MiR193a Modulation and Podocyte Phenotype †
Abstract
:1. Introduction
2. Methods
2.1. Human Podocytes
2.2. Silencing of YY1, Sox2, and WT1
2.3. Transfection of a Specific miR193a Inhibitor and miR193a Expression Plasmid
2.4. RNA Isolation and qPCR Studies
- GAPDH fw 5′ CCC ATC ACC ATC TTC CAG GAG 3′; rev 5′ GTT GTC ATG GAT GAC CTT GGC 3′,
- WT1 fw 5′ CGAGAGCGATAACCACACAACG 3′; rev 5′ GTCTCAGATGCCGACCGTACAA 3′,
- APOL1 fw 5′ ATCTCAGCTGAAAGCGGTGAAC 3′; rev 5′ TGACTTTGCCCCCTCATGTAAG 3′,
- CD2AP fw 5′ CTGTCAGCTGCAGAGAAGAAA 3′; rev 5′ TTGGGTTGGAGAATGTCCAC 3′.
- Nephrin fw 5′CCCCTCTATGATGAAGTACAAATGGA3′; rev and 5′GTACGGATTTCCTCAGGTCTTCT3′
- VDR fw 5′-CTTCAGGCGAAGCATGAAGC-3′; rev 5′-CCTTCATCATGCCGATGTCC-3′
2.5. MicroRNA Assay
2.6. Western Blot Studies
2.7. Immunoprecipitation (IP)
2.8. miR193a Gene and Transcription Factor Binding
2.9. Homology Modeling and Docking Studies
2.10. DNA-Protein Interactions
2.11. Visualization
2.12. APOL1 3′UTR and miR193a Sequence Identification
2.13. APOL1 3′ UTR and miR193a Secondary Structure
2.14. APOL1 3′ UTR and miR193a Tertiary Structure Prediction and Docking
2.15. Molecular Dynamics (MD) Simulations
2.16. Statistical Analyses
3. Results
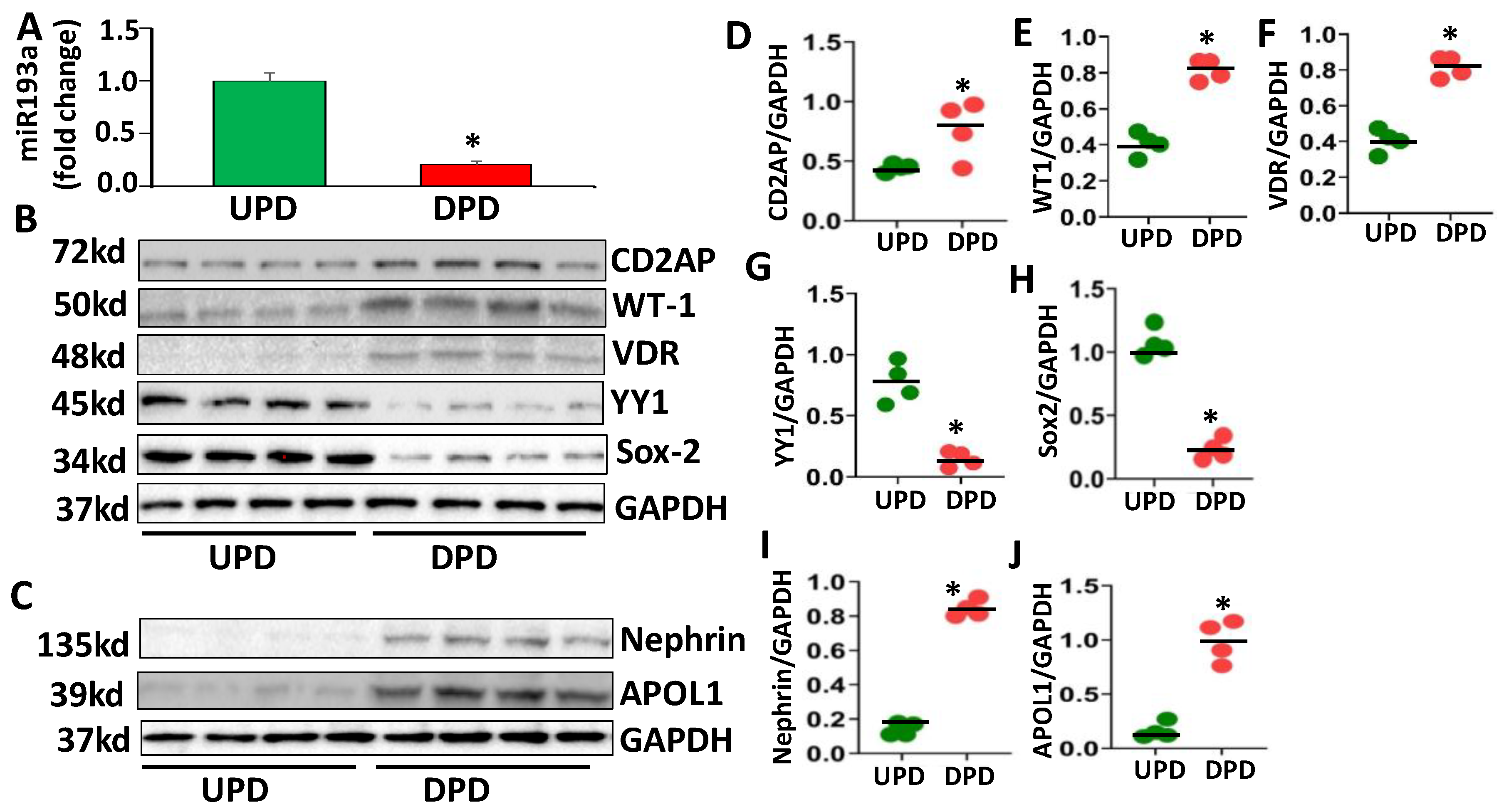
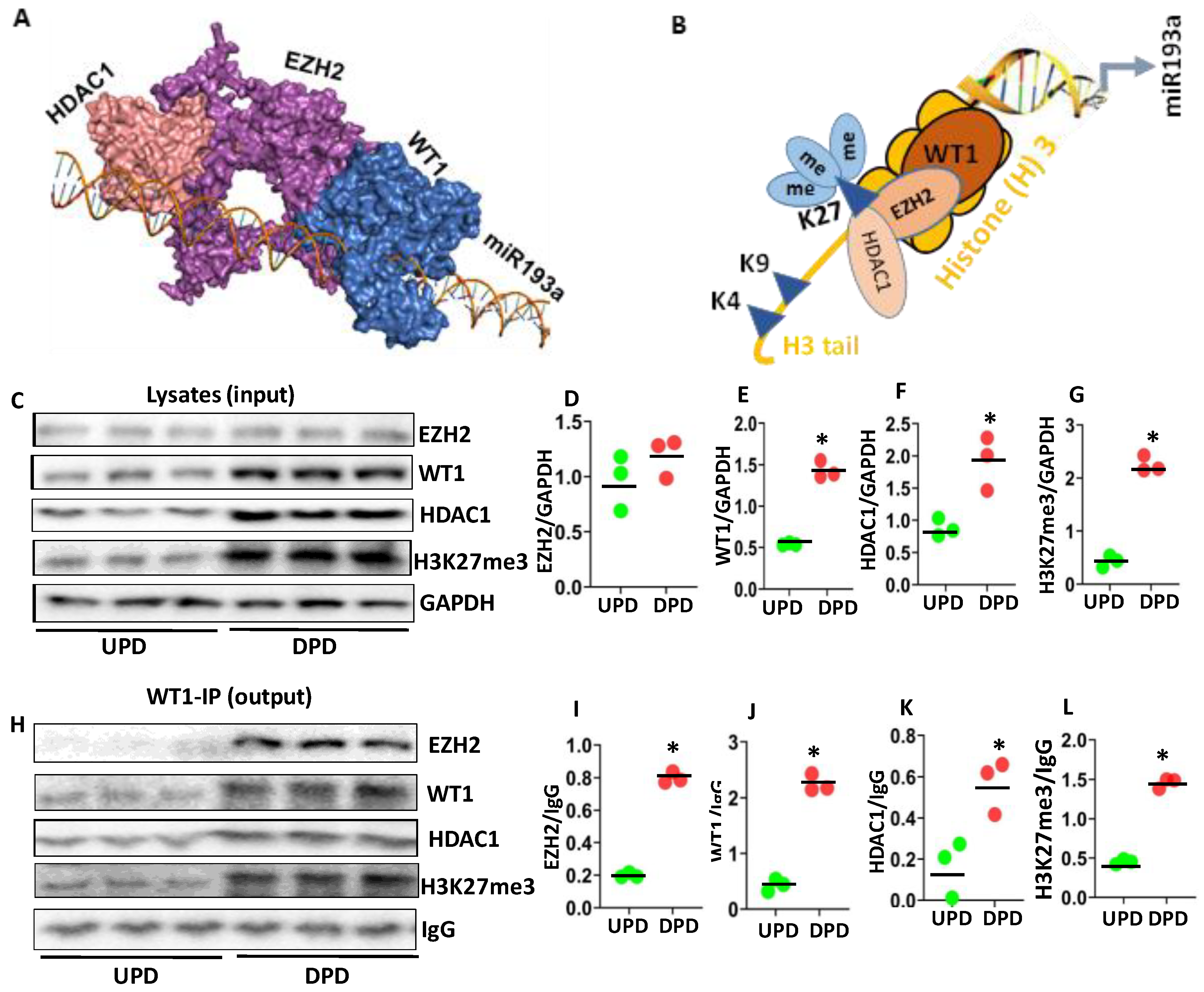
3.1. Evaluation of Molecular Profiles of Undifferentiated (UPD) and Differentiated Podocytes (DPD)
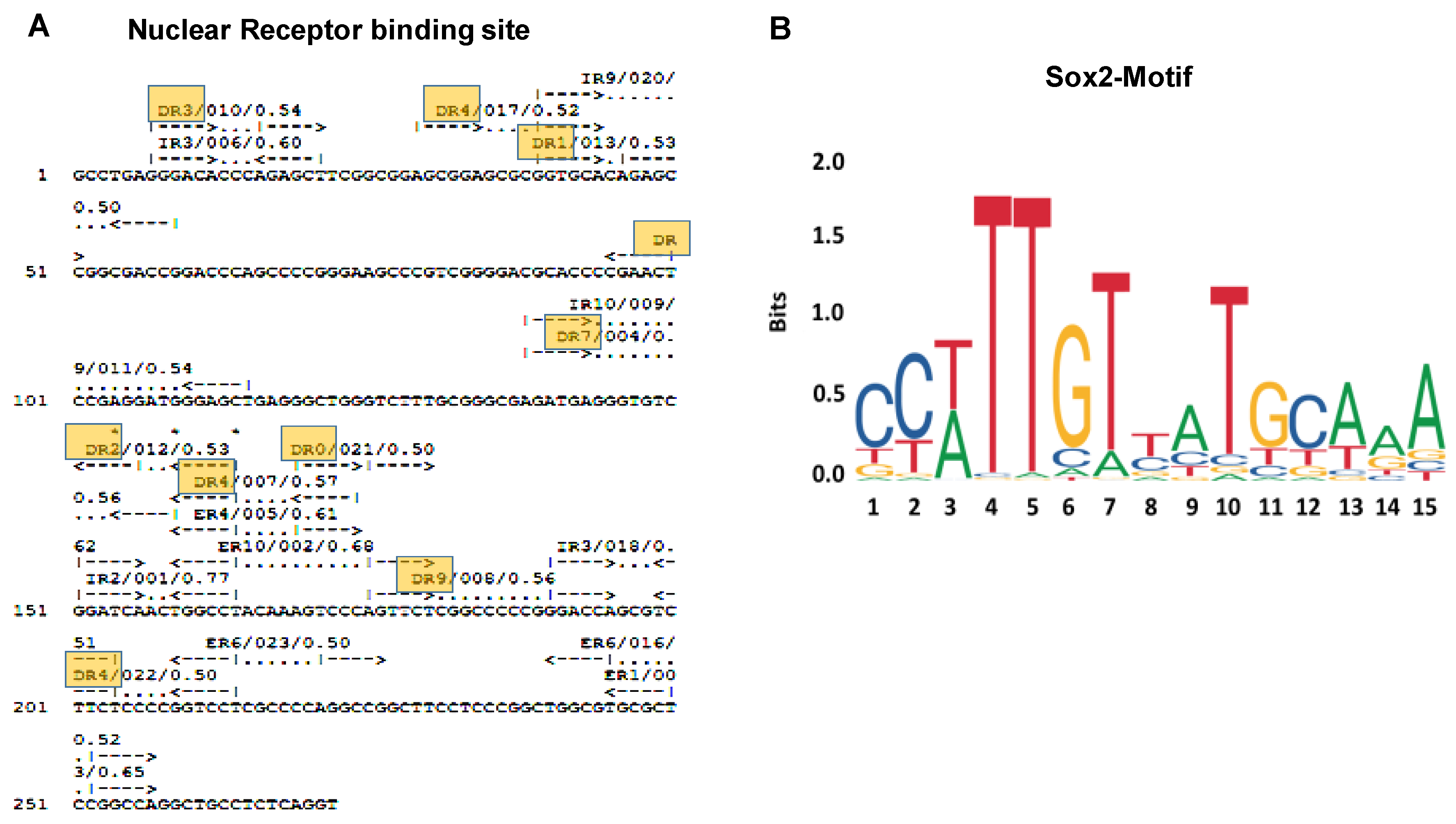
3.2. Prediction of Binding Sites for Transcription Factors
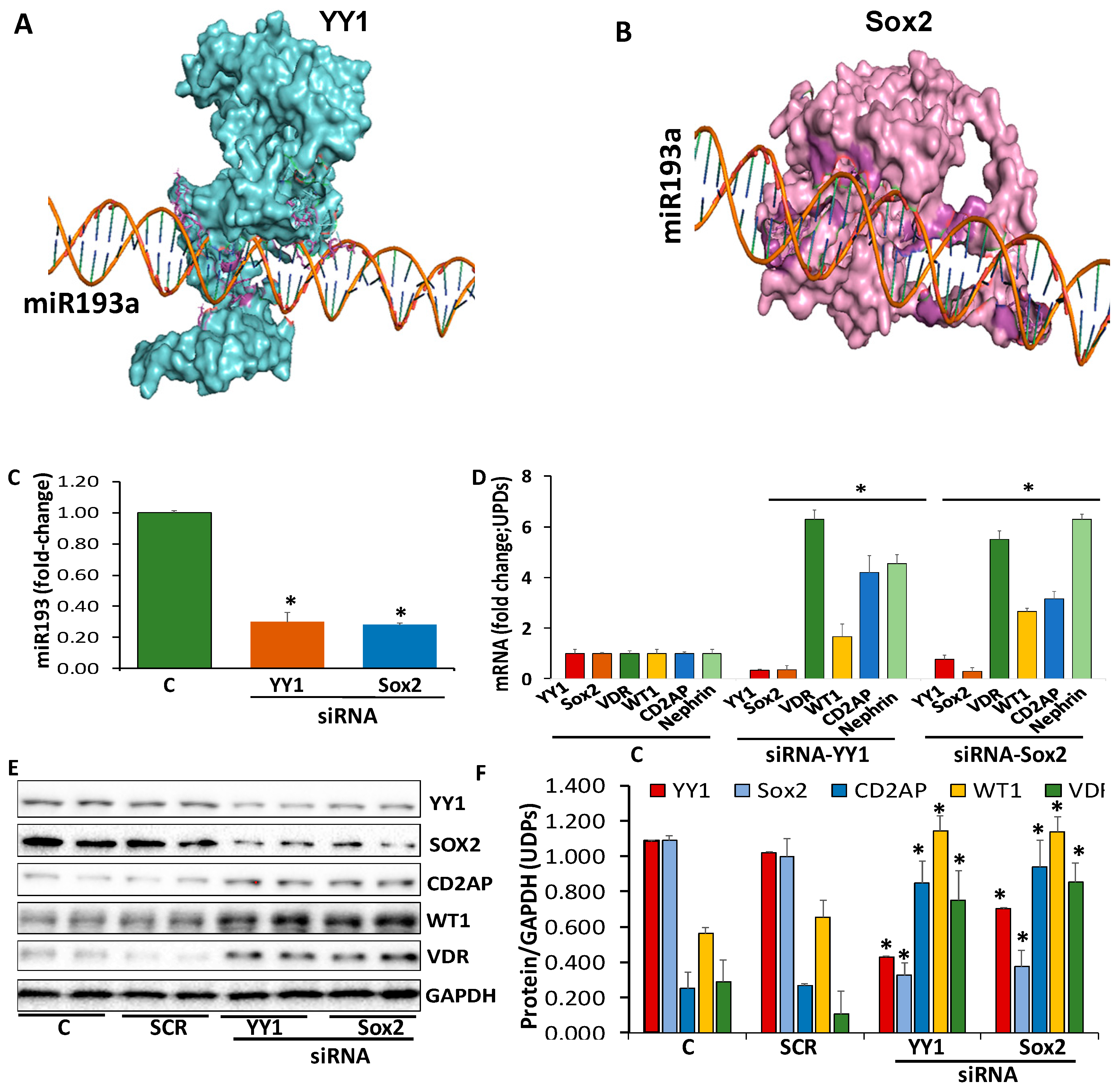
3.3. Evaluation of the Transcription Factors Enhancing miR193a Expression and Associated Downstream Signaling
3.4. Determination of Repressors Contributing to the Downing of miR193a Expression
3.5. Molecular Dynamics (MD) Simulation of miR193a Promoter and Transcription Factors
3.5.1. YY1 and miR193a
3.5.2. Sox2 and miR193a
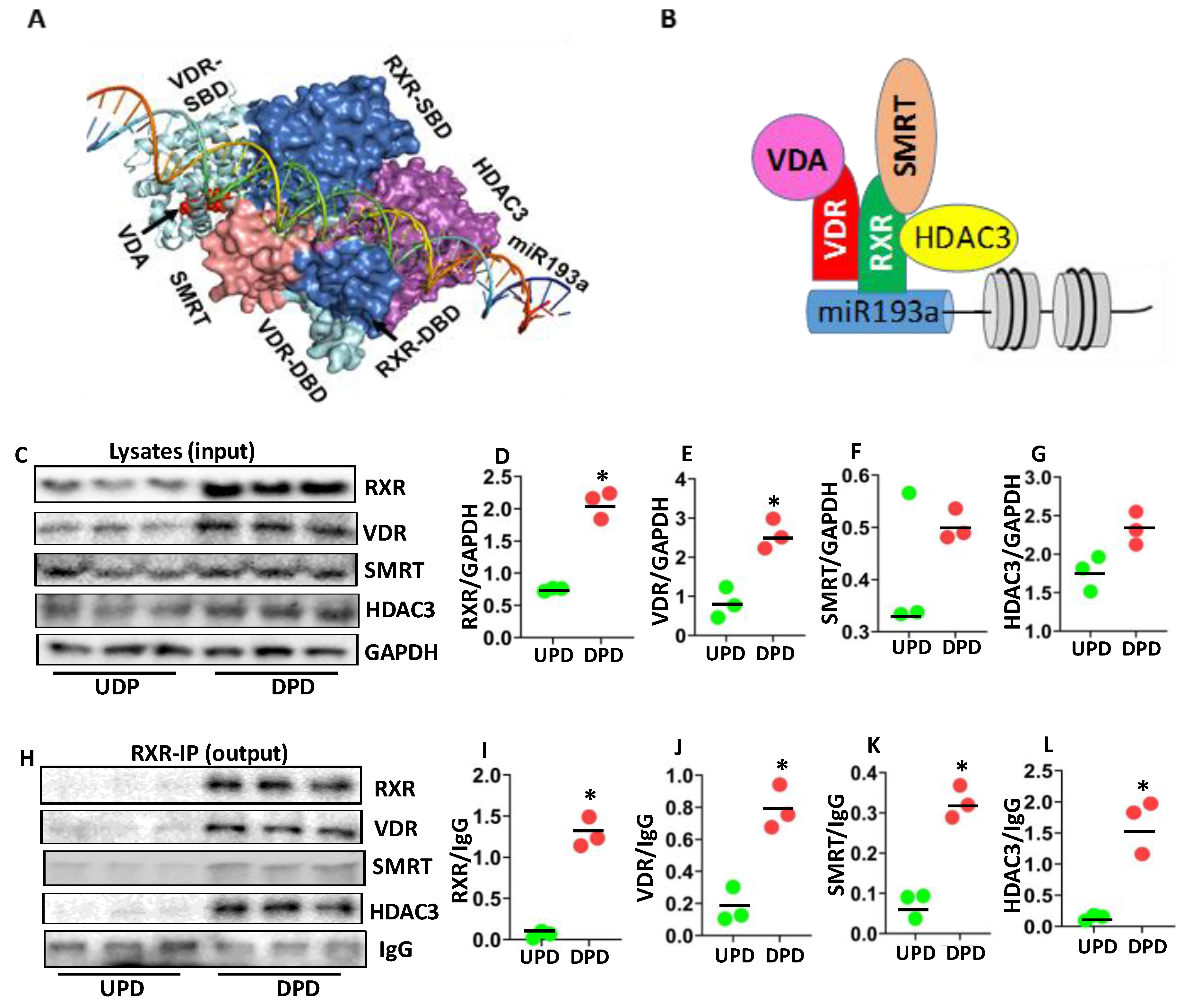
3.5.3. VDR-RXR and miR193a
3.5.4. WT1 and miR193a
3.6. Analysis of VDR-RXR Repressor Contributing to the Downing of miR193a Expression
3.7. Analysis of WT1 Repressor Complex Contributing to the Downing of miR193a
3.8. Comparison of miR193a-5p Targets
3.9. Molecular Genetics of APOL1
3.10. APOL1 3′ UTR and miR193a Secondary Structures
3.11. miR193a Binding with 3′ UTR Region of APOL1
3.12. Tertiary Structure of APOL1 3′ UTR and miR193a
3.13. Evaluation of the Effect of the Downing of miR193a and Overexpression of miR193a on the APOL1 Expression and Podocyte Molecular Phenotype
3.14. Molecular Dynamics (MD) Simulation of miR193a-5p and APOL1 3′ UTR
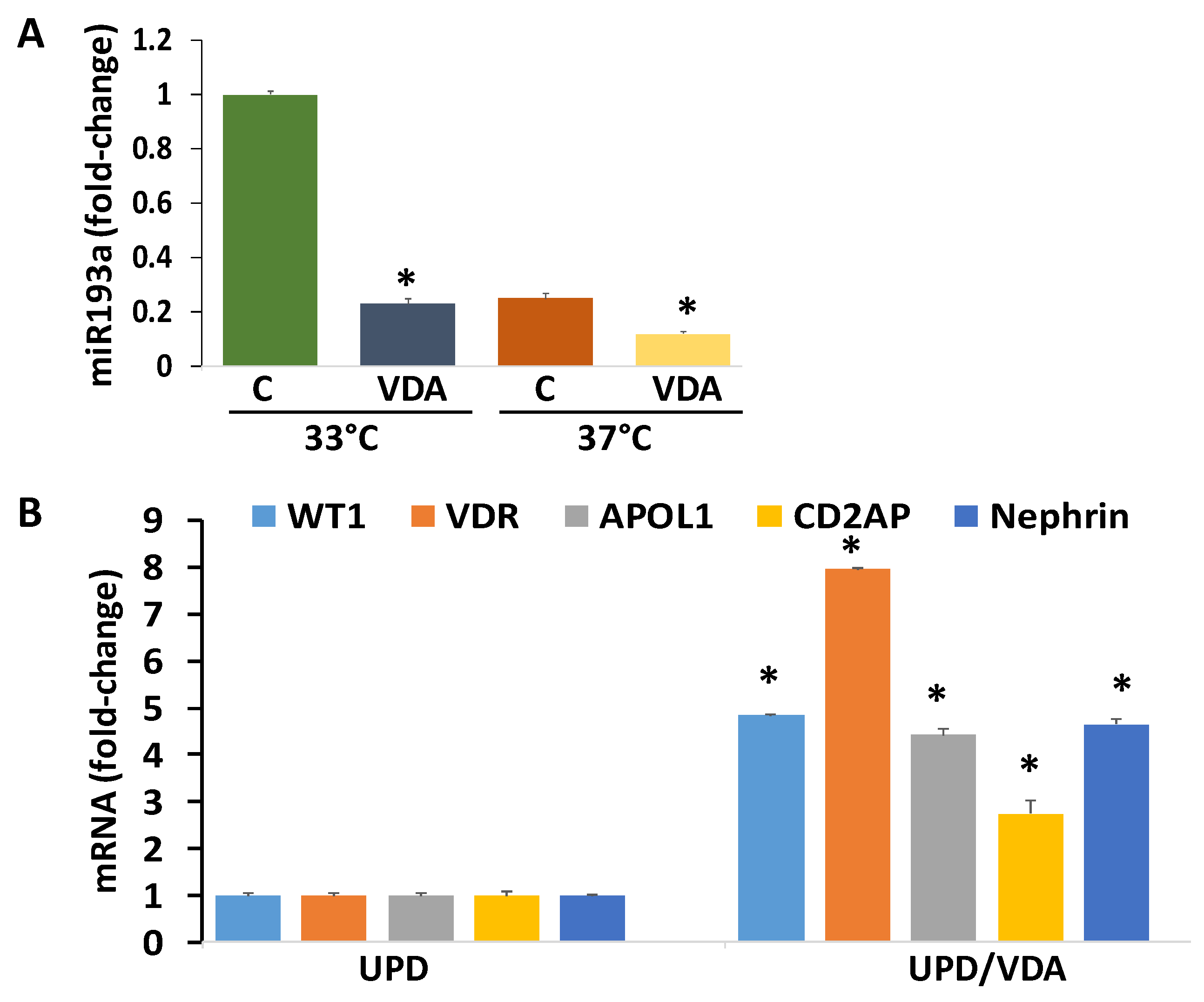
3.15. Evaluation of the Effect of VDA (Vitamin D Receptor Agonist) on Downing of miR193a and Associated Expression of Podocyte Molecular Markers
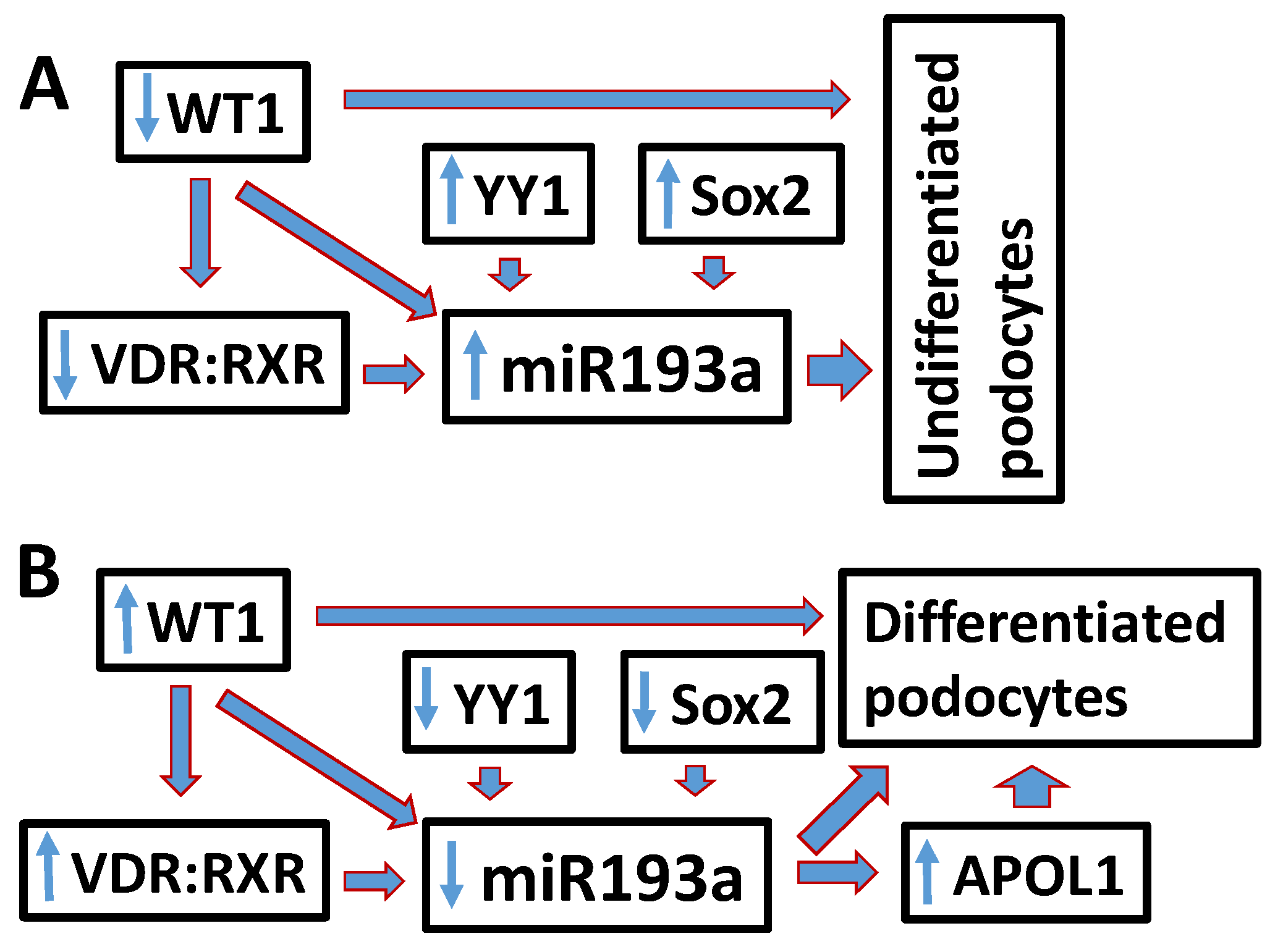
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Huang, Y.; Shen, X.J.; Zou, Q.; Wang, S.P.; Tang, S.M.; Zhang, G.Z. Biological functions of microRNAs: A review. J. Physiol. Biochem. 2011, 67, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Zhao, Y.; Yang, S.; Zhang, H.; Chen, F. Integrative analysis of miRNA-mRNA and miRNA-miRNA interactions. Biomed Res. Int. 2014, 2014, 8. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [Green Version]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs Are conserved targets of MicroRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Vashistha, H.; Lan, X.; Chandel, N.; Ayasolla, K.; Marashi Shoshtari, S.; Aslam, R.; Paliwal, N.; Abbruscato, F.; Mikulak, J.; et al. Role of APOL1 in human parietal epithelial cell transition. Am. J. Pathol. 2018, 188, 2508–2528. [Google Scholar] [CrossRef] [Green Version]
- Kietzmann, L.; Guhr, S.S.; Meyer, T.N.; Ni, L.; Sachs, M.; Panzer, U.; Stahl, R.A.; Saleem, M.A.; Kerjaschki, D.; Gebeshuber, C.A.; et al. MicroRNA-193a regulates the transdifferentiation of human parietal epithelial cells toward a podocyte phenotype. J. Am. Soc. Nephrol. 2015, 26, 1389–1401. [Google Scholar] [CrossRef] [Green Version]
- Lazzeri, E.; Romagnani, P. Podocyte biology: Differentiation of parietal epithelial cells into podocytes. Nat. Rev. Nephrol. 2015, 11, 7–8. [Google Scholar] [CrossRef]
- Mishra, A.; Ayasolla, K.; Kumar, V.; Lan, X.; Vashistha, H.; Aslam, R.; Hussain, A.; Chowdhary, S.; Marashi Shoshtari, S.; Paliwal, N. Modulation of apolipoprotein L1-microRNA-193a axis prevents podocyte dedifferentiation in high-glucose milieu. Am. J. Physiol. Ren. Physiol. 2018, 314, F832–F843. [Google Scholar] [CrossRef]
- Kumar, V.; Paliwal, N.; Ayasolla, K.; Vashistha, H.; Jha, A.; Chandel, N.; Chowdhary, S.; Saleem, M.A.; Malhotra, A.; Chander, P.N.; et al. Disruption of APOL1-miR193a axis induces disorganization of podocyte actin cytoskeleton. Sci. Rep. 2019, 9, 3582. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Ayasolla, K.; Jha, A.; Mishra, A.; Vashistha, H.; Lan, X.; Qayyum, M.; Chinnapaka, S.; Purohit, R.; Mikulak, J.; et al. Disrupted apolipoprotein L1-miR193a axis dedifferentiates podocytes through autophagy blockade in an APOL1 risk milieu. Am. J. Physiol. Cell Physiol. 2019, 317, C209–C225. [Google Scholar] [CrossRef]
- Kumar, V.; Singhal, P.C. APOL1 and kidney cell function. Am. J. Physiol. Ren. Physiol. 2019, 317, 463–477. [Google Scholar] [CrossRef]
- Rauch, C.; Feifel, E.; Kern, G.; Murphy, C.; Meier, F.; Parson, W.; Beilmann, M.; Jennings, P.; Gstraunthaler, G.; Wilmes, A. Differentiation of human iPSCs into functional podocytes. PLoS ONE 2018, 13, e0203869. [Google Scholar] [CrossRef] [Green Version]
- Knauss, J.L.; Miao, N.; Kim, S.N.; Nie, Y.; Shi, Y.; Wu, T.; Pinto, H.B.; Donohoe, M.E.; Sun, T. Long noncoding RNA Sox2ot and transcription factor YY1 co-regulate the differentiation of cortical neural progenitors by repressing Sox2. Cell Death Dis. 2018, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Podvinec, M.; Kaufmann, M.R.; Handschin, C.; Meyer, U.A. NUBIScan, an in silico approach for prediction of nuclear receptor response elements. Mol. Endocrinol. 2002, 16, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- TFSITESCAN. Available online: http://www.ifti.org/cgi-bin/ifti/Tfsitescan.pl (accessed on 9 November 2019).
- Messeguer, X.; Escudero, R.; Farré, D.; Núñez, O.; Martínez, J.; Albà, M.M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002, 18, 333–334. [Google Scholar] [CrossRef]
- Bailey, T.L.; Bodén, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [Green Version]
- Dijk, M.V.; Bonvin, A.M.J.J. 3D-DART: A DNA structure modeling server. Nucleic Acids Res. 2009, 37, W235–W239. [Google Scholar] [CrossRef] [PubMed]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein structure refinement driven by side chain repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierce, B.G.; Wiehe, K.; Hwang, H.; Kim, B.H.; Vreven, T.; Weng, Z. ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 2014, 30, 1771–1773. [Google Scholar] [CrossRef] [PubMed]
- Charles, E.V.; Zdobnov, E.M. miRmap: Comprehensive prediction of microRNA target repression strength. Nucleic Acids Res. 2012, 40, 11673–11683. [Google Scholar]
- Reuter, J.S.; Mathews, D.H. RNA structure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef] [Green Version]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Boniecki, M.J.; Lach, G.; Dawson, W.K.; Tomala, K.; Lukasz, P.; Soltysinski, T.; Rother, K.M.; Bujnick, J.M. SimRNA: A coarse-grained method for RNA folding simulations and 3D structure prediction. Nucleic Acids Res. 2016, 44, e63. [Google Scholar] [CrossRef]
- Biesiada, M.; Wieczorek, K.P.; Adamiak, R.W.; Purzycka, K.J. RNAComposer and RNA 3D structure prediction for nanotechnology. Methods 2016, 103, 120–127. [Google Scholar] [CrossRef]
- Jiahua, H.; Jun, W.; Huanyu, T.; Yi, X.; Sheng-You, H. HNADOCK: A nucleic acid docking server for modeling RNA/DNA–RNA/DNA 3D complex structures. Nucleic Acids Res. 2019, 47, W35–W42. [Google Scholar]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–779. [Google Scholar] [CrossRef]
- Sagendorf, J.M.; Berman, H.M.; Rohs, R. DNAproDB: An interactive tool for structural analysis of DNA–protein complexes. Nucleic Acids Res. 2017, 45, W89–W97. [Google Scholar] [CrossRef] [PubMed]
- Darnell, S.J.; Page, D.; Mitchell, J.C. Automated decision-tree approach to predicting protein-protein interaction hot spots. Proteins 2007, 68, 813–823. [Google Scholar] [CrossRef] [PubMed]
- The PyMOL Molecular Graphics System, Version 2.0; Schrodinger, LLC: New York, NY, USA, 2017.
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunhwan, J.; Kim, T.; Iyar, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 11, 1859–1865. [Google Scholar]
- Berendsen, H.J.C.; Spoel, D.V.D.; Drunen, R.V. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Saleem, M.A.; O’Hare, M.J.; Reiser, J.; Coward, R.J.; Inward, C.D.; Farren, T.; Xing, C.Y.; Ni, L.; Mathieson, P.W.; Mundel, P. Conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J. Am. Soc. Nephrol. 2002, 13, 630–638. [Google Scholar]
- Stormo, G.D. Modeling the specificity of protein-DNA interactions. Quant. Biol. 2013, 1, 115–130. [Google Scholar] [CrossRef] [Green Version]
- Mahony, S.; Benos, P.V. STAMP: A web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 2007, 35, W253–W258. [Google Scholar] [CrossRef] [Green Version]
- Mann, M.; Wright, P.R.; Backofen, R. IntaRNA 2.0: Enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017, 45, W435–W439. [Google Scholar] [CrossRef]
- Gebeshuber, C.A.; Kornauth, C.; Dong, L.; Sierig, R.; Seibler, J.; Reiss, M.; Tauber, S.; Bilban, M.; Wang, S.; Kain, R.; et al. Focal segmental glomerulosclerosis is induced by microRNA-193a and its downregulation of WT1. Nat. Med. 2013, 19, 481–487. [Google Scholar] [CrossRef]
- Li, Y.C.; Kong, J.; Wei, M.; Chen, Z.F.; Liu, S.Q.; Cao, L.P. 1,25-Dihydroxyvitamin D(3) is a negative endocrine regulator of the renin-angiotensin system. J. Clin. Investig. 2002, 110, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Singh, T.; Ayasolla, K.; Rai, P.; Chandel, N.; Haque, S.; Lederman, R.; Husain, M.; Vethantham, V.; Chawla, A.; Vashistha, H.; et al. AT1R blockade in adverse milieus: Role of SMRT and corepressor complexes. Am. J. Physiol. Ren. Physiol. 2015, 309, F189–F203. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Complex | Surface Area (Å2) | ΔGint (kcal/mol) | No. of Hydrogen bonds (NHB) | No. of Salt Bridges (NSB) | ΔGdiss (kcal/mol) | TΔSdiss (kcal/mol) |
|---|---|---|---|---|---|---|
| miR193a-YY1 | 2745.9 | −50.4 | 160 | 0 | 107.3 | 14.2 |
| miR193a-WT1 | 2812.8 | −52.2 | 161 | 0 | 109.5 | 14.2 |
| miR193a-Sox2 | 3165.8 | −58.1 | 161 | 0 | 115.4 | 14.2 |
| miR193a-VDR-RXR | 11576.9 | −76.4 | 161 | 3 | 5.8 | 12.6 |
| Complexes | Interacting Residues |
|---|---|
| miR193a-YY1 | Glu19, Ile20, Pro32, Val33, Glu37, Thr38, Val40, Val41, Glu46, Asp49, Lys259, Asp269, Leu270, Ser271, Lys301, Gly302, Cys303, Thr304, Lys305, Glu336, Ser338, Lys339, Arg342, Leu366, Asp367, Phe368, and Lys401 |
| miR193a-Sox2 | Lys97, Arg100, Met104, Tyr110, Asn166, Ala194, Gln195, Met196, Gln197, Ser230, Thr234, Pro235, Gly236, Met237, Ser252, Ser253, Pro254, Pro255, Val256, and Arg264 |
| miR193a-WT1 | Ser315, Glu316, Arg321, Lys322, Tyr334, Phe335, His339, Lys346, Phe383, Gln384, Cys385, Lys386, Leu398, Lys399, Thr402, Met442, Thr443, and Lys444 |
| miR193a-RXR(DBD) - miR193a-VDR-RXR heterodimer | Cys135, Ala136, Ile137, Leu167, Thr168, Tyr169, Thr170, Lys175, Asp176, Leu178, Ile179, Tyr189, Tyr192, Gln193, Lys194, Leu196, Ala197, and Met198. |
| miR193a-RXR(SBD) - miR193a-VDR-RXR heterodimer | Lys289, Ser295, Glu296, Leu297, Pro298, Leu299, Asp300, Gln302, Gly387, Ser389, Glu393, Met459, Leu460, Glu461, and Ala462. |
| miR193a-VDR(DBD) - miR193a-VDR-RXR heterodimer | Arg18, Asn19, Val20, Pro21, Arg22, Ile23, Cys24, Gly25, Cys27, Gly28, Asp29, Arg30, Met39, Arg67, Ile68, Thr69, Lys70, Asp71, Asn72, and Arg83. |
| miR193a-VDR(SBD) - miR193a-VDR-RXR heterodimer | Thr146, Gly289, Asn290, and Gln291 |
| miRNA | Gene | ΔG Open | ΔG Total | ΔG Duplex | ΔG Binding | ΔG Duplex Seed | ΔG Binding Seed | 3′UTR Position | AU Content | 3′ Pairing |
|---|---|---|---|---|---|---|---|---|---|---|
| hsa-mir-193a-5p | APOL1 | 20.37 | 3.57 | −16.80 | −17.90 | −8.70 | −9.11 | 456.00 | 0.42 | 2.00 |
| hsa-mir-193a-5p | BLID | 19.96 | 5.16 | −14.80 | −15.97 | −9.70 | −10.17 | 92.00 | 0.79 | 1.00 |
| hsa-mir-193a-5p | GPR17 | 19.42 | 7.33 | −16.60 | −17.70 | −8.70 | −9.11 | 125.00 | 0.34 | 2.00 |
| hsa-mir-193a-5p | SYNPO2L | 17.92 | 5.02 | −12.90 | −13.95 | −9.70 | −10.70 | 894.00 | 0.74 | 1.00 |
| hsa-mir-193a-5p | NKIRAS1 | 15.35 | 0.15 | −15.20 | −16.23 | −8.70 | −9.11 | 641.00 | 0.51 | 2.00 |
| hsa-mir-193a-5p | EIF2S1 | 12.90 | −1.50 | −14.40 | −15.06 | −9.70 | −10.17 | 467.00 | 0.83 | 1.50 |
| Complex | Surface Area (Å2) | ΔGint (kcal/mol) | No. of Hydrogen bonds (NHB) | No. of Salt Bridges (NSB) | ΔGdiss (kcal/mol) | TΔSdiss (kcal/mol) |
|---|---|---|---|---|---|---|
| APOL1 3′ UTR- miR193a-5p | 1341.3 | −27.3 | 32 | 0 | 29.7 | 11.8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jha, A.; Saha, S.; Ayasolla, K.; Vashistha, H.; Malhotra, A.; Skorecki, K.; Singhal, P.C. MiR193a Modulation and Podocyte Phenotype. Cells 2020, 9, 1004. https://doi.org/10.3390/cells9041004
Jha A, Saha S, Ayasolla K, Vashistha H, Malhotra A, Skorecki K, Singhal PC. MiR193a Modulation and Podocyte Phenotype. Cells. 2020; 9(4):1004. https://doi.org/10.3390/cells9041004
Chicago/Turabian StyleJha, Alok, Shourav Saha, Kamesh Ayasolla, Himanshu Vashistha, Ashwani Malhotra, Karl Skorecki, and Pravin C. Singhal. 2020. "MiR193a Modulation and Podocyte Phenotype" Cells 9, no. 4: 1004. https://doi.org/10.3390/cells9041004






