Lamin A and Prelamin A Counteract Migration of Osteosarcoma Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures, Transfection and Treatments
2.2. Cell Proliferation
2.3. Wound Healing Assay
2.4. Annexin V-FITC/PI Staining
2.5. Western Blot Analysis
2.6. qRT-PCR
2.7. Immunofluorescence Microscopy
2.8. Statistical Analysis
3. Results
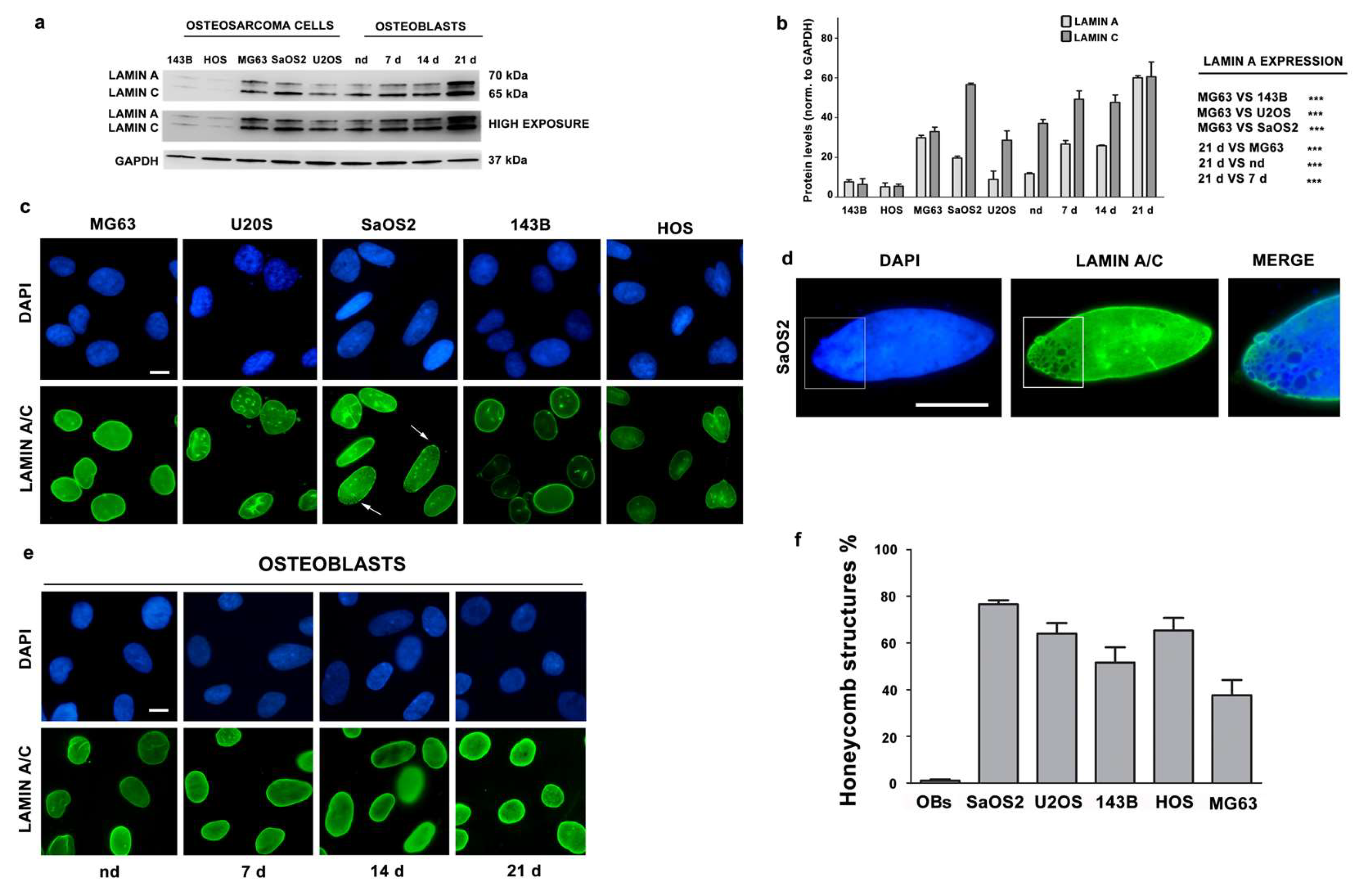
3.1. Lamin A/C Expression Increases in Differentiating Osteoblasts
3.2. Lamin A/C Expression is Reduced in Osteosarcoma Cells
3.3. Migration and Proliferation Ability are Increased in OS Cells with Low LMNA Expression
3.4. Prelamin A Accumulation Reduces Migration Potential of OS Cells
3.5. Mevinolin Effects in OS Cells Are Related to Prelamin A Accumulation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marina, N.; Gebhardt, M.; Teot, L.; Gorlick, R. Biology and therapeutic advances for pediatric osteosarcoma. Oncologist 2004, 9, 422–441. [Google Scholar] [CrossRef] [PubMed]
- Taran, S.J.; Taran, R.; Malipatil, N.B. Pediatric osteosarcoma: An updated review. Indian J. Med. Paediatr. Oncol. 2017, 38, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Manara, M.C.; Valente, S.; Cristalli, C.; Nicoletti, G.; Landuzzi, L.; Zwergel, C.; Mazzone, R.; Stazi, G.; Arimondo, P.B.; Pasello, M.; et al. A Quinoline-Based DNA methyltransferase inhibitor as a possible adjuvant in osteosarcoma therapy. Mol. Cancer Ther. 2018, 17, 1881–1892. [Google Scholar] [CrossRef] [PubMed]
- Ando, K.; Heymann, M.F.; Stresing, V.; Mori, K.; Redini, F.; Heymann, D. Current therapeutic strategies and novel approaches in osteosarcoma. Cancers 2013, 5, 591–616. [Google Scholar] [CrossRef]
- Martin, J.W.; Squire, J.A.; Zielenska, M. The genetics of osteosarcoma. Sarcoma 2012, 2012, 627254. [Google Scholar] [CrossRef]
- Dani, N.; Olivero, M.; Mareschi, K.; van Duist, M.M.; Miretti, S.; Cuvertino, S.; Patane, S.; Calogero, R.; Ferracini, R.; Scotlandi, K.; et al. The MET oncogene transforms human primary bone-derived cells into osteosarcomas by targeting committed osteo-progenitors. J. Bone Miner. Res. 2012, 27, 1322–1334. [Google Scholar] [CrossRef]
- Lye, K.L.; Nordin, N.; Vidyadaran, S.; Thilakavathy, K. Mesenchymal stem cells: From stem cells to sarcomas. Cell Biol. Int. 2016, 40, 610–618. [Google Scholar] [CrossRef]
- Xiao, W.; Mohseny, A.B.; Hogendoorn, P.C.; Cleton-Jansen, A.M. Mesenchymal stem cell transformation and sarcoma genesis. Clin. Sarcoma Res. 2013, 3, 10. [Google Scholar] [CrossRef]
- Alhudiri, I.M.; Nolan, C.C.; Ellis, I.O.; Elzagheid, A.; Rakha, E.A.; Green, A.R.; Chapman, C.J. Expression of Lamin A/C in early-stage breast cancer and its prognostic value. Breast Cancer Res. Treat. 2019, 174, 661–668. [Google Scholar] [CrossRef]
- Kaspi, E.; Frankel, D.; Guinde, J.; Perrin, S.; Laroumagne, S.; Robaglia-Schlupp, A.; Ostacolo, K.; Harhouri, K.; Tazi-Mezalek, R.; Micallef, J.; et al. Low lamin A expression in lung adenocarcinoma cells from pleural effusions is a pejorative factor associated with high number of metastatic sites and poor performance status. PLoS ONE 2017, 12, e0183136. [Google Scholar] [CrossRef]
- Bell, E.S.; Lammerding, J. Causes and consequences of nuclear envelope alterations in tumour progression. Eur. J. Cell Biol. 2016, 95, 449–464. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.; Chen, P.; Li, L.; Tan, H.; Zhou, J.; Zhou, Y.; Yang, X.; Wu, X. Loss of lamin A but not lamin C expression in epithelial ovarian cancer cells is associated with metastasis and poor prognosis. Pathol. Res. Pract. 2015, 211, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Sakthivel, K.M.; Sehgal, P. A novel role of lamins from genetic disease to cancer biomarkers. Oncol. Rev. 2016, 10, 309. [Google Scholar] [CrossRef] [PubMed]
- Davidson, P.M.; Lammerding, J. Broken nuclei—Lamins, nuclear mechanics, and disease. Trends Cell Biol. 2014, 24, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K.W. Cancer genes and the pathways they control. Nat. Med. 2004, 10, 789–799. [Google Scholar] [CrossRef]
- Prokocimer, M.; Margalit, A.; Gruenbaum, Y. The nuclear lamina and its proposed roles in tumorigenesis: Projection on the hematologic malignancies and future targeted therapy. J. Struct. Biol. 2006, 155, 351–360. [Google Scholar] [CrossRef]
- Wu, Z.; Wu, L.; Weng, D.; Xu, D.; Geng, J.; Zhao, F. Reduced expression of lamin A/C correlates with poor histological differentiation and prognosis in primary gastric carcinoma. J. Exp. Clin. Cancer Res. 2009, 28, 8. [Google Scholar] [CrossRef]
- Belt, E.J.; Fijneman, R.J.; van den Berg, E.G.; Bril, H.; Delis-van Diemen, P.M.; Tijssen, M.; van Essen, H.F.; de Lange-de Klerk, E.S.; Belien, J.A.; Stockmann, H.B.; et al. Loss of lamin A/C expression in stage II and III colon cancer is associated with disease recurrence. Eur J. Cancer 2011, 47, 1837–1845. [Google Scholar] [CrossRef]
- Wazir, U.; Ahmed, M.H.; Bridger, J.M.; Harvey, A.; Jiang, W.G.; Sharma, A.K.; Mokbel, K. The clinicopathological significance of lamin A/C, lamin B1 and lamin B receptor mRNA expression in human breast cancer. Cell. Mol. Biol. Lett. 2013, 18, 595–611. [Google Scholar] [CrossRef]
- Matsumoto, A.; Hieda, M.; Yokoyama, Y.; Nishioka, Y.; Yoshidome, K.; Tsujimoto, M.; Matsuura, N. Global loss of a nuclear lamina component, lamin A/C, and LINC complex components SUN1, SUN2, and nesprin-2 in breast cancer. Cancer Med. 2015, 4, 1547–1557. [Google Scholar] [CrossRef]
- Agrelo, R.; Setien, F.; Espada, J.; Artiga, M.J.; Rodriguez, M.; Perez-Rosado, A.; Sanchez-Aguilera, A.; Fraga, M.F.; Piris, M.A.; Esteller, M. Inactivation of the lamin A/C gene by CpG island promoter hypermethylation in hematologic malignancies, and its association with poor survival in nodal diffuse large B-cell lymphoma. J. Clin. Oncol. 2005, 23, 3940–3947. [Google Scholar] [CrossRef] [PubMed]
- Jakobisiak, M.; Golab, J. Statins can modulate effectiveness of antitumor therapeutic modalities. Med. Res. Rev. 2010, 30, 102–135. [Google Scholar] [CrossRef] [PubMed]
- Aljada, A.; Doria, J.; Saleh, A.M.; Al-Matar, S.H.; AlGabbani, S.; Shamsa, H.B.; Al-Bawab, A.; Ahmed, A.A. Altered Lamin A/C splice variant expression as a possible diagnostic marker in breast cancer. Cell. Oncol. 2016, 39, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Lattanzi, G. Prelamin A-mediated nuclear envelope dynamics in normal and laminopathic cells. Biochem. Soc. Trans. 2011, 39, 1698–1704. [Google Scholar] [CrossRef]
- Camozzi, D.; Capanni, C.; Cenni, V.; Mattioli, E.; Columbaro, M.; Squarzoni, S.; Lattanzi, G. Diverse lamin-dependent mechanisms interact to control chromatin dynamics. Focus on laminopathies. Nucleus 2014, 5, 427–440. [Google Scholar] [CrossRef]
- Benini, S.; Baldini, N.; Manara, M.C.; Chano, T.; Serra, M.; Rizzi, S.; Lollini, P.L.; Picci, P.; Scotlandi, K. Redundancy of autocrine loops in human osteosarcoma cells. Int. J. Cancer 1999, 80, 581–588. [Google Scholar] [CrossRef]
- Ramazzotti, G.; Fiume, R.; Chiarini, F.; Campana, G.; Ratti, S.; Billi, A.M.; Manzoli, L.; Follo, M.Y.; Suh, P.G.; McCubrey, J.; et al. Phospholipase C-β1 interacts with cyclin E in adipose-derived stem cells osteogenic differentiation. Adv. Biol. Regul. 2019, 71, 1–9. [Google Scholar] [CrossRef]
- Mattioli, E.; Andrenacci, D.; Mai, A.; Valente, S.; Robijns, J.; De Vos, W.H.; Capanni, C.; Lattanzi, G. Statins and histone deacetylase inhibitors affect Lamin A/C—Histone deacetylase 2 interaction in human cells. Front. Cell Dev. Biol. 2019, 7, 6. [Google Scholar] [CrossRef]
- Lonetti, A.; Cappellini, A.; Bertaina, A.; Locatelli, F.; Pession, A.; Buontempo, F.; Evangelisti, C.; Orsini, E.; Zambonin, L.; Neri, L.M.; et al. Improving nelarabine efficacy in T cell acute lymphoblastic leukemia by targeting aberrant PI3K/AKT/mTOR signaling pathway. J. Hematol. Oncol. 2016, 9, 114. [Google Scholar] [CrossRef]
- Huang, S.J.; Fu, R.H.; Shyu, W.C.; Liu, S.P.; Jong, G.P.; Chiu, Y.W.; Wu, H.S.; Tsou, Y.A.; Cheng, C.W.; Lin, S.Z. Adipose-Derived stem cells: Isolation, characterization, and differentiation potential. Cell Transplant. 2013, 22, 701–709. [Google Scholar] [CrossRef]
- Luu, H.H.; Kang, Q.; Park, J.K.; Si, W.; Luo, Q.; Jiang, W.; Yin, H.; Montag, A.G.; Simon, M.A.; Peabody, T.D.; et al. An orthotopic model of human osteosarcoma growth and spontaneous pulmonary metastasis. Clin. Exp. Metastasis 2005, 22, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Bermudez, A.; Vallejo, C.G.; Vicente-Blanco, R.J.; Gallardo, M.E.; Fernandez-Moreno, M.A.; Quintanilla, M.; Garesse, R. Enhanced tumorigenicity by mitochondrial DNA mild mutations. Oncotarget 2015, 6, 13628–13643. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, E.; Columbaro, M.; Capanni, C.; Maraldi, N.M.; Cenni, V.; Scotlandi, K.; Marino, M.T.; Merlini, L.; Squarzoni, S.; Lattanzi, G. Prelamin A-mediated recruitment of SUN1 to the nuclear envelope directs nuclear positioning in human muscle. Cell Death Differ. 2011, 18, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Bikkul, M.U.; Clements, C.S.; Godwin, L.S.; Goldberg, M.W.; Kill, I.R.; Bridger, J.M. Farnesyltransferase inhibitor and rapamycin correct aberrant genome organisation and decrease DNA damage respectively, in Hutchinson-Gilford progeria syndrome fibroblasts. Biogerontology 2018, 19, 579–602. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, E.; Columbaro, M.; Capanni, C.; Santi, S.; Maraldi, N.M.; D’Apice, M.R.; Novelli, G.; Riccio, M.; Squarzoni, S.; Foisner, R.; et al. Drugs affecting prelamin A processing: Effects on heterochromatin organization. Exp. Cell Res. 2008, 314, 453–462. [Google Scholar] [CrossRef]
- Zucchini, C.; Manara, M.C.; Cristalli, C.; Carrabotta, M.; Greco, S.; Pinca, R.S.; Ferrari, C.; Landuzzi, L.; Pasello, M.; Lollini, P.L.; et al. ROCK2 deprivation leads to the inhibition of tumor growth and metastatic potential in osteosarcoma cells through the modulation of YAP activity. J. Exp. Clin. Cancer Res. 2019, 38, 503. [Google Scholar] [CrossRef]
- Pinca, R.S.; Manara, M.C.; Chiadini, V.; Picci, P.; Zucchini, C.; Scotlandi, K. Targeting ROCK2 rather than ROCK1 inhibits Ewing sarcoma malignancy. Oncol. Rep. 2017, 37, 1387–1393. [Google Scholar] [CrossRef][Green Version]
- Konstantinopoulos, P.A.; Karamouzis, M.V.; Papavassiliou, A.G. Post-Translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat. Rev. Drug Discov. 2007, 6, 541–555. [Google Scholar] [CrossRef]
- Rauschert, I.; Aldunate, F.; Preussner, J.; Arocena-Sutz, M.; Peraza, V.; Looso, M.; Benech, J.C.; Agrelo, R. Promoter hypermethylation as a mechanism for Lamin A/C silencing in a subset of neuroblastoma cells. PLoS ONE 2017, 12, e0175953. [Google Scholar] [CrossRef]
- Tsukune, N.; Naito, M.; Kubota, T.; Ozawa, Y.; Nagao, M.; Ohashi, A.; Sato, S.; Takahashi, T. Lamin A overexpression promotes osteoblast differentiation and calcification in the MC3T3-E1 preosteoblastic cell line. Biochem. Biophys. Res. Commun. 2017, 488, 664–670. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, Y.; Keyimu, R.; Hao, J.; Zhao, Z.; Ye, R. The role of lamin A/C in mesenchymal stem cell differentiation. J. Physiol. Biochem. 2019, 75, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Rauner, M.; Sipos, W.; Goettsch, C.; Wutzl, A.; Foisner, R.; Pietschmann, P.; Hofbauer, L.C. Inhibition of lamin A/C attenuates osteoblast differentiation and enhances RANKL-dependent osteoclastogenesis. J. Bone Miner. Res. 2009, 24, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Istvan, E.S.; Deisenhofer, J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science 2001, 292, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Fromigue, O.; Hamidouche, Z.; Marie, P.J. Blockade of the RhoA-JNK-c-Jun-MMP2 cascade by atorvastatin reduces osteosarcoma cell invasion. J. Biol. Chem. 2008, 283, 30549–30556. [Google Scholar] [CrossRef] [PubMed]
- Kany, S.; Woschek, M.; Kneip, N.; Sturm, R.; Kalbitz, M.; Hanschen, M.; Relja, B. Simvastatin exerts anticancer effects in osteosarcoma cell lines via geranylgeranylation and c-Jun activation. Int. J. Oncol. 2018, 52, 1285–1294. [Google Scholar] [CrossRef] [PubMed]
- de la Rosa, J.; Freije, J.M.; Cabanillas, R.; Osorio, F.G.; Fraga, M.F.; Fernandez-Garcia, M.S.; Rad, R.; Fanjul, V.; Ugalde, A.P.; Liang, Q.; et al. Prelamin A causes progeria through cell-extrinsic mechanisms and prevents cancer invasion. Nat. Commun. 2013, 4, 2268. [Google Scholar] [CrossRef] [PubMed]
- Matralis, A.N.; Xanthopoulos, D.; Huot, G.; Lopes-Paciencia, S.; Cole, C.; de Vries, H.; Ferbeyre, G.; Tsantrizos, Y.S. Molecular tools that block maturation of the nuclear lamin A and decelerate cancer cell migration. Bioorg. Med. Chem. 2018, 26, 5547–5554. [Google Scholar] [CrossRef]
- Harada, T.; Swift, J.; Irianto, J.; Shin, J.W.; Spinler, K.R.; Athirasala, A.; Diegmiller, R.; Dingal, P.C.; Ivanovska, I.L.; Discher, D.E. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell Biol. 2014, 204, 669–682. [Google Scholar] [CrossRef]
- Osmanagic-Myers, S.; Dechat, T.; Foisner, R. Lamins at the crossroads of mechanosignaling. Genes Dev. 2015, 29, 225–237. [Google Scholar] [CrossRef]
- Urciuoli, E.; Petrini, S.; D’Oria, V.; Leopizzi, M.; Rocca, C.D.; Peruzzi, B. Nuclear Lamins and Emerin are differentially expressed in osteosarcoma cells and scale with tumor aggressiveness. Cancers 2020, 12, 443. [Google Scholar] [CrossRef]
- Haque, F.; Mazzeo, D.; Patel, J.T.; Smallwood, D.T.; Ellis, J.A.; Shanahan, C.M.; Shackleton, S. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J. Biol. Chem. 2010, 285, 3487–3498. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Chi, Y.H.; Mutalif, R.A.; Starost, M.F.; Myers, T.G.; Anderson, S.A.; Stewart, C.L.; Jeang, K.T. Accumulation of the inner nuclear envelope protein Sun1 is pathogenic in progeric and dystrophic laminopathies. Cell 2012, 149, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Luo, Q.; Sun, J.; Song, G. Cytoskeletal control of nuclear morphology and stiffness are required for OPN-induced bone-marrow-derived mesenchymal stem cell migration. Biochem. Cell Biol. 2019, 97, 463–470. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Evangelisti, C.; Paganelli, F.; Giuntini, G.; Mattioli, E.; Cappellini, A.; Ramazzotti, G.; Faenza, I.; Maltarello, M.C.; Martelli, A.M.; Scotlandi, K.; et al. Lamin A and Prelamin A Counteract Migration of Osteosarcoma Cells. Cells 2020, 9, 774. https://doi.org/10.3390/cells9030774
Evangelisti C, Paganelli F, Giuntini G, Mattioli E, Cappellini A, Ramazzotti G, Faenza I, Maltarello MC, Martelli AM, Scotlandi K, et al. Lamin A and Prelamin A Counteract Migration of Osteosarcoma Cells. Cells. 2020; 9(3):774. https://doi.org/10.3390/cells9030774
Chicago/Turabian StyleEvangelisti, Camilla, Francesca Paganelli, Gaia Giuntini, Elisabetta Mattioli, Alessandra Cappellini, Giulia Ramazzotti, Irene Faenza, Maria Cristina Maltarello, Alberto M. Martelli, Katia Scotlandi, and et al. 2020. "Lamin A and Prelamin A Counteract Migration of Osteosarcoma Cells" Cells 9, no. 3: 774. https://doi.org/10.3390/cells9030774
APA StyleEvangelisti, C., Paganelli, F., Giuntini, G., Mattioli, E., Cappellini, A., Ramazzotti, G., Faenza, I., Maltarello, M. C., Martelli, A. M., Scotlandi, K., Chiarini, F., & Lattanzi, G. (2020). Lamin A and Prelamin A Counteract Migration of Osteosarcoma Cells. Cells, 9(3), 774. https://doi.org/10.3390/cells9030774









