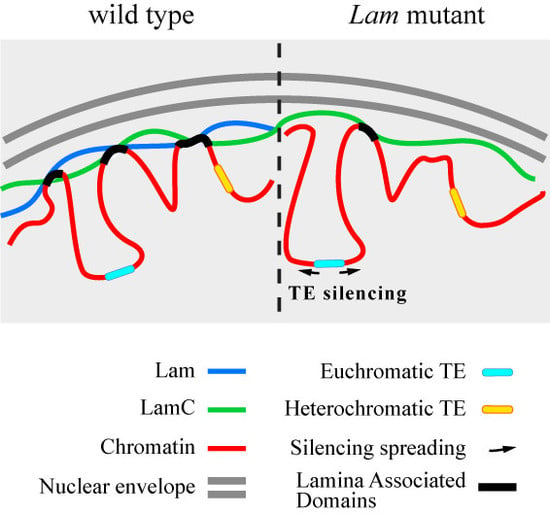
Silencing of Euchromatic Transposable Elements as a Consequence of Nuclear Lamina Dysfunction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Drosophila Stocks
2.2. Mortality Determination
2.3. β-Galactosidase Staining
2.4. Quantitative RT-PCR
2.5. Microscopy Analysis of Adult Eyes
3. Results
3.1. Pharate Mortality Induced by Lam Loss-of-Function Mutations is Linked to Gypsy Retrotransposon Silencing
3.2. Silencing of TEs Located Near Euchromatic Genes in Lam Mutant Somatic Tissues
3.3. The Effect of Lam Inactivation on TE Expression is Dependent on the Genetic Background
3.4. The Silencing of Euchromatic TEs Induced by Lam Inactivation Spreads to Neighbor Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvak, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef] [PubMed]
- Nefedova, L.; Kim, A. Mechanisms of LTR-Retroelement Transposition: Lessons from Drosophila melanogaster. Viruses 2017, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.S. Non-long terminal repeat (non-LTR) retrotransposons: Mechanisms, recent developments, and unanswered questions. Mob. DNA 2010, 1, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchon, N.; Vaury, C. RNAi: A defensive RNA-silencing against viruses and transposable elements. Heredity 2006, 96, 195–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castel, S.E.; Martienssen, R.A. RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013, 14, 100–112. [Google Scholar] [CrossRef]
- Driver, C.J.; McKechnie, S.W. Transposable elements as a factor in the aging of Drosophila melanogaster. Ann. N. Y. Acad. Sci. 1992, 673, 83–91. [Google Scholar] [CrossRef]
- Shevelyov, Y.Y.; Ulianov, S.V. The Nuclear Lamina as an Organizer of Chromosome Architecture. Cells Basel 2019, 8. [Google Scholar] [CrossRef] [Green Version]
- De Leeuw, R.; Gruenbaum, Y.; Medalia, O. Nuclear Lamins: Thin Filaments with Major Functions. Trends Cell Biol. 2018, 28, 34–45. [Google Scholar] [CrossRef]
- Adam, S.A.; Goldman, R.D. Insights into the differences between the A- and B-type nuclear lamins. Adv. Biol. Regul. 2012, 52, 108–113. [Google Scholar] [CrossRef] [Green Version]
- Camozzi, D.; Capanni, C.; Cenni, V.; Mattioli, E.; Columbaro, M.; Squarzoni, S.; Lattanzi, G. Diverse lamin-dependent mechanisms interact to control chromatin dynamics. Focus on laminopathies. Nucl. Phila. 2014, 5, 427–440. [Google Scholar] [CrossRef] [Green Version]
- Cenni, V.; D’Apice, M.R.; Garagnani, P.; Columbaro, M.; Novelli, G.; Franceschi, C.; Lattanzi, G. Mandibuloacral dysplasia: A premature ageing disease with aspects of physiological ageing. Ageing Res. Rev. 2018, 42, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ditaranto, R.; Boriani, G.; Biffi, M.; Lorenzini, M.; Graziosi, M.; Ziacchi, M.; Pasquale, F.; Vitale, G.; Berardini, A.; Rinaldi, R.; et al. Differences in cardiac phenotype and natural history of laminopathies with and without neuromuscular onset. Orphanet J. Rare Dis. 2019, 14, 263. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, E.; Columbaro, M.; Capanni, C.; Maraldi, N.M.; Cenni, V.; Scotlandi, K.; Marino, M.T.; Merlini, L.; Squarzoni, S.; Lattanzi, G. Prelamin A-mediated recruitment of SUN1 to the nuclear envelope directs nuclear positioning in human muscle. Cell Death Differ. 2011, 18, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, C.; Columbaro, M.; Schena, E.; Prencipe, S.; Andrenacci, D.; Iozzo, P.; Angela Guzzardi, M.; Capanni, C.; Mattioli, E.; Loi, M.; et al. Altered adipocyte differentiation and unbalanced autophagy in type 2 Familial Partial Lipodystrophy: An in vitro and in vivo study of adipose tissue browning. Exp. Mol. Med. 2019, 51, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohnekamp, J.; Cryderman, D.E.; Thiemann, D.A.; Magin, T.M.; Wallrath, L.L. Using Drosophila for Studies of Intermediate Filaments. Methods Enzymol. 2016, 568, 707–726. [Google Scholar] [CrossRef]
- Bossie, C.A.; Sanders, M.M. A Cdna from Drosophila-Melanogaster Encodes a Lamin C-Like Intermediate Filament Protein. J. Cell Sci. 1993, 104, 1263–1272. [Google Scholar]
- Palka, M.; Tomczak, A.; Grabowska, K.; Machowska, M.; Piekarowicz, K.; Rzepecka, D.; Rzepecki, R. Laminopathies: What can humans learn from fruit flies. Cell. Mol. Biol. Lett. 2018, 23, 32. [Google Scholar] [CrossRef] [Green Version]
- Riemer, D.; Weber, K. The Organization of the Gene for Drosophila Lamin-C—Limited Homology with Vertebrate Lamin Genes and Lack of Homology Versus the Drosophila Lamin Dmo Gene. Eur. J. Cell Biol. 1994, 63, 299–306. [Google Scholar]
- Smith, D.E.; Gruenbaum, Y.; Berrios, M.; Fisher, P.A. Biosynthesis and interconversion of Drosophila nuclear lamin isoforms during normal growth and in response to heat shock. J. Cell Biol. 1987, 105, 771–790. [Google Scholar] [CrossRef]
- Riemer, D.; Stuurman, N.; Berrios, M.; Hunter, C.; Fisher, P.A.; Weber, K. Expression of Drosophila Lamin-C Is Developmentally-Regulated -Analogies with Vertebrate a-Type Lamins. J. Cell Sci. 1995, 108, 3189–3198. [Google Scholar]
- Munoz-Alarcon, A.; Pavlovic, M.; Wismar, J.; Schmitt, B.; Eriksson, M.; Kylsten, P.; Dushay, M.S. Characterization of lamin Mutation Phenotypes in Drosophila and Comparison to Human Laminopathies. PLoS ONE 2007, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenz-Bohme, B.; Wismar, J.; Fuchs, S.; Reifegerste, R.; Buchner, E.; Betz, H.; Schmitt, B. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J. Cell Biol. 1997, 137, 1001–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worman, H.J.; Bonne, G. “Laminopathies”: A wide spectrum of human diseases. Exp. Cell Res. 2007, 313, 2121–2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osouda, S.; Nakamura, Y.; de Saint Phalle, B.; McConnell, M.; Horigome, T.; Sugiyama, S.; Fisher, P.A.; Furukawa, K. Null mutants of Drosophila B-type lamin Dm(0) show aberrant tissue differentiation rather than obvious nuclear shape distortion or specific defects during cell proliferation. Dev. Biol. 2005, 284, 219–232. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Zheng, X.; Xiao, D.; Zheng, Y. Age-associated de-repression of retrotransposons in the Drosophila fat body, its potential cause and consequence. Aging Cell 2016, 15, 542–552. [Google Scholar] [CrossRef] [Green Version]
- Vazquez, B.N.; Thackray, J.K.; Simonet, N.G.; Chahar, S.; Kane-Goldsmith, N.; Newkirk, S.J.; Lee, S.; Xing, J.; Verzi, M.P.; An, W.; et al. SIRT7 mediates L1 elements transcriptional repression and their association with the nuclear lamina. Nucleic Acids Res. 2019, 47, 7870–7885. [Google Scholar] [CrossRef] [Green Version]
- Pelisson, A.; Mejlumian, L.; Robert, V.; Terzian, C.; Bucheton, A. Drosophila germline invasion by the endogenous retrovirus gypsy: Involvement of the viral env gene. Insect Biochem. Mol. Biol. 2002, 32, 1249–1256. [Google Scholar] [CrossRef]
- Li, W.; Prazak, L.; Chatterjee, N.; Gruninger, S.; Krug, L.; Theodorou, D.; Dubnau, J. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci. 2013, 16, 529–531. [Google Scholar] [CrossRef] [Green Version]
- Guillemin, K.; Williams, T.; Krasnow, M.A. A nuclear lamin is required for cytoplasmic organization and egg polarity in Drosophila. Nat. Cell Biol. 2001, 3, 848–851. [Google Scholar] [CrossRef]
- Patterson, K.; Molofsky, A.B.; Robinson, C.; Acosta, S.; Cater, C.; Fischer, J.A. The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol. Biol. Cell 2004, 15, 600–610. [Google Scholar] [CrossRef] [Green Version]
- Guida, V.; Cernilogar, F.M.; Filograna, A.; De Gregorio, R.; Ishizu, H.; Siomi, M.C.; Schotta, G.; Bellenchi, G.C.; Andrenacci, D. Production of Small Noncoding RNAs from the flamenco Locus Is Regulated by the gypsy Retrotransposon of Drosophila melanogaster. Genetics 2016, 204, 631–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghildiyal, M.; Seitz, H.; Horwich, M.D.; Li, C.; Du, T.; Lee, S.; Xu, J.; Kittler, E.L.; Zapp, M.L.; Weng, Z.; et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 2008, 320, 1077–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desset, S.; Vaury, C. Transcriptional interference mediated by retrotransposons within the genome of their host: Lessons from alleles of the white gene from Drosophila melanogaster. Cytogenet. Genome Res. 2005, 110, 209–214. [Google Scholar] [CrossRef]
- Searles, L.L.; Ruth, R.S.; Pret, A.M.; Fridell, R.A.; Ali, A.J. Structure and transcription of the Drosophila melanogaster vermilion gene and several mutant alleles. Mol. Cell. Biol. 1990, 10, 1423–1431. [Google Scholar] [CrossRef] [Green Version]
- Shevelyov, Y.Y.; Lavrov, S.A.; Mikhaylova, L.M.; Nurminsky, I.D.; Kulathinal, R.J.; Egorova, K.S.; Rozovsky, Y.M.; Nurminsky, D.I. The B-type lamin is required for somatic repression of testis-specific gene clusters. Proc. Natl. Acad. Sci. USA 2009, 106, 3282–3287. [Google Scholar] [CrossRef] [Green Version]
- Meuleman, W.; Peric-Hupkes, D.; Kind, J.; Beaudry, J.B.; Pagie, L.; Kellis, M.; Reinders, M.; Wessels, L.; van Steensel, B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013, 23, 270–280. [Google Scholar] [CrossRef] [Green Version]
- Zullo, J.M.; Demarco, I.A.; Pique-Regi, R.; Gaffney, D.J.; Epstein, C.B.; Spooner, C.J.; Luperchio, T.R.; Bernstein, B.E.; Pritchard, J.K.; Reddy, K.L.; et al. DNA Sequence-Dependent Compartmentalization and Silencing of Chromatin at the Nuclear Lamina. Cell 2012, 149, 1474–1487. [Google Scholar] [CrossRef] [Green Version]
- De Cecco, M.; Ito, T.; Petrashen, A.P.; Elias, A.E.; Skvir, N.J.; Criscione, S.W.; Caligiana, A.; Brocculi, G.; Adney, E.M.; Boeke, J.D.; et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 2019, 566, 73–78. [Google Scholar] [CrossRef]
- Jorgensen, R.A. Cosuppression, flower color patterns, and metastable gene expression States. Science 1995, 268, 686–691. [Google Scholar] [CrossRef] [Green Version]
- Andrenacci, D.; Cavaliere, V.; Lattanzi, G. The role of transposable elements activity in aging and their possible involvement in laminopathic diseases. Ageing Res. Rev. 2020, 57, 100995. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.C. The Role of piRNA-Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in Drosophila melanogaster. PLoS Genet. 2015, 11, e1005269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.C.G.; Karpen, G.H. Pervasive epigenetic effects of Drosophila euchromatic transposable elements impact their evolution. eLife 2017, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sienski, G.; Donertas, D.; Brennecke, J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 2012, 151, 964–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shpiz, S.; Ryazansky, S.; Olovnikov, I.; Abramov, Y.; Kalmykova, A. Euchromatic Transposon Insertions Trigger Production of Novel Pi- and Endo-siRNAs at the Target Sites in the Drosophila Germline. PLoS Genet. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Maggi, L.; Carboni, N.; Bernasconi, P. Skeletal Muscle Laminopathies: A Review of Clinical and Molecular Features. Cells (Basel) 2016, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cavaliere, V.; Lattanzi, G.; Andrenacci, D. Silencing of Euchromatic Transposable Elements as a Consequence of Nuclear Lamina Dysfunction. Cells 2020, 9, 625. https://doi.org/10.3390/cells9030625
Cavaliere V, Lattanzi G, Andrenacci D. Silencing of Euchromatic Transposable Elements as a Consequence of Nuclear Lamina Dysfunction. Cells. 2020; 9(3):625. https://doi.org/10.3390/cells9030625
Chicago/Turabian StyleCavaliere, Valeria, Giovanna Lattanzi, and Davide Andrenacci. 2020. "Silencing of Euchromatic Transposable Elements as a Consequence of Nuclear Lamina Dysfunction" Cells 9, no. 3: 625. https://doi.org/10.3390/cells9030625
APA StyleCavaliere, V., Lattanzi, G., & Andrenacci, D. (2020). Silencing of Euchromatic Transposable Elements as a Consequence of Nuclear Lamina Dysfunction. Cells, 9(3), 625. https://doi.org/10.3390/cells9030625