.; Ma, J.; Wang, S. Long non-coding RNAs in the regulation of myeloid cells. J. Hematol. Oncol. 2019, 9, 99. [Google Scholar] [CrossRef] [Green Version]
- Amarante, M.K.; De Lucca, F.L.; De Oliveira, C.E.C.; Fungaro, M.H.P.; Reiche, E.M.V.; Muxel, S.M.; Watanabe, M.A.E. Expression of noncoding mRNA in human blood cells activated with synthetic peptide of HIV. Blood Cells Mol. Dis. 2005, 35, 286–290. [Google Scholar] [CrossRef]
- Liu, A.Y.T.; Torchia, B.S.; Migeon, B.R.; Siliciano, R.F. The human NTT gene: Identification of a novel 17-kb noncoding nuclear RNA expressed in activated CD4+ T cells. Genomics 1997, 39, 171–184. [Google Scholar] [CrossRef]
- Yang, C.-A.; Li, J.-P.; Yen, J.-C.; Lai, I.-L.; Ho, Y.-C.; Chen, Y.-C.; Lan, J.-L.; Chang, J.-G. LncRNA NTT/PBOV1 Axis Promotes Monocyte Differentiation and Is Elevated in Rheumatoid Arthritis. Int. J. Mol. Sci. 2018, 19, 2806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krawczyk, M. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-kappaB complexes. eLife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Sierra-Filardi, E.; Puig-Kröger, A.; Blanco, F.-J.; Nieto, C.; Bragado, R.; Palomero, M.I.; Bernabeu, C.; Vega, M.A.; Cor´bi, A.L. Activin A skews macrophage polarization by promoting a proinflammatory phenotype and inhibiting the acquisition of anti-inflammatory macrophage markers. Blood 2011, 117, 5092–5101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salogni, L.; Musso, T.; Bosisio, D.; Mirolo, M.; Jala, V.R.; Haribabu, B.; Locati, M.; Sozzani, S. Activin A induces dendritic cell migration through the polariz ed release of CXC chemokine ligands 12 and 14. Blood 2009, 113, 5848–5856. [Google Scholar] [CrossRef] [Green Version]
- Escribese, M.M.; Sierra-Filardi, E.; Nieto, C.; Samaniego, R.; Sanchez-Torres, C.; Matsuyama, T.; Calderón-Gómez, E.; Vega, M.A.; Salas, A.; Sánchez-Mateos, P.; et al. The Prolyl Hydroxylase PHD3 Identifies Proinflammatory Macrophages and Its Expression Is Regulated by Activin A. J. Immunol. 2012, 189, 1946–1954. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.-T.; Lin, H.-S.; Shen, C.; Ma, Y.-N.; Wang, F.; Zhao, H.-L.; Yu, J.; Zhang, J.-W. PU.1-Regulated Long Noncoding RNA lnc-MC Controls Human Monocyte/Macrophage Differentiation through Interaction with MicroRNA 199a-5p. Mol. Cell. Biol. 2015, 35, 3212–3224. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tian, J.; Wang, S. The potential therapeutic role of myeloid-derived suppressor cells in autoimmune arthritis. Semin. Arthritis Rheum. 2016, 45, 490–495. [Google Scholar] [CrossRef]
- Reddy, M.A.; Chen, Z.; Park, J.T.; Wang, M.; Lanting, L.; Zhang, Q.; Bhatt, K.; Leung, A.; Wu, X.; Putta, S.; et al. Regulation of Inflammatory Phenotype in Macrophages by a Diabetes-Induced Long Noncoding RNA. Diabetes 2014, 63, 4249–4261. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Luo, Q.; Yao, F.; Qing, C.; Ye, J.; Deng, Y.; Li, J. Identification of Differentially Expressed Long Non-coding RNAs in Polarized Macrophages. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Obaid, M.; Udden, S.M.N.; Deb, P.; Shihabeddin, N.; Zaki, H.; Mandal, S.S. LncRNA HOTAIR regulates lipopolysaccharide-induced cytokine expression and inflammatory response in macrophages. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Xue, Y.; Han, Y.; Lin, L.; Wu, C.; Xu, S.; Jiang, Z.; Xu, J.; Liu, Q.; Cao, X. The STAT3-Binding Long Noncoding RNA lnc-DC Controls Human Dendritic Cell Differentiation. Science 2014, 344, 310–313. [Google Scholar] [CrossRef] [PubMed]
- Xin, J.; Li, J.; Feng, Y.; Wang, L.; Zhang, Y.; Yang, R. Downregulation of long noncoding RNA HOTAIRM1 promotes monocyte/dendritic cell differentiation through competitively binding to endogenous miR-3960. OncoTargets Ther. 2017, 10, 1307–1315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Zhang, H.; Zheng, Y.; Jin, X.; Liu, M.; Li, S.; Zhao, Q.; Liu, X.; Wang, Y.; Shi, M.; et al. The Long Noncoding RNA MALAT1 Induces Tolerogenic Dendritic Cells and Regulatory T Cells via miR155/Dendritic Cell-Specific Intercellular Adhesion Molecule-3 Grabbing Nonintegrin/IL10 Axis. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Zheng, Y.; Sun, Y.; Li, S.; Chen, L.; Jin, X.; Hou, X.; Liu, X.; Chen, Q.; Li, J.; et al. Knockdown of NEAT1 induces tolerogenic phenotype in dendritic cells by inhibiting activation of NLRP3 inflammasome. Theranostics 2019, 9, 3425–3442. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Shuang, L.; Zheng, Y.; Zhang, M.; Zhang, H.; Sun, Y.; Yu, B. Silencing of lncRNA NEAT1 induces tolerogenic dendritic cells and immune tolerance in heart transplantation. J. Immunol. 2017, 198, 82.27. [Google Scholar]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 39. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Wu, L.; Qian, J.; Qu, B.; Xia, S.; La, T.; Wu, Y.; Ma, J.; Zeng, J.; Guo, Q.; et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J. Autoimmun. 2016, 75, 96–104. [Google Scholar] [CrossRef]
- Riva, P.; Ratti, A.; Venturin, M. The Long Non-Coding RNAs in Neurodegenerative Diseases: Novel Mechanisms of Pathogenesis. Curr. Alzheimer Res. 2016, 13, 1219–1231. [Google Scholar] [CrossRef]
- Chen, T.; Wang, H.; Yang, P.; He, Z.-Y. Prognostic role of long noncoding RNA NEAT1 in various carcinomas: A meta-analysis. OncoTargets Ther. 2017, 10, 993–1000. [Google Scholar] [CrossRef] [Green Version]
- Sutterwala, F.S.; Ogura, Y.; Szczepanik, M.; Lara-Tejero, M.; Lichtenberger, G.S.; Grant, E.P.; Bertin, J.; Coyle, A.J.; Galán, J.E.; Askenase, P.W.; et al. Critical Role for NALP3/CIAS1/Cryopyrin in Innate and Adaptive Immunity through Its Regulation of Caspase-1. Immunity 2006, 24, 317–327. [Google Scholar] [CrossRef] [Green Version]
- Brahic, M.; Bureau, J.F.; Michiels, T. The genetics of the persistent infection and demyelinating disease caused by Theiler’s virus. Annu. Rev. Microbiol. 2005, 59, 279–298. [Google Scholar] [CrossRef] [PubMed]
- Vigneau, S.; Levillayer, F.; Crespeau, H.; Cattolico, L.; Caudron, B.; Bihl, F.; Robert, C.; Brahic, M.; Weissenbach, J.; Bureau, J.-F. Homology between a 173-kb Region from Mouse Chromosome 10, Telomeric to the Ifng Locus, and Human Chromosome 12q15. Genomics 2001, 78, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Vigneau, S.; Rohrlich, P.S.; Brahic, M.; Bureau, J.F. Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J. Virol. 2003, 77, 5632–5638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collier, S.P.; Collins, P.L.; Williams, C.L.; Boothby, M.R.; Aune, T.M. Cutting edge: Influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J. Immunol. 2012, 189, 2084–2088. [Google Scholar] [CrossRef] [Green Version]
- Gomez, J.A.; Wapinski, O.L.; Yang, Y.W.; Bureau, J.F.; Gopinath, S.; Monack, D.M.; Chang, H.Y.; Brahic, M.; Kirkegaard, K. The NeST long ncRNA controls micro-bial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 2013, 152, 743–754. [Google Scholar] [CrossRef] [Green Version]
- Collier, S.P.; Henderson, M.A.; Tossberg, J.T.; Aune, T.M. Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. J. Immunol. 2014, 193, 3959–3965. [Google Scholar] [CrossRef] [Green Version]
- Koh, B.H.; Hwang, S.S.; Kim, J.Y.; Lee, W.; Kang, M.-J.; Lee, C.G.; Park, J.-W.; Flavell, R.A.; Lee, G.R. Th2 LCR is essential for regulation of Th2 cytokine genes and for pathogenesis of allergic asthma. Proc. Natl. Acad. Sci. USA 2010, 107, 10614–10619. [Google Scholar] [CrossRef] [Green Version]
- Spurlock, C.F.; Tossberg, J.T.; Guo, Y.; Collier, S.P.; Crooke, P.S.; Aune, T.M. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef]
- Hu, G.; Tang, Q.; Sharma, S.; Yu, F.; Escobar, T.M.; Muljo, S.A.; Zhu, J.; Zhao, K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 2013, 14, 1190–1198. [Google Scholar] [CrossRef] [Green Version]
- Ranzani, V.; Rossetti, G.; Panzeri, I.; Arrigoni, A.; Bonnal, R.J.P.; Curti, S.; Gruarin, P.; Provasi, E.; Sugliano, E.; Marconi, M.; et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat. Immunol. 2015, 16, 318–325. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Liu, G.; Wei, C.; Gao, C.; Hao, J. Linc-MAF-4 regulates Th1/Th2 differentiation and is associated with the pathogenesis of multiple sclerosis by targeting MAF. FASEB J. 2017, 31, 519–525. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhong, H.; Xie, X.; Chen, C.Y.; Huang, D.; Shen, L.; Zhang, H.; Chen, Z.W.; Zeng, G. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc. Natl. Acad. Sci. USA 2015, 112, E3883–E3892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willingham, A.T. A Strategy for Probing the Function of Noncoding RNAs Finds a Repressor of NFAT. Science 2005, 309, 1570–1573. [Google Scholar] [CrossRef] [PubMed]
- Okamura, H.; Aramburu, J.; García-Rodríguez, C.; Viola, J.P.; Raghavan, A.; Tahiliani, M.; Zhang, X.; Qin, J.; Hogan, P.G.; Rao, A. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol. Cell 2000, 6, 539–550. [Google Scholar] [CrossRef]
- Sharma, S.; Findlay, G.M.; Bandukwala, H.S.; Oberdoerffer, S.; Baust, B.; Li, Z.; Schmidt, V.; Hogan, P.G.; Sacks, D.B.; Rao, A. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc. Natl. Acad. Sci. USA 2011, 108, 11381–11386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Lee, J.; Krummey, S.; Lu, W.; Cai, H.; Lenardo, M.J. The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat. Immunol. 2011, 12, 1063–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondělková, K.; Vokurková, D.; Krejsek, J.; Borská, L.; Fiala, Z.; Andrýs, C. Regulatory T cells (Treg) and Their Roles in Immune System with Respect to Immunopathological Disorders. Acta Med. 2010, 53, 73–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zemmour, D.; Pratama, A.; Loughhead, S.M.; Mathis, D.; Benoist, C. Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc. Natl. Acad. Sci. USA 2017, 114, E3472–E3480. [Google Scholar] [CrossRef] [Green Version]
- Brajic, A.; Franckaert, D.; Burton, O.; Bornschein, S.; Calvanese, A.L.; Demeyer, S.; Cools, J.; Dooley, J.; Schlenner, S.; Liston, A. The Long Non-coding RNA Flatr Anticipates Foxp3 Expression in Regulatory T Cells. Front. Immunol. 2018, 9, 25. [Google Scholar] [CrossRef]
- Hystad, M.E.; Myklebust, J.H.; Bø, T.H.; Sivertsen, E.A.; Rian, E.; Forfang, L.; Munthe, E.; Rosenwald, A.; Chiorazzi, M.; Jonassen, I.; et al. Characterization of early stages of human B cell development by gene expression profiling. J. Immunol. 2007, 179, 3662–3671. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, R.; Seidl, T.; Neeb, M.; Rolink, A.; Melchers, F. Changes in Gene Expression Profiles in Developing B Cells of Murine Bone Marrow. Genome Res. 2002, 12, 98–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maizels, N. IMMUNOGLOBULIN GENE DIVERSIFICATION. Annu. Rev. Genet. 2005, 39, 23–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petri, A.; Dybkær, K.; Bøgsted, M.; Thrue, C.A.; Hagedorn, P.H.; Schmitz, A.; Bødker, J.S.; Johnsen, H.E.; Kauppinen, S. Long Noncoding RNA Expression during Human B-Cell Development. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nutt, S.L.; Heavey, B.; Rolink, A.G.; Busslinger, M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 1999, 7, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Verma-Gaur, J.; Torkamani, A.; Schaffer, L.; Head, S.R.; Schork, N.J.; Feeney, A.J. Noncoding transcription within the Igh distal V(H) region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc. Natl. Acad. Sci. USA 2012, 109, 17004–17009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodoen, M.B.; Lanier, L.L. Natural killer cells as an initial defense against pathogens. Curr. Opin. Immunol. 2006, 18, 391–398. [Google Scholar] [CrossRef]
- Poli, A.; Michel, T.; Thérésine, M.; Andrès, E.; Hentges, F.; Zimmer, J. CD56bright natural killer (NK) cells: An important NK cell subset. Immunology 2009, 126, 458–465. [Google Scholar] [CrossRef]
- Moretta, L. Dissecting CD56dim human NK cells. Blood 2010, 116, 3689–3691. [Google Scholar] [CrossRef] [Green Version]
- Long, E.O.; Kim, H.S.; Liu, N.; Peterson, M.E.; Rajagopalan, S. Controlling natural killer cell responses: Integration of signals for activation and inhibition. Annu. Rev. Immunol. 2013, 31, 227–258. [Google Scholar] [CrossRef] [Green Version]
- Van Acker, H.H.; Capsomidis, A.; Smits, E.L.; Van Tendeloo, V.F. CD56 in the Immune System: More Than a Marker for Cytotoxicity? Front. Immunol. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Zhang, R.; Ni, F.; Fu, B.; Wu, Y.; Sun, R.; Tian, Z.; Wei, H. A long noncoding RNA positively regulates CD56 in human natural killer cells. Oncotarget 2016, 7, 72546–72558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, N.; Berhani, O.; Schmiedel, D.; Duev-Cohen, A.; Seidel, E.; Kol, I.; Tsukerman, P.; Hecht, M.; Reches, A.; Gamliel, M.; et al. IFNG-AS1 Enhances Interferon Gamma Production in Human Natural Killer Cells. iScience 2019, 11, 466–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricaño-Ponce, I.; Wijmenga, C. Mapping of Immune-Mediated Disease Genes. Annu. Rev. Genom. Hum. Genet. 2013, 14, 325–353. [Google Scholar] [CrossRef] [PubMed]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpenter, S. Long noncoding RNA: Novel links between gene expression and innate immunity. Virus Res. 2016, 212, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Westra, H.-J.; Karjalainen, J.; Zhernakova, D.V.; Esko, T.; Hrdlickova, B.; Almeida, R.; Zhernakova, A.; Reinmaa, E.; Võsa, U.; et al. Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding RNA Expression. PLoS Genet. 2013, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Kim, D.; Han, J.; Kim, Y.; Lee, M.; Jin, E.J. PBMC and exosome derived hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin. Exp. Med. 2015, 15, 121–126. [Google Scholar] [CrossRef]
- Stuhlmüller, B.; Kunisch, E.; Franz, J.; Martínez-Gamboa, L.; Hernández, M.M.; Pruss, A.; Ulbrich, N.; Erdmann, V.A.; Burmester, G.R.; Kinne, R.W. Detection of Oncofetal H19 RNA in Rheumatoid Arthritis Synovial Tissue. Am. J. Pathol. 2003, 163, 901–911. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Zhang, Z.; Yu, A.M.; Wang, W.; Wei, Z.; Akhter, E.; Maurer, K.; Reis, P.C.; Song, L.; Petri, M.; et al. The SLE Transcriptome Exhibits Evidence of Chronic Endotoxin Exposure and Has Widespread Dysregulation of Non-Coding and Coding RNAs. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Hrdlickova, B.; Kumar, V.; Kanduri, K.; Zhernakova, D.V.; Tripathi, S.; Karjalainen, J.; Lund, R.J.; Li, Y.; Ullah, U.; Modderman, R.; et al. Expression profiles of long non-coding RNAs located in autoimmune disease associated regions reveal immune cell-type specificity. Genome Med. 2014, 6, 88. [Google Scholar] [CrossRef]
- Chang, D.; Clayton, D. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science 1987, 235, 1178–1184. [Google Scholar] [CrossRef] [PubMed]
- Maida, Y.; Yasukawa, M.; Furuuchi, M.; Lassmann, T.; Possemato, R.; Okamoto, N.; Kasim, V.; Hayashizaki, Y.; Hahn, W.C.; Masutomi, K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature 2009, 461, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Liu, S.; Fan, J.; Jin, Y.; Tian, B.; Zheng, X.; Fu, H. Nuclear retention of the lnc RNA SNHG 1 by doxorubicin attenuates hnRNPC–p53 protein interactions. EMBO Rep. 2017, 18, 536–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrieri, C.; Cimatti, L.; Biagioli, M.; Beugnet, A.; Zucchelli, S.; Fedele, S.; Pesce, E.; Ferrer, I.; Collavin, L.; Santoro, C.; et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012, 491, 454–457. [Google Scholar] [CrossRef]
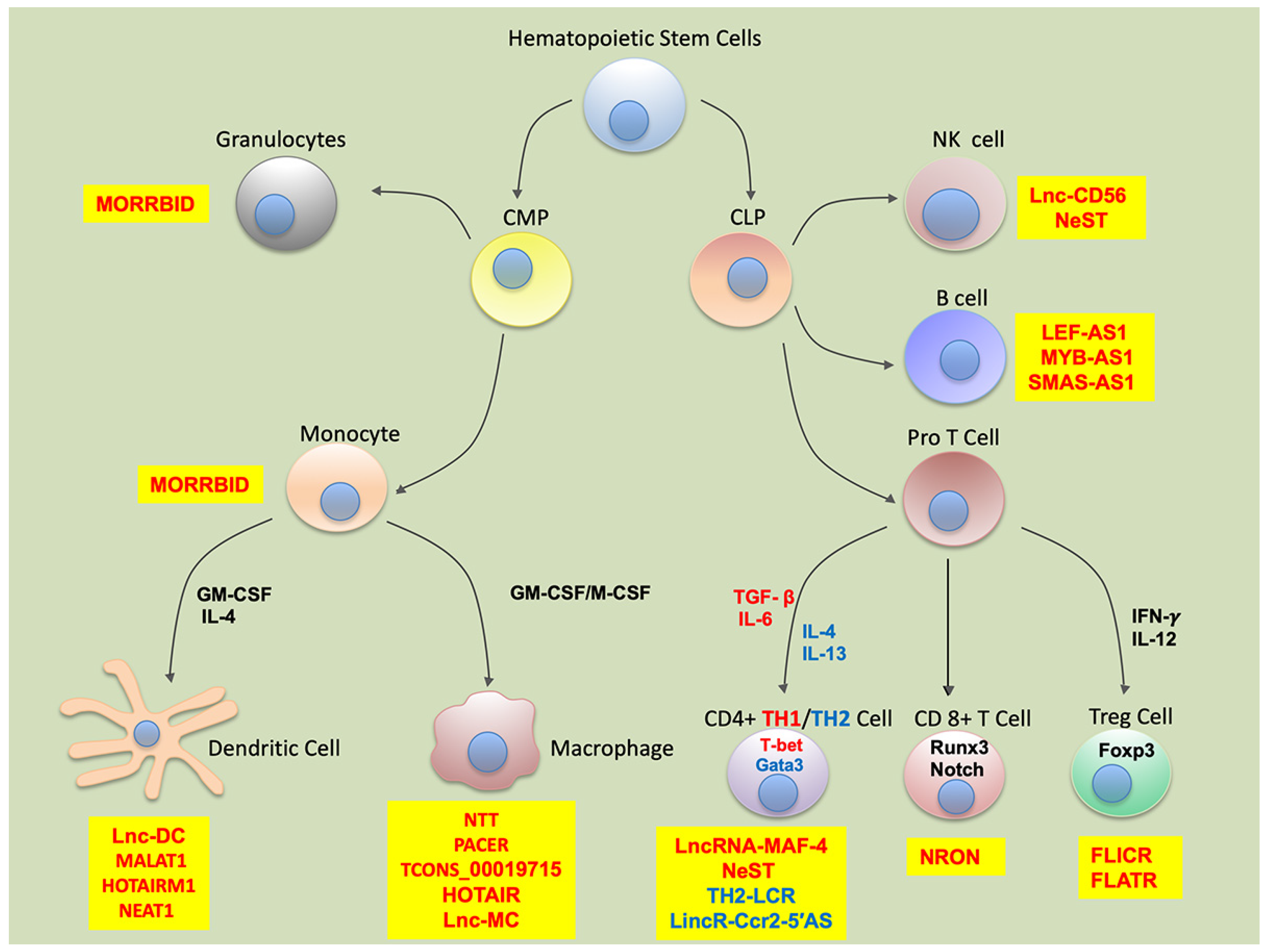


| Myeloid Cells | |||
|---|---|---|---|
| LncRNA | Cell Type | Function | Refs. |
| Morrbid | Myeloid cells | Controls the survival of short-lived myeloid cells by cis regulation of Bcl2l11 expression | Kotzin, 2016 |
| NTT | Monocytes | Binds to promoter of PBOV1 via hnRNP-, U promotes cell cycle arrest, differentiation into M0M2, increase in IL-10, CXCL10 mRNA levels, and upregulation of the costimulatory molecules | Yang, 2018 |
| PACER | Monocytes | Binds to and titrates the repressive NF-B1 homodimer away from the COX2 promoter, thereby facilitating binding of the activating RELA/NF-B1 heterodimer and subsequent formation of transcription preinitiation complexes | Krawczyk, 2014 |
| Lnc-MC | Monocytes | Facilitates the differentiation of monocytes by enhancing the effect of PU.1 and sequestering miR-199a-5p and increasing the expression of ACVR1B | Chen, 2015 |
| TCONS_00019715 | Monocytes | Promotes macrophage polarization towards pro-inflammatory (M1) phenotype | Huang, 2016 |
| HOTAIR | Monocytes | Enhances proinflammatory NFκB signaling by promoting IκBα degradation | Obaid, 2018 |
| Lnc-DC | Dendritic cells | Promotes STAT3 signaling by interacting with the C terminus of STAT3 to prevent the dephosphorylation of STAT3 Y705 by SHP1 | Wang, 2014 |
| HOTAIRM1 | Dendritic cells | Promotes monocyte/dendritic cell differentiation through competitively binding to endogenous miR-3960 | Xin, 2017 |
| NEAT1 | Dendritic cells | Induces tolerogenic phenotype in DC and promotes Treg polarization by inhibiting NLRP3 via sequestering miR-3076-3p | Zhang, 2019 Wu, 2017 |
| Malat1 | Dendritic cells | Induces tolerogenic phenotype in DC and promote Treg polarization by sponging miR-155 and upregulating PU.1 expression | Wu, 2018 |
| Lymphoid Cells | |||
| LncRNA | Cell Type | Function | Refs. |
| NeST | CD8+ T cell and TH1 cell | Binds to WDR5 and regulate the expression of IFN-gamma by recruiting transcription activation complex to IFN-gamma promoter | Gomez, 2013 Collier, 2014 |
| NRON | T cell | Sequester phosphorylated NFAT in cytoplasm of resting T cells | Willingham, 2005; Okamura, 2002; Sharma, 2001 |
| TH2-LCR | TH2 cell | Regulates the expression of TH2 cell cytokines, including IL-4, IL-5, and IL-13 | Koh, 2010; Spurlock, 2015 |
| LncRNA-CD244 | CD8+ T cell | Inhibits expression of IFNG and TNF by recruiting EZH2 to their promoters | Wang, 2015 |
| Linc-MAF-4 | TH1 cell | Regulates the expression of MAF and promote TH1 differentiation | Ranzani, 2015 Zhang, 2017 |
| LincR-Ccr2-5′AS | TH2 cell | Regulate the expression of TH2 cytokines | Hu, 2013 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, I.; Valverde, A.; Ahmad, F.; Naqvi, A.R. Long Noncoding RNA in Myeloid and Lymphoid Cell Differentiation, Polarization and Function. Cells 2020, 9, 269. https://doi.org/10.3390/cells9020269
Ahmad I, Valverde A, Ahmad F, Naqvi AR. Long Noncoding RNA in Myeloid and Lymphoid Cell Differentiation, Polarization and Function. Cells. 2020; 9(2):269. https://doi.org/10.3390/cells9020269
Chicago/Turabian StyleAhmad, Imran, Araceli Valverde, Fayek Ahmad, and Afsar Raza Naqvi. 2020. "Long Noncoding RNA in Myeloid and Lymphoid Cell Differentiation, Polarization and Function" Cells 9, no. 2: 269. https://doi.org/10.3390/cells9020269
APA StyleAhmad, I., Valverde, A., Ahmad, F., & Naqvi, A. R. (2020). Long Noncoding RNA in Myeloid and Lymphoid Cell Differentiation, Polarization and Function. Cells, 9(2), 269. https://doi.org/10.3390/cells9020269





