Noninvasive Diagnosis of NAFLD and NASH
Abstract
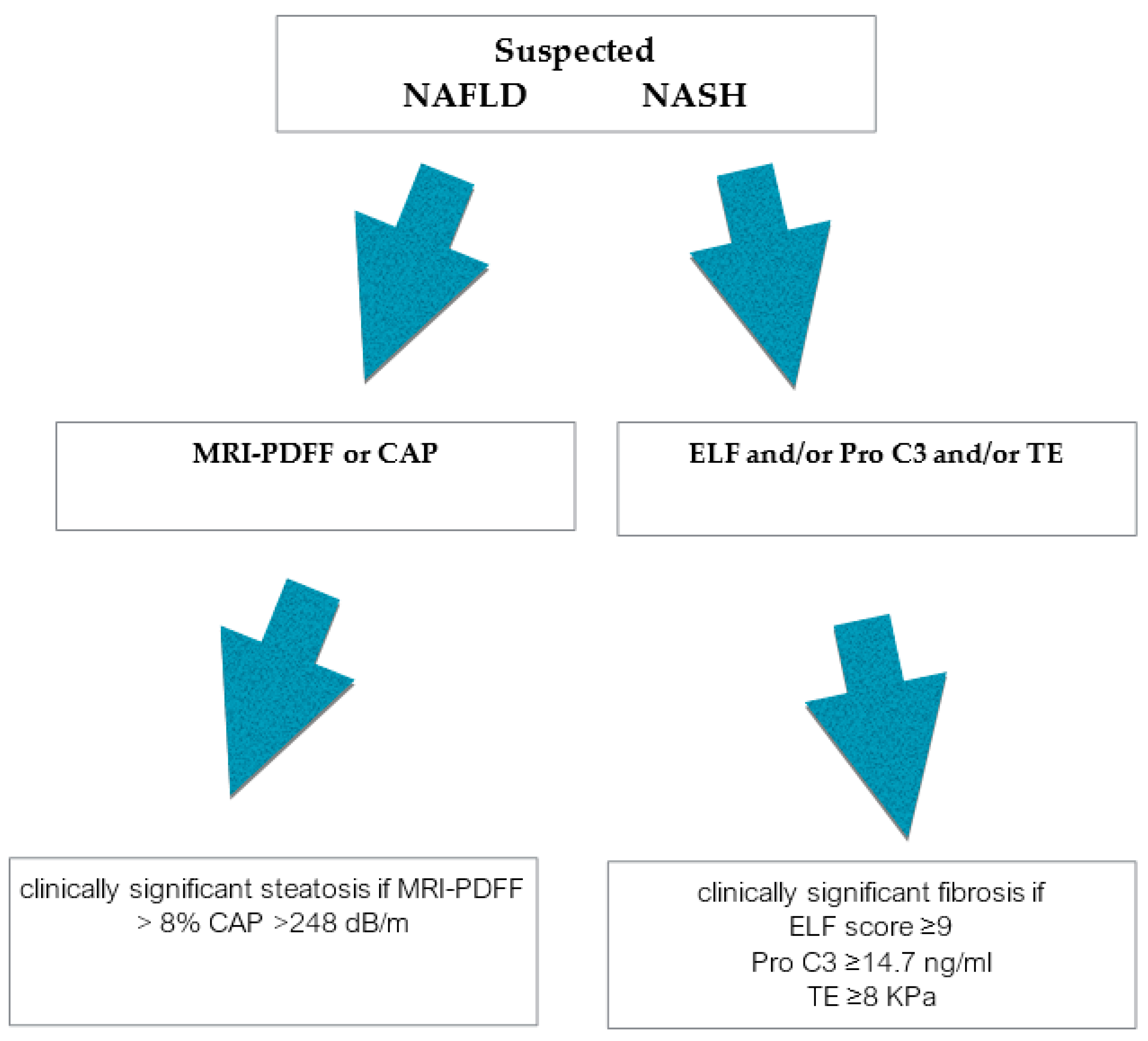
:1. Introduction
2. Non- or Minimally Invasive Methods
3. Non- or Minimally Invasive Differentiation of Simple Steatosis from NASH
3.1. Historical Biomarkers
3.2. Emerging Noninvasive Imaging Methods for NAFLD Diagnosis
3.2.1. Magnetic Resonance Imaging
3.2.2. CAP
4. Methods for NASH Diagnosis
4.1. Panels for Diagnosis of NASH and Fibrosis Including Minimally Invasive Blood Markers
4.2. Emerging Minimally Invasive Blood Markers in NASH Diagnosis
4.3. Emerging Noninvasive Imaging Markers in NASH Diagnosis
5. Genetic Biomarkers
6. OMICS-Based Markers
7. Emerging Circulating Biomarkers
8. Microbiota-Related Biomarkers
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| A2M | alpha2macroglobulin |
| AMEe | S-Adenosylmethionine |
| APO-F | Plasmatic apolipoprotein |
| AUROC | Area Under the Receiver Operating Characteristics |
| ALT | alanine aminotransferase |
| AST | aspartate aminotransferase |
| BCAA | Brached chain aminoacid |
| BMI | Body mass index |
| CAP | Controlled attenuated parameter |
| CHI3L | chitinase 3-like protein 1 |
| CK18 | Cytokeratin 18 |
| ELF | Enhanced Liver Fibrosis panel |
| ECM | Extracellular matrix turnover |
| FIB-4 | Fibrosis Index-4 |
| F3/F4 | Fibrosis stage 3 /4 |
| FGF21 gene | Fibroblast growth factor gene |
| FLI | Fatty liver Index |
| FXR | Farnesoid X receptor |
| GGT | gammaglutamil transferase |
| HA | Hyaluronic acid |
| HIS | Hepatic Steatosis Index |
| H-MRS | Magnetic resonance spectroscopy |
| KPa | KiloPascal |
| IL-8 | interleukin 8 |
| LC-MS | Liquid chromatography-Mass spectrometry |
| LFQP | label free quantitative proteomics approach |
| LncRNA | long non codingRNA |
| MAF | Minor allele frequency |
| MBOAT7 | Membrane-bound O-acyltransferase domain-containing protein 7 |
| MRI-PDFF | Magnetic resonance imaging-proton density fat fraction |
| MRE | Magnetic Resonance elastography |
| MRI | Magnetic Resonance Imaging |
| miRNA | microRNA |
| NAFLD | Non alcoholic fatty liver disease |
| NLFS | NAFLD Liver Fat score |
| NASH | Non alcoholic steatohepatitis |
| NASHTest | Non alcoholic steatohepatitis test |
| NPV | negative predictive value |
| PLT | platelets |
| PNPLA3 | Patatin-like phospholipase domain-containing protein 3 |
| PPV | Positive predictive value |
| pSWE | Point shear wave elastography |
| Pro-C3 | Pro-ollagen III |
| sCD | soluble macrophage activation marker |
| T2DM | type 2 diabetes mellitus |
| TE | Transient Elastography |
| TIMP1 | Metallopeptidase inhibitor 1 |
| TGs | tryglicerides |
| TM6F2 | Transmembrane 6 superfamily mamber 2 |
| TNF | tumor necrosis factor |
| TyG | Triglyceride and Glucose Index |
| US | ultrasound |
| VAI | Visceral Adiposity Index |
References
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baranova, A.; Younossi, Z.M. The future is around the corner: Noninvasive diagnosis of progressive nonalcoholic steatohepatitis. Hepatology 2008, 47, 374–375. [Google Scholar] [CrossRef] [PubMed]
- Skelly, M.M.; James, P.D.; Ryder, S.D. Findings on liver biopsy to investigate abnormal liver function tests in the absence of diagnostic serology. J. Hepatol. 2001, 35, 195–199. [Google Scholar] [CrossRef]
- Pendino, G.M.; Mariano, A.; Surace, P.; Caserta, C.A.; Fiorillo, M.T.; Amante, A.; Bruno, S.; Mangano, C.; Polito, I.; Amato, F.; et al. Prevalence and etiology of altered liver tests: A population-based survey in a Mediterranean town. Hepatology 2005, 41, 1151–1159. [Google Scholar] [CrossRef] [PubMed]
- Browning, J.D.; Szczepaniak, L.S.; Dobbins, R.; Nuremberg, P.; Horton, J.D.; Cohen, J.C.; Grundy, S.M.; Hobbs, H.H. Prevalence of hepatic steatosis in an urban population in the United States: Impact of ethnicity. Hepatology 2004, 40, 1387–1395. [Google Scholar] [CrossRef] [PubMed]
- Neuschwander-Tetri, B.A.; Caldwell, S.H. Nonalcoholic steatohepatitis: Summary of an AASLD single topic conference. Hepatology 2003, 37, 1202–1219. [Google Scholar] [CrossRef]
- Sorrentino, P.; Tarantino, G.; Conca, P.; Perrella, A.; Terracciano, M.L.; Vecchione, R.; Gargiulo, G.; Gennarelli, N.; Lobello, R. Silent non-alcoholic fatty liver disease-a clinical-histological study. J. Hepatol. 2004, 41, 751–757. [Google Scholar] [CrossRef]
- Fracanzani, A.L.; Valenti, L.; Bugianesi, E.; Andreoletti, M.; Colli, A.; Vanni, E.; Bertelli, C.; Fatta, E.; Bignamini, D.; Marchesini, G.; et al. Risk of severe liver disease in nonalcoholic fatty liver disease with normal aminotransferase levels: A role for insulin resistance and diabetes. Hepatology 2008, 48, 792–798. [Google Scholar] [CrossRef]
- Changzhou, C.; Yiming, L.; Chaohui, Y. Circulating miRNAs as novel diagnostic biomarkers in nonalcoholic fatty liver disease: A systematic review and meta-analysis. Can. J. Gastroenterol. Hepatol. 2019, 2019, 2096161. [Google Scholar] [CrossRef]
- Kotronen, A.; Peltonen, M.; Hakkarainen, A.; Sevastianova, K.; Bergholm, R.; Johansson, L.M.; Lundbom, N.; Rissanen, A.; Ridderstråle, M.; Groop, L.; et al. Prediction of non-alcoholic fatty liver disease and liver fat using metabolic and genetic factors. Gastroenterology 2009, 137, 865–872. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, D.; Kim, H.J.; Lee, C.H.; Yang, J.I.; Kim, W.; Kim, Y.J.; Yoon, J.H.; Cho, S.H.; Sung, M.W.; et al. Hepatic steatosis index: A simple screening tool reflecting nonalcoholic fatty liver disease. Dig. Liver Dis. 2010, 42, 503–508. [Google Scholar] [CrossRef]
- Bedogni, G.; Bellentani, S.; Miglioli, L.; Masutti, F.; Passalacqua, M.; Castiglione, A.; Tiribelli, C. The Fatty Liver Index: A simple and accurate predictor of hepatic steatosis in the general population. BMC Gastroenterol. 2006, 6, 33. [Google Scholar] [CrossRef] [Green Version]
- Kahal, S.; Strabburger, K.; Nowotny, B.; Livingstone, R.; Klüppelholz, B.; Keßel, K.; Hwang, J.-H.; Giani, G.; Hoffmann, B.; Pacini, G.; et al. Comparison of liver fat indices for the diagnosis of hepatic steatosis and insulin resistance. PLoS ONE 2014, 4, e94059. [Google Scholar]
- Pascot, A.; Lemieux, S.; Lemieux, I.; Prud’homme, D.; Tremblay, A.; Bouchard, C.; Nadeau, A.; Couillard, C.; Tchernof, A.; Bergeron, J.; et al. Age-related increase in visceral adipose tissue and body fat and the metabolic risk profile of premenopausal women. Diabetes Care 1999, 22, 1471–1478. [Google Scholar] [CrossRef]
- Zhang, S.; Du, T.; Zhang, J.; Lu, H.; Lin, X.; Xie, J.; Yang, Y.; Yu, X. The triglyceride and glucose index (TyG) is an effective biomarker to identify nonalcoholic fatty liver disease. Lipids Health Dis. 2017, 16, 15. [Google Scholar] [CrossRef] [Green Version]
- Fedchuk, L.; Nascimbeni, F.; Pais, R.; Charlotte, F.; Housset, C.; Ratziu, V. Performance and limitations of steatosis biomarkers in patients with nonalcoholic fatty liver disease. Alimentary 2014, 40, 1209–1222. [Google Scholar] [CrossRef] [PubMed]
- Poynard, T.; Ratziu, V.; Naveau, S.; Thabut, D.; Charlotte, F.; Messous, D.; Capron, D.; Abella, A.; Massard, J.; Ngo, Y.; et al. The diagnostic value of biomarkers (SteatoTest) for the prediction of liver steatosis. Comp. Hepatol. 2005, 4, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoo, T.; Byddar, M.; Hamilton, G.; Middleton, M.S.; Gamst, A.C.; Wolfson, T.; Hassanein, T.; Patton, H.M.; Lavine, J.E.; Schwimmer, J.B.; et al. Nonalcoholic Fatty Liver Disease: Diagnostic and Fat-Grading Accuracy of Low-Flip-Angle Multiecho Gradient-Recalled-Echo MR Imaging at 1.5 T. Radiology 2009, 251, 67–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, G.; Cruite, I.; Shiehmorteza, M.; Wolfson, T.; Gamst, A.C.; Hamilton, G.; Bydder, M.; Middleton, M.S.; Sirlin, C.B. Reproducibility of MRI-determined proton density fat fraction across two different MR scanner platforms. J. Magn. Reson. Imaging 2011, 34, 928–934. [Google Scholar] [CrossRef] [Green Version]
- Tang, A.; Tan, J.; Sun, M.; Hamilton, G.; Bydder, M.; Wolfson, T.; Gamst, A.C.; Middleton, M.; Brunt, E.M.; Loomba, R.; et al. Nonalcoholic fatty liver disease: MR imaging of liver proton density fat fraction to assess hepatic steatosis. Radiology 2013, 267, 422–431. [Google Scholar] [CrossRef] [Green Version]
- Negrete, L.; Middleton, M.S.; Clark, L.; Wolfson, T.; Gamst, A.C.; Lam, J.; Changchien, C.; Deyoung-Dominguez, I.M.; Hamilton, G.; Loomba, R.; et al. Inter-examination precision of magnitude-based MRI for estimation of segmental hepatic proton density fat fraction in obese subjects. J. Magn. Reson. Imaging 2014, 39, 1265–1271. [Google Scholar] [CrossRef] [Green Version]
- Middleton, M.S.; Heba, E.R.; Hooker, C.A.; Bashir, M.R.; Fowler, K.J.; Sandrasegaran, K.; Brunt, E.M.; Kleiner, D.E.; Doo, E.; Van Natta, M.L.; et al. Agreement between magnetic resonance imaging proton density fat fraction measurements and pathologist-assigned steatosis grades of liver biopsies from adults with non-alcoholic steatohepatitis. Gastroenterology 2017, 153, 753–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, J.; Bettencourt, R.; Cui, J.; Salotti, J.; Hooker, J.; Bhatt, A.; Hernandez, C.; Nguyen, P.; Aryafar, H.; Valasek, M.; et al. Association of non invasive quantitative decline in liver fat content on MRI with histologic response in nonalcoholic steatohepatitis. Ther. Adv. Gastroenterol. 2016, 9, 692–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loomba, R.; Neuschwander, B.A.; Sanyal, A.; Chalasani, N.; Diehl, A.M.; Terrault, N.; Kowdley, K.; Dasarathy, S.; Kleiner, D.; Behling, C.; et al. Multicenter validation of association between decline in MRI-PDFF and histologic response in nonalcoholic stetohepatitis. Hepatology 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Sasso, M.; Tengher-Barna, I.; Ziol, M.; Miette, V.; Fournier, C.; Sandrin, L.; Poupon, R.; Cardoso, A.C.; Marcellin, P.; Douvin, C.; et al. Novel controlled attenuation parameter for noninvasive assessment of steatosis using Fibroscan (R): Validation in chronic hepatitis C. J. Viral Hepat. 2012, 19, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Karlas, T.; Petroff, D.; Sasso, M.; Fan, J.G.; Mi, Y.Q.; de Lédinghen, V.; Kumar, M.; Lupsor-Platon, M.; Han, K.H.; Cardoso, A.C.; et al. Individual patients data meta-analysis of controlled attenuation parameter (CAP) technology for assessing steatosis. J. Hepatol. 2017, 66, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Myers, R.P.; Pollett, A.; Kirsch, R.; Pomier-Layrargues, G.; Beaton, M.; Levstik, M.; Duarte-Rojo, A.; Wong, D.; Crotty, P.; Elkashab, M. Controlled Attenuation Parameter (CAP): A non-invasive method for the detection of hepatic steatosis based on transient elastography. Liver Int. 2012, 32, 902–910. [Google Scholar] [CrossRef]
- De Ledinghen, V.; Wong, G.; Vergniol, J.; Chan, H.L.; Hiriart, J.B.; Chan, A.W.; Chermak, F.; Choi, P.C.; Foucher, J.; Chan, C.K.; et al. Controlled attenuation parameter for the diagnosis of steatosis in nonalcoholic fatty liver disease. J. Gastroenterol. Hepatol. 2016, 31, 848–855. [Google Scholar] [CrossRef]
- Boursier, J.; Fraysse, J.; Lafuma, A.; Torreton, E.; Ozbay, A.B. Increased healthcare resource utilization and costs in non-acoholic fatty liver disease/non-acoholic steatohepatitis patients with liver disease progression: A multivariate analysis of french national hospital care. J. Hepatol. 2019, 70, e1–e44. [Google Scholar]
- Cusi, K.; Chang, Z.; Harrison, S.; Lomonaco, R.; Bril, F.; Orsak, B.; Ortiz-Lopez, C.; Hecht, J.; Feldstein, A.E.; Webb, A.; et al. Limited value of plasma cytokeratin-18 as a biomarker for NASH and fibrosis in aptients with non-alcoholic fatty liver disease. J. Hepatol. 2014, 60, 167–174. [Google Scholar] [CrossRef]
- Kazankov, K.; Barrera, F.; Jon Møller, H.; Chiara Rosso, C.; Bugianesi, E.; David, E.; Ibrahim Kamal Jouness, R.; Esmaili, S.; Eslam, M.; McLeod, D.; et al. The macrophage activation marker sCD163 is associated with morphological disease stages patients with non-alcoholic fatty liver disease. Liver Int. 2016, 36, 1549–1557. [Google Scholar] [CrossRef] [PubMed]
- Palekar, N.A.; Naus, R.; Larson, S.P.; Ward, J.; Harrison, S.A. Clinical model for distinguishing nonalcoholic steatohepatitis from simple steatosis in patients with nonalcoholic fatty liver disease. Liver Int. 2006, 26, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Poynard, T.; Ratziu, V.; Charlotte, F.; Messous, D.; Munteanu, M.; Imbert-Bismut, F.; Massard, J.; Bonyhay, L.; Tahiri, M.; Thabut, D.; et al. Diagnostic value of biochemical markers (NAShTest) for the prediction of nonalcoholic steatohepatitis in patients with non alcoholic fatty liver 2006. BMC Gastroenterol. 2006, 10, 6–34. [Google Scholar]
- Dowman, J.K.; Tomlinson, J.W.; Newsome, P.N. Systematic review: The diagnosis and staging of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis. Aliment. Pharmacol. Ther. 2011, 33, 525–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, L.A.; Feldstein, A.E. Non-invasive diagnosis of nonalcoholic fatty liver and nonalcoholic steatohepatitis. J. Dig. Dis. 2011, 12, 10–16. [Google Scholar] [CrossRef]
- Shah, A.G.; Lydecker, A.; Murray, K.; Tetri, B.N.; Contos, M.J.; Sanyal, A.J. Comparison of noninvasive markers of fibrosis in patients with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2009, 7, 1104–1112. [Google Scholar] [CrossRef] [Green Version]
- Younossi, Z.M.; Jarrar, M.; Nugent, C.; Randhawa, M.; Afendy, M.; Stepanova, M.; Rafiq, N.; Goodman, Z.; Chandhoke, V.; Baranova, A. A novel diagnostic biomarker panel for obesity-related nonalcholic steatohepatitis (NASH). Obes. Surg. 2008, 18, 1430–1437. [Google Scholar] [CrossRef]
- Loomba, R.; Jain, A.; Diehl, A.M.; Guy, C.D.; Portenier, D.; Sudan, R.; Singh, S.; Faulkner, C.; Richards, L.; Hester, K.D.; et al. Validation of Serum Test for Advanced Liver Fibrosis in Patients with Nonalcoholic Steatohepatitis. Clin. Gastroenterol. Hepatol. 2019, 17, 1867–1876. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, W.M.; Voelker, M.; Thiel, R.; Becka, M.; Burt, A.; Schuppan, D.; Hubscher, S.; Roskams, T.; Pinzani, M.; Arthur, M.J. Serum markers detect the presence of liver fibrosis: A cohort study. Gastroenterology 2004, 127, 1704–1713. [Google Scholar] [CrossRef] [Green Version]
- Guha, I.N.; Parkes, J.; Roderick, P.; Chattopadhyay, D.; Cross, R.; Harris, S.; Kaye, P.; Burt, A.D.; Ryder, S.D.; Aithal, G.P.; et al. Noninvasive markers of fibrosis in nonalcoholic fatty liver disease: Validating the European Liver Fibrosis panel and exploring simple markers. Hepatology 2008, 47, 455–460. [Google Scholar] [CrossRef]
- Pinzani, M. The ELF panel: A new crystal ball in hepatology? Gut 2010, 59, 1165–1166. [Google Scholar] [CrossRef]
- Nielsen, M.J.; Nedergaard, A.F.; Sun, S.; Veidal, S.S.; Larsen, L.; Zheng, Q.; Suett, C.; Henriksen, K.; Christiansen, C.; Karsdal, M.A.; et al. The neo-epitope specific PRO-C3 ELISA measures true formation of type III collagen associated with liver and muscle parameters. Am. J. Transl. Res. 2013, 5, 303–315. [Google Scholar]
- Tanwar, S.; Trembling, P.M.; Guha, I.N.; Parkes, J.; Kaye, P.; Burt, A.D.; Ryder, S.D.; Aithal, G.P.; Day, C.P.; Rosenberg, W.M. Validation of terminal peptide of Procollagen III for the detection and assessment of nonalcoholic steatohepatitis in patients with nonalcoholic fatty liver disease. Hepatology 2013, 57, 103–111. [Google Scholar] [CrossRef]
- Boyle, M.; Tiniakos, D.; Schattenberg, J.M.; Ratziu, V.; Bugianessi, E.; Petta, S.; Oliveira, C.P.; Govaere, O.; Younes, R.; McPherson, S.; et al. Performance of the PRO-C3 collagen neo-epitope biomarker in non-alcoholic fatty liver disease. J. Hepatol. 2019, 1, 188–198. [Google Scholar] [CrossRef]
- Sanyal, A.J.; Hanf, R.; Harrison, S.A.; Bedossa, P.; Anstee, Q.M.; Ratziu, V.; Francque, S.M.; Majd, Z.; Sudrick, F.B.; Praca, E.; et al. A NIS4 for detection of active NASH (NAS ≥ 4) and significant fibrosis (F ≥ 2) in 714 patients at risk of NASH: Diagnostic metrics are not affected by age, gender, type 2 diabetes or obesity. Hepatology 2018, 68, 89A. [Google Scholar]
- Iruarrizaga-Lejarreta, M.; Martínez-Arranz, I.; Morrison, M.C.; Varela-Rey, M.; Ramos, D.F.; Delacruz-Villar, L.; Noureddin, M.; Martínez-Chantar, M.L.; Kleemann, R.; Alonso, C.; et al. Targeting the NAFLD metabolome and the shaping of precision medicine for patients with NASH. J. Hepatol. 2018, 68, S362–S363. [Google Scholar] [CrossRef]
- Mayo, R.; Crespo, J.; Martínez-Arranz, I.; Banales, J.M.; Arias, M.; Mincholé, I.; Aller de la Fuente, R.; Jimenez-Agüero, R.; Alonso, C.; de Luis, D.A.; et al. Metabolomic-based noninvasive serum test to diagnose nonalcoholic Steatohepatitis: Results from discovery and validation cohorts. Hepatol. Commun. 2018, 7, 807–818. [Google Scholar] [CrossRef]
- European Association for Study of Liver; Asociacion Latinoamericana para el Estudio del Higado; Castera, L.; Chan, H.; Arrese, M.; Afdhal, N.; Bedossa, P.; Friedrich-Rust, M.; Han, K.H.; Pinzani, M. EASL-ALEH Clinical practice guidelines tests for evaluation of on-invasive tests for liver disease severity and prognosis. J. Hepatol. 2015, 63, 237–264. [Google Scholar]
- Boursier, J.; Vergniol, J.; Guillet, A.; Hiriart, J.B.; Lannes, A.; Le Bail, B.; Michalak, S.; Chermak, F.; Bertrais, S.; Foucher, J.; et al. Diagnostic accuracy and prognostic significance of blood fibrosis tests and liver stiffness measurement by FibroScan in non alcoholic fatty liver disease. J. Hepatol. 2016, 65, 570–578. [Google Scholar] [CrossRef]
- Tapper, E.B.; Challies, T.; Imad Nasser, I.; Afdhal, N.H.; Lai, M. The performance of vibration controlled transient elastography in a US cohort of patients with nonalcoholic fatty liver disease. Am. J. Gastroenterol. 2016, 111, 677–684. [Google Scholar] [CrossRef] [Green Version]
- Bamber, J.; Cosgrove, D.; Dietrich, C.F.; Fromageau, J.; Bojunga, J.; Calliada, F.; Cantisani, V.; Correas, J.M.; D’Onofrio, M.; Drakonaki, E.E.; et al. EFSUMB guidelines and recommendations on the clinical use of ultrasound elastography. Part 1 Basic principles and technologies. Uktraschall Med. 2013, 34, 169–184. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Venkatesh, S.K.; Wang, Z.; Miller, F.H.; Motosugi, U.; Low, R.N.; Hassanein, T.; Asbach, P.; Godfrey, E.M.; Yin, M.; et al. Diagnostic performances of magnetic resonance elastography in staging liver fibrosis: A systematic review and meta-analysis of individual participant data. Clin. Gastroenterol. Hepatol. 2015, 13, 440–451. [Google Scholar] [CrossRef] [Green Version]
- Loomba, R.; Cui, J.; Wolfson, T.; Haufe, W.; Hooker, J.; Szeverenyi, N.; Ang, B.; Bhatt, A.; Wang, K.; Aryafar, H.; et al. Novel 3D magnetic resonance elastography for the noninvasive diagnosis of advanced fibrosis in NAFLD: A prospective study. Am. J. Gastroenterol. 2016, 111, 986–994. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, R.; Pavlides, M.; Tunnicliffe, E.M.; Piechnik, S.K.; Sarania, N.; Philips, R.; Collier, J.D.; Booth, J.C.; Schneider, J.E.; Wang, L.M.; et al. Multiparametric magnetic resonance for the non-invasive diagnosis of liver disease. J. Hepatol. 2014, 60, 69–77. [Google Scholar] [CrossRef] [Green Version]
- Pavlides, M.; Banerjee, R.; Sellwood, J.; Kelly, C.J.; Robson, M.D.; Booth, J.C.; Collier, J.; Neubauer, S.; Barnes, E. Multiparametric magnetic resonance imaging predicts clinical outcomes in a patients with chronic liver disease. J. Hepatol. 2016, 64, 308–315. [Google Scholar] [CrossRef] [Green Version]
- Romeo, S.; Kozlitina, J.; Xing, C.; Pertsemlidis, A.; Cox, D.; Pennacchio, L.A.; Boerwinkle, E.; Cohen, J.C.; Hobbs, H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008, 40, 1461–1465. [Google Scholar] [CrossRef] [Green Version]
- Kozlitina, J.; Smagris, E.; Stender, S.; Nordestgaard, B.G.; Zhou, H.H.; Tybjærg-Hansen, A.; Vogt, T.F.; Hobbs, H.H.; Cohen, J.C. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2014, 46, 352–356. [Google Scholar] [CrossRef] [Green Version]
- Mancina, R.; Dongiovanni, P.; Petta, S.; Pingitore, P.; Meroni, M.; Rametta, R.; Borén, J.; Montalcini, T.; Pujia, A.; Wiklund, O.; et al. The MBOAT7-TMC4 Variant rs641738 Increases Risk of Nonalcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology 2016, 150, 1219–1230. [Google Scholar] [CrossRef] [Green Version]
- Eslam, M.; Valenti, L.; Romeo, S. Genetics and epigenetics of NAFLD and NASH. J. Hepatol. 2018, 68, 268–279. [Google Scholar] [CrossRef]
- Eslam, M.; Hashem, A.M.; Romero-Gomez, M.; Berg, T.; Dore, G.J.; Mangia, A.; Chan, H.L.Y.; Irving, W.L.; Sheridan, D.; Abate, M.L.; et al. Fibrogene a gene-based model for staging liver fibrosis. J. Hepatol. 2016, 64, 390–398. [Google Scholar] [CrossRef] [Green Version]
- Kirpich, I.A.; Gobejishvili, L.N.; Bon Homme, M.; Waigel, S.; Cave, M.; Arteel, G.; Barve, S.S.; McClain, C.J.; Deaciuc, I.V. Integrated hepatic transcriptome and proteome analysis of mice with high-fat diet-induced nonalcoholic fatty liver disease. J. Nutr. Biochem. 2011, 22, 38–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Younossi, Z.M.; Baranova, A.; Ziegler, K.; Del Giacco, L.; Schlauch, K.; Born, T.L.; Elariny, H.; Gorreta, F.; VanMeter, A.; Younoszai, A.; et al. A genomic and proteomic study of the spectrum of non-alcoholic fatty liver disease. Hepatology 2005, 25, 760–771. [Google Scholar]
- Charlton, M.; Viker, K.; Krishnan, A.; Sanderson, S.; Veldt, B.; Kaalsbeek, A.J.; Kendrick, M.; Thompson, G.; Que, F.; Swain, J.; et al. Differential expression of lumican and fatty acid binding protein-1-New insights into the histologic spectrum of non-alcoholic fatty liver disease. Hepatology 2009, 49, 1375–1384. [Google Scholar] [CrossRef] [Green Version]
- Bell, L.N.; Theodorakis, J.L.; Vuppalanchi, R.; Saxena, R.; Bemis, K.G.; Wang, M.; Chalasani, N. Serum proteomics and biomarker discovery across the spectrum of nonalcoholic fatty liver disease. Hepatology 2010, 51, 111–120. [Google Scholar] [CrossRef] [Green Version]
- Bell, L.N.; Vuppalanchi, R.; Watkins, P.B.; Bonkovsky, H.L.; Serrano, J.; Fontana, R.J.; Wang, M.; Rochon, J.; Chalasani, N. Serum proteomic profiling in patiets with drug-induced liver injury. Aliment. Pharmacol. Ther. 2012, 35, 600–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.; Xu, C.; Xu, L.; Yu, J.; Miao, M.; Li, Y. Serum proteomic analy- sis revealed diagnostic value of hemoglobin for nonalcoholic fatty liver disease. J. Hepatol. 2012, 56, 241–247. [Google Scholar] [CrossRef]
- Suwen, Q.; Depeng, X.; Qiaoliang, L.; Xie, N.; Xia, J.; Huo, Q.; Li, P.; Chen, Q. Metabolomics screening of serum identifies pyroglutamate as a diagnostic biomarker for nonalcoholic steatohepatitis. Clin. Chim. Acta 2017, 473, 89–95. [Google Scholar]
- Kalhan, S.C.; Guo, L.; Edmison, J.; Dasarathy, S.; McCullough, A.J.; Hanson, R.W.; Milburn, M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metab. Clin. Exp. 2011, 60, 404–413. [Google Scholar] [CrossRef] [Green Version]
- Ferslew, B.C.; Xie, G.; Johnston, C.K.; Su, M.; Stewart, P.W.; Jia, W.; Brouwer, K.L.; Barritt, A.S. Altered bile acid metabolome in patients with nonalcoholic steatohepatitis. Dig. Dis. Sci. 2015, 60, 3318–3328. [Google Scholar] [CrossRef] [Green Version]
- Puri, P.; Wiest, M.; Cheung, O.; Mirshahi, F.; Sargeant, C.; Min, H.K.; Contos, M.J.; Sterling, R.K.; Fuchs, M.; Zhou, H. The plasma lipidomic signature of non alcoholic steatohepatitis. Hepatology 2009, 50, 1827–1838. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Oresic, M.; Leivonen, M.; Gopalacharyulu, P.; Hyysalo, J.; Arola, J.; Verrijken, A.; Francque, S.; Van Gaal, L.; Hyötyläinen, T.; et al. Non invasive detection of nonalcoholic steatohepatitis using clinical markers and circulating levels of lipids and metabolites. Clin. Gastroenterol. Hepatol. 2016, 14, 1463–1472. [Google Scholar] [CrossRef] [Green Version]
- Mahli, H. Emerging role of extracellular vescicles in liver diseases. Am. J. Physiol.-Gastrointest. Liver Physiol. 2019, 317, G739–G749. [Google Scholar] [CrossRef]
- Kornek, M.; Lynch, M.; Mehta, S.H.; Lai, M.; Exley, M.; Afdhal, N.H.; Schuppan, D. Circulating microparticles as disease-specific biomarkers of severity of in in patients with hepatitis C or nonalcoholic steatohepatitis. Gastroenterology 2012, 143, 448–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Brenner, D.A.; Kisseleva, T. Combatting fibrosis: Exosome-based therapies in the regression of liver fibrosis. Hepatol. Commun. 2019, 3, 180–192. [Google Scholar] [CrossRef]
- Mann, J.; Reeves, H.; Feldestein, A.E. Liquid biopsy for liver disease. Gut 2018, 67, 2204–2212. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Kim, S.Y.; Ko, E.; Lee, J.H.; Yi, H.S.; Yoo, Y.J.; Je, J.; Suh, S.J.; Jung, Y.K.; Kim, J.H.; et al. Exosomes derived from palmitic acid-treated hepatocytes induce fibrotic activation of hepatic stellate cells. Sci. Rep. 2017, 7, 3710. [Google Scholar] [CrossRef] [PubMed]
- Hardy, T.; Zyebel, M.; Day, C.P.; Dipper, C.; Masson, S.; McPherson, S.; Henderson, E.; Tiniakos, D.; White, S.; French, J.; et al. Plasma DNA methylation: A potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut 2017, 66, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Zeybel, M.; Hardy, T.; Wong, Y.K.; Mathers, J.C.; Fox, C.R.; Gackowska, A.; Oakley, F.; Burt, A.D.; Wilson, C.L.; Anstee, Q.M.; et al. Multigenerational epigenetic adaptation of the hepatic wound-healing response. Nat. Med. 2012, 18, 1369–1377. [Google Scholar] [CrossRef] [Green Version]
- Lambrecht, J.; Verhulst, S.; Mannaerts, I.; Reynaert, H.; van Grunsven, L.A. Prospects in non-invasive assessment of liver fibrosis: Liquid biopsy as the future gold standard? BBA-Mol. Basis Dis. 2018, 1864, 1024–1036. [Google Scholar] [CrossRef]
- Sun, C.; Liu, X.; Yi, Z.; Xiao, X.; Yang, M.; Hu, G.; Liu, H.; Liao, L.; Huang, F. Genome-wide analysis of long noncoding RNA expression profiles in patients with non-alcoholic fatty liver disease. IUBMB Life 2015, 67, 847–852. [Google Scholar] [CrossRef]
- Chen, G.; Yu, D.; Nian, X.; Liu, J.; Koenig, R.J.; Xu, B.; Sheng, L. LncRNA SRA promotes hepatic steatosis through repressing the expression of adipose trygliceride lipase (ATGL). Sci. Rep. 2016, 6, 35531. [Google Scholar] [CrossRef]
- Di Mauro, S.; Scamporrino, A.; Petta, S.; Urbano, F.; Filippello, A.; Ragusa, M.; Di Martino, M.T.; Scionti, F.; Grimaudo, S.; Pipitone, R.M.; et al. Serum coding and non-coding RNAs as biomarkers of NAFLD and fibrosis severity. Liver Int. 2019, 39, 1742–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szabo, G.; Csak, T. Role of microRNAs in NAFLD/NASH. Dig. Dis. Sci. 2016, 61, 1314–1324. [Google Scholar] [CrossRef] [PubMed]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revelaed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, O.; Puri, P.; Eicken, C.; Contos, M.J.; Mirshahi, F.; Maher, J.W.; Kellum, J.M.; Min, H.; Luketic, V.A.; Sanyal, A.J. Nonalcoholic steatohepatitis is associated with altered hepatic microRNA expression. Hepatology 2008, 48, 1810–1820. [Google Scholar] [CrossRef] [Green Version]
- Boursier, J.; Diehl, A.M. Implication of gut microbiota in nonalcoholic fatty liver disease. PLoS Pathog. 2015, 11, e1004559. [Google Scholar] [CrossRef] [Green Version]
- Loomba, R.; Seguritan, V.; Li, W.; Long, T.; Klitgord, N.; Bhatt, A.; SingDulai, P.; Caussy, C.; Battencourt, R.; Highlander, S.; et al. Gut microbiome-derived biomarkers for the detection of advanced fibrosis in NAFLD. Cell Metab. 2017, 25, 1054–1062. [Google Scholar] [CrossRef]
| A. Blood Liver Funtion Tests | Parameter Measured | Pros | Cons | AUROC |
|---|---|---|---|---|
| ELF panel [39] | Hyaluronic acid (HA), Tissue inhibitor metalloproteinase 1 (TIMP1), and Aminoterminal peptide of procollagen 3 (PIIINP). | Feasible in large number of subjects Good outcome correlation | Commercial test not routinely available | 0.93 in adults 0.99 in pediatric patients |
| Pro-C3 [42] | Pro collagen III | Able to discriminate simple fatty liver from NASH and different stages of fibrosis | Commercial test | 0.86 |
| NASH NIS4 [44] | MicroRNA 34a-5p; alpha2 macrogobulin (A2M), Haemooglobin A1c (HbA1c), and Chitinase-3-like protein 1 (CHI3L1 also known as YKL40) | This tool can enrich the selection of patients—candidate to experimental trials—with active NASH and significant fibrosis | Commercial test; performances might vary according to the baseline characteristics of the studied population | 0.82 |
| Lipidomic serum test § (OWLiver) [45] | Two subsequent analyses of 11 and 20 triglycerides panel to be used in adults with BMI > 25 | Able to discriminate normal liver form NAFLD and NAFLD from NASH | Commercial test performed in a centralized laboratory | 0.79 or 0.81 (according to inclusion or exclusion of patients with glucose >136 mg/dl) |
| B.US-Based Physical Tests | Parameter Measured | Pros | Cons | AUROC |
| TE [47,48] | Liver stiffness | Short processing time and outpatient clinic setting | Measurement failures reported in up to 20% and XL probe required in obese patients | 0.95 for F4 0.93 for F3 0.84 for F2 fibrosis |
| Point shear wave elastgraphy (ARFI) [49] | Liver stiffness | Short processing and outclinic setting | Quality criteria not well defined, lack of large-scale studies | 0.78–0.89 for F4 0.74–0.97 for F3 0.70–0.83 for F2 fibrosis |
| B. Not US-Based physical tests | Parameter Measured | Pros | Cons | AUROC |
| MRE [50,51] | Liver stiffness | Not influenced by BMI and inflammation | Long processing, expensive, and not largely available | 0.88–0.97 for F4 0.89–0.96 for F3 0.86–0.89 for F2 |
| LiverMultiScan (multiparametric resonance) [52] | Fibrosis and inflammation mapping | Quick and no contrast agent required | Further validation studies required | 0.85 for F4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piazzolla, V.A.; Mangia, A. Noninvasive Diagnosis of NAFLD and NASH. Cells 2020, 9, 1005. https://doi.org/10.3390/cells9041005
Piazzolla VA, Mangia A. Noninvasive Diagnosis of NAFLD and NASH. Cells. 2020; 9(4):1005. https://doi.org/10.3390/cells9041005
Chicago/Turabian StylePiazzolla, Valeria Annarita, and Alessandra Mangia. 2020. "Noninvasive Diagnosis of NAFLD and NASH" Cells 9, no. 4: 1005. https://doi.org/10.3390/cells9041005
APA StylePiazzolla, V. A., & Mangia, A. (2020). Noninvasive Diagnosis of NAFLD and NASH. Cells, 9(4), 1005. https://doi.org/10.3390/cells9041005






