A Novel Function for KLF4 in Modulating the De-Differentiation of EpCAM−/CD133− nonStem Cells into EpCAM+/CD133+ Liver Cancer Stem Cells in HCC Cell Line HuH7
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of LCSCs and Non-stem Cells from HuH-7
2.2. Overexpression of KLF4 in EpCAM-/CD133- Non-stem Cells
2.3. RT-qPCR
2.4. Chromatin Immunoprecipitation Assay (ChIP) and ChIP-qPCR
2.5. Immunofluorescence Staining
2.6. Colony Forming and Spheroid Formation Assays
2.7. MTT Assay
2.8. Migration and Invasion Assay
2.9. Transfection and Luciferase Reporter Assay
2.10. Xenotransplantation (In vivo Tumorigenicity Assay)
2.11. Ethics Statement
2.12. IHC Staining
2.13. Statistical Analysis
3. Results
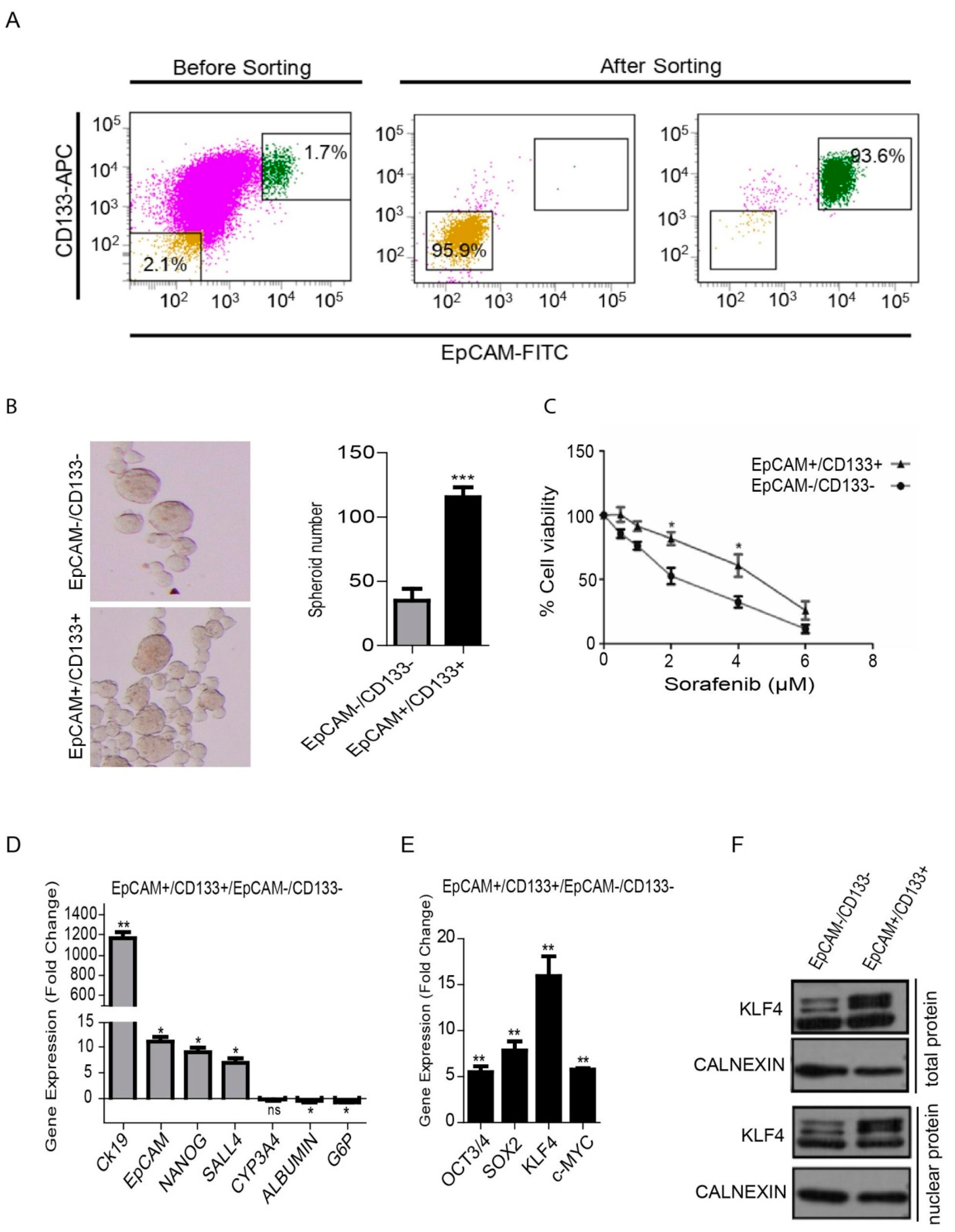
3.1. Characterization of Liver Cancer Stem Cells (LCSCs) and Non-Stem Cells in HuH-7
3.2. KLF4 Increases Stemness Phenotype via Transcriptional Regulation of EpCAM
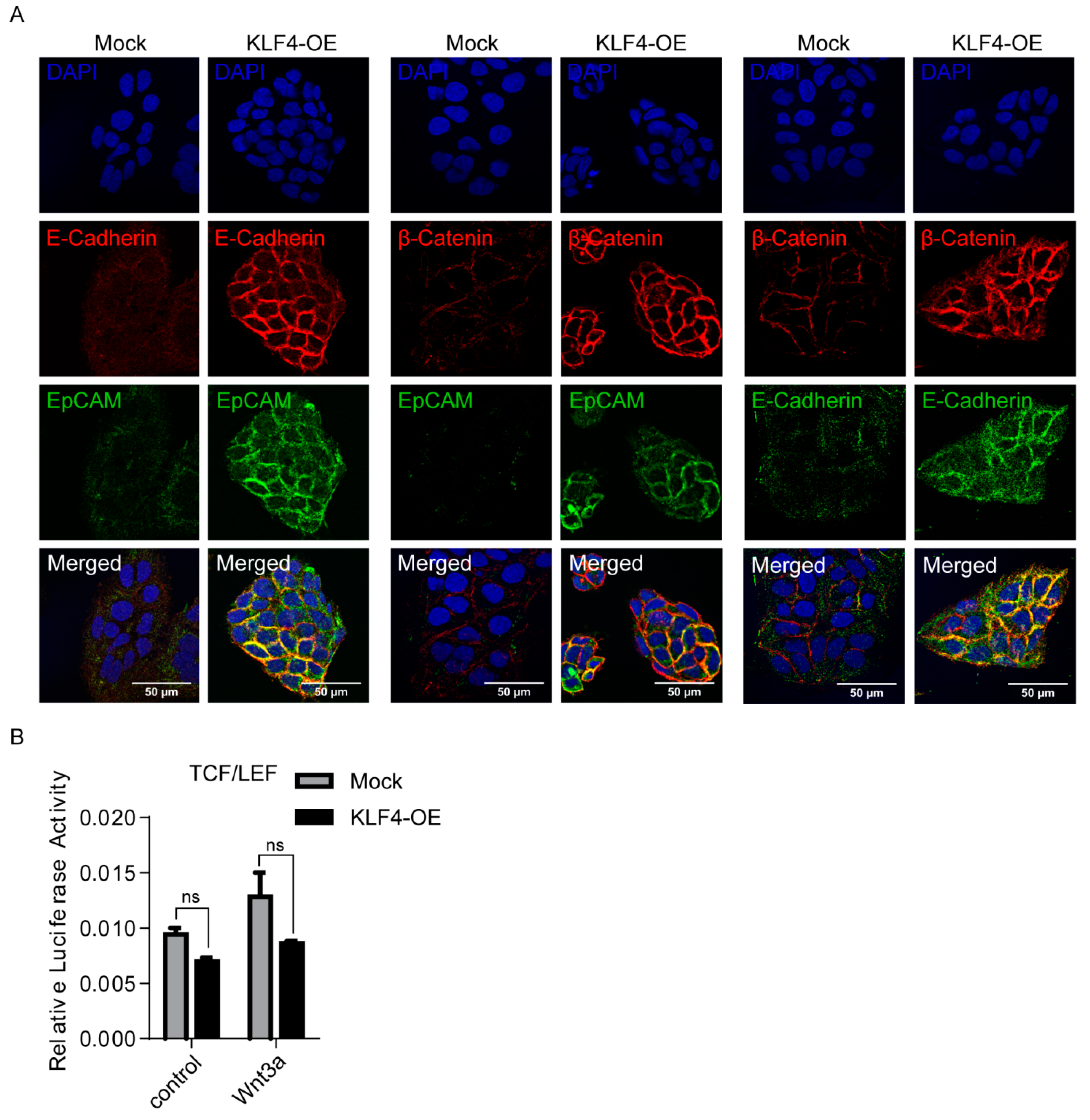
3.3. KLF4 Increases EpCAM, E-CAD and β-CAT Cell Membrane Expressions
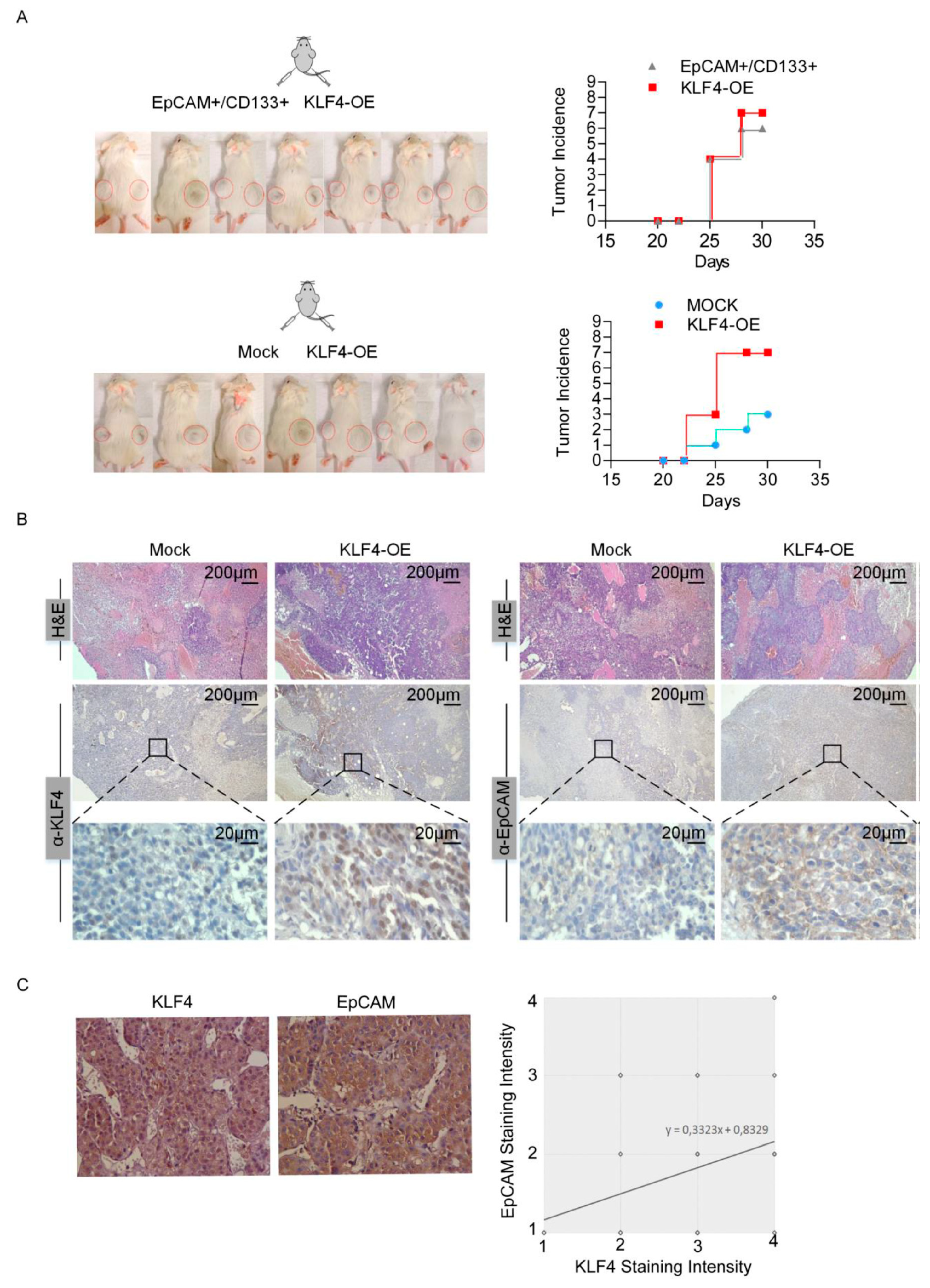
3.4. KLF4 Restores Tumorigenic Potential of Non-stem Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Global Burden of Disease Cancer, C.; Fitzmaurice, C.; Allen, C.; Barber, R.M.; Barregard, L.; Bhutta, Z.A.; Brenner, H.; Dicker, D.J.; Chimed-Orchir, O.; Dandona, R.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2017, 3, 524–548. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; He, M.; Chan, A.W.H.; Song, Q.X.; Sze, S.C.; Chen, H.; Man, M.K.H.; Man, K.; Chan, S.L.; Lai, P.B.S.; et al. Genomic and Epigenomic Features of Primary and Recurrent Hepatocellular Carcinomas. Gastroenterology 2019, 157, 1630–1645 e1636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeng, K.S.; Chang, C.F.; Jeng, W.J.; Sheen, I.S.; Jeng, C.J. Heterogeneity of hepatocellular carcinoma contributes to cancer progression. Crit. Rev. Oncol. Hematol. 2015, 94, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Losic, B.; Craig, A.J.; Villacorta-Martin, C.; Martins-Filho, S.N.; Akers, N.; Chen, X.; Ahsen, M.E.; von Felden, J.; Labgaa, I.; D’Avola, D.; et al. Intratumoral heterogeneity and clonal evolution in liver cancer. Nat. Commun. 2020, 11, 291. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Zhao, X.; Yang, X.R.; Li, F.Q.; Zhou, X.L.; Wu, K.; Zhang, X.; Sun, Q.M.; Cao, Y.; Zhu, H.M.; et al. Circumventing intratumoral heterogeneity to identify potential therapeutic targets in hepatocellular carcinoma. J. Hepatol. 2017, 67, 293–301. [Google Scholar] [CrossRef]
- Vermeulen, L.; de Sousa e Melo, F.; Richel, D.J.; Medema, J.P. The developing cancer stem-cell model: Clinical challenges and opportunities. Lancet Oncol. 2012, 13, e83–e89. [Google Scholar] [CrossRef]
- Nguyen, L.V.; Vanner, R.; Dirks, P.; Eaves, C.J. Cancer stem cells: An evolving concept. Nat. Rev. Cancer 2012, 12, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Medema, J.P. Cancer stem cells: The challenges ahead. Nat. Cell Biol. 2013, 15, 338–344. [Google Scholar] [CrossRef]
- Sottoriva, A.; Verhoeff, J.J.; Borovski, T.; McWeeney, S.K.; Naumov, L.; Medema, J.P.; Sloot, P.M.; Vermeulen, L. Cancer stem cell tumor model reveals invasive morphology and increased phenotypical heterogeneity. Cancer Res. 2010, 70, 46–56. [Google Scholar] [CrossRef] [Green Version]
- Tang, D.G. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012, 22, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Peitzsch, C.; Tyutyunnykova, A.; Pantel, K.; Dubrovska, A. Cancer stem cells: The root of tumor recurrence and metastases. Semin. Cancer Biol. 2017, 44, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lewis, M.T.; Huang, J.; Gutierrez, C.; Osborne, C.K.; Wu, M.F.; Hilsenbeck, S.G.; Pavlick, A.; Zhang, X.; Chamness, G.C.; et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J. Natl. Cancer Inst. 2008, 100, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Kusumbe, A.P.; Bapat, S.A. Cancer stem cells and aneuploid populations within developing tumors are the major determinants of tumor dormancy. Cancer Res. 2009, 69, 9245–9253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Li, Y.; Yu, T.S.; McKay, R.M.; Burns, D.K.; Kernie, S.G.; Parada, L.F. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 2012, 488, 522–526. [Google Scholar] [CrossRef] [Green Version]
- De Angelis, M.L.; Francescangeli, F.; La Torre, F.; Zeuner, A. Stem Cell Plasticity and Dormancy in the Development of Cancer Therapy Resistance. Front Oncol. 2019, 9, 626. [Google Scholar] [CrossRef] [Green Version]
- Dirks, P. Cancer stem cells: Invitation to a second round. Nature 2010, 466, 40–41. [Google Scholar] [CrossRef]
- Vicente-Duenas, C.; Hauer, J.; Ruiz-Roca, L.; Ingenhag, D.; Rodriguez-Meira, A.; Auer, F.; Borkhardt, A.; Sanchez-Garcia, I. Tumoral stem cell reprogramming as a driver of cancer: Theory, biological models, implications in cancer therapy. Semin. Cancer Biol. 2015, 32, 3–9. [Google Scholar] [CrossRef]
- Das, P.K.; Pillai, S.; Rakib, M.A.; Khanam, J.A.; Gopalan, V.; Lam, A.K.Y.; Islam, F. Plasticity of Cancer Stem Cell: Origin and Role in Disease Progression and Therapy Resistance. Stem. Cell Rev. Rep. 2020, 16, 397–412. [Google Scholar] [CrossRef]
- Strauss, R.; Hamerlik, P.; Lieber, A.; Bartek, J. Regulation of stem cell plasticity: Mechanisms and relevance to tissue biology and cancer. Mol. Ther. 2012, 20, 887–897. [Google Scholar] [CrossRef] [Green Version]
- Govaere, O.; Roskams, T. Pathogenesis and prognosis of hepatocellular carcinoma at the cellular and molecular levels. Clin. Liver Dis. 2015, 19, 261–276. [Google Scholar] [CrossRef] [PubMed]
- Dolle, L.; Theise, N.D.; Schmelzer, E.; Boulter, L.; Gires, O.; van Grunsven, L.A. EpCAM and the biology of hepatic stem/progenitor cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G233–G250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terris, B.; Cavard, C.; Perret, C. EpCAM, a new marker for cancer stem cells in hepatocellular carcinoma. J. Hepatol. 2010, 52, 280–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okabe, M.; Tsukahara, Y.; Tanaka, M.; Suzuki, K.; Saito, S.; Kamiya, Y.; Tsujimura, T.; Nakamura, K.; Miyajima, A. Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse liver. Development 2009, 136, 1951–1960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmelzer, E.; Reid, L.M. EpCAM expression in normal, non-pathological tissues. Front Biosci. 2008, 13, 3096–3100. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, T.; Forgues, M.; Wang, W.; Kim, J.W.; Ye, Q.; Jia, H.; Budhu, A.; Zanetti, K.A.; Chen, Y.; Qin, L.X.; et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008, 68, 1451–1461. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, T.; Ji, J.; Budhu, A.; Forgues, M.; Yang, W.; Wang, H.Y.; Jia, H.; Ye, Q.; Qin, L.X.; Wauthier, E.; et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 2009, 136, 1012–1024. [Google Scholar] [CrossRef] [Green Version]
- Suetsugu, A.; Nagaki, M.; Aoki, H.; Motohashi, T.; Kunisada, T.; Moriwaki, H. Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem. Biophys. Res. Commun. 2006, 351, 820–824. [Google Scholar] [CrossRef]
- Yin, S.; Li, J.; Hu, C.; Chen, X.; Yao, M.; Yan, M.; Jiang, G.; Ge, C.; Xie, H.; Wan, D.; et al. CD133 positive hepatocellular carcinoma cells possess high capacity for tumorigenicity. Int. J. Cancer 2007, 120, 1444–1450. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, D.; Zhang, H.; He, H.; Zhang, C.; Zhao, W.; Shao, R.G. CD133(+)EpCAM(+) phenotype possesses more characteristics of tumor initiating cells in hepatocellular carcinoma Huh7 cells. Int. J. Biol. Sci. 2012, 8, 992–1004. [Google Scholar] [CrossRef] [Green Version]
- Went, P.; Vasei, M.; Bubendorf, L.; Terracciano, L.; Tornillo, L.; Riede, U.; Kononen, J.; Simon, R.; Sauter, G.; Baeuerle, P.A. Frequent high-level expression of the immunotherapeutic target Ep-CAM in colon, stomach, prostate and lung cancers. Br. J. Cancer 2006, 94, 128–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osta, W.A.; Chen, Y.; Mikhitarian, K.; Mitas, M.; Salem, M.; Hannun, Y.A.; Cole, D.J.; Gillanders, W.E. EpCAM is overexpressed in breast cancer and is a potential target for breast cancer gene therapy. Cancer Res. 2004, 64, 5818–5824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baeuerle, P.A.; Gires, O. EpCAM (CD326) finding its role in cancer. Br. J. Cancer 2007, 96, 417–423. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007, 445, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.T.; Berry, P.A.; Hyde, C.; Stower, M.J.; Maitland, N.J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005, 65, 10946–10951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eramo, A.; Lotti, F.; Sette, G.; Pilozzi, E.; Biffoni, M.; Di Virgilio, A.; Conticello, C.; Ruco, L.; Peschle, C.; De Maria, R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008, 15, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Rowland, B.D.; Bernards, R.; Peeper, D.S. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat. Cell Biol. 2005, 7, 1074–1082. [Google Scholar] [CrossRef] [PubMed]
- Rowland, B.D.; Peeper, D.S. KLF4, p21 and context-dependent opposing forces in cancer. Nat. Rev. Cancer 2006, 6, 11–23. [Google Scholar] [CrossRef]
- Ghaleb, A.M.; Yang, V.W. Kruppel-like factor 4 (KLF4): What we currently know. Gene 2017, 611, 27–37. [Google Scholar] [CrossRef]
- Leng, Z.; Li, Y.; Zhou, G.; Lv, X.; Ai, W.; Li, J.; Hou, L. Kruppel-like factor 4 regulates stemness and mesenchymal properties of colorectal cancer stem cells through the TGF-beta1/Smad/snail pathway. J. Cell Mol. Med. 2020, 24, 1866–1877. [Google Scholar] [CrossRef] [Green Version]
- Murgai, M.; Ju, W.; Eason, M.; Kline, J.; Beury, D.W.; Kaczanowska, S.; Miettinen, M.M.; Kruhlak, M.; Lei, H.; Shern, J.F.; et al. KLF4-dependent perivascular cell plasticity mediates pre-metastatic niche formation and metastasis. Nat. Med. 2017, 23, 1176–1190. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.T.; Li, Y.L.; Zhang, Y.Q.; Xu, T.; Lu, B.; Fang, L.; Gao, J.Q.; Yu, L.S.; Zhu, D.F.; Yang, B.; et al. KLF4 functions as an oncogene in promoting cancer stem cell-like characteristics in osteosarcoma cells. Acta Pharmacol. Sin. 2019, 40, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Li, J.; Chen, H.; Fu, J.; Ray, S.; Huang, S.; Zheng, H.; Ai, W. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene 2011, 30, 2161–2172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.Y.; Wang, L.; Luan, S.H.; Zhang, H.S.; Huang, W.T.; Wang, N.H. The PGI-KLF4 pathway regulates self-renewal of glioma stem cells residing in the mesenchymal niches in human gliomas. Neoplasma 2014, 61, 401–410. [Google Scholar] [CrossRef] [Green Version]
- Firtina Karagonlar, Z.; Koc, D.; Sahin, E.; Avci, S.T.; Yilmaz, M.; Atabey, N.; Erdal, E. Effect of adipocyte-secreted factors on EpCAM+/CD133+ hepatic stem cell population. Biochem. Biophys. Res. Commun. 2016, 474, 482–490. [Google Scholar] [CrossRef]
- Papapetrou, E.P.; Tomishima, M.J.; Chambers, S.M.; Mica, Y.; Reed, E.; Menon, J.; Tabar, V.; Mo, Q.; Studer, L.; Sadelain, M. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 12759–12764. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Peng, Z.; Tang, H.; Xie, D.; Jia, Z.; Zhong, L.; Zhao, S.; Ma, Z.; Gao, Y.; Zeng, L.; et al. Loss of KLF4 and consequential downregulation of Smad7 exacerbate oncogenic TGF-beta signaling in and promote progression of hepatocellular carcinoma. Oncogene 2017, 36, 2957–2968. [Google Scholar] [CrossRef] [Green Version]
- Firtina Karagonlar, Z.; Koc, D.; Iscan, E.; Erdal, E.; Atabey, N. Elevated hepatocyte growth factor expression as an autocrine c-Met activation mechanism in acquired resistance to sorafenib in hepatocellular carcinoma cells. Cancer Sci. 2016, 107, 407–416. [Google Scholar] [CrossRef]
- Rodriguez, F.J.; Lewis-Tuffin, L.J.; Anastasiadis, P.Z. E-cadherin’s dark side: Possible role in tumor progression. Biochim. Biophys. Acta 2012, 1826, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Padmanaban, V.; Krol, I.; Suhail, Y.; Szczerba, B.M.; Aceto, N.; Bader, J.S.; Ewald, A.J. E-cadherin is required for metastasis in multiple models of breast cancer. Nature 2019, 573, 439–444. [Google Scholar] [CrossRef]
- Hu, Q.P.; Kuang, J.Y.; Yang, Q.K.; Bian, X.W.; Yu, S.C. Beyond a tumor suppressor: Soluble E-cadherin promotes the progression of cancer. Int. J. Cancer 2016, 138, 2804–2812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yori, J.L.; Johnson, E.; Zhou, G.; Jain, M.K.; Keri, R.A. Kruppel-like factor 4 inhibits epithelial-to-mesenchymal transition through regulation of E-cadherin gene expression. J. Biol. Chem. 2010, 285, 16854–16863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiwari, N.; Meyer-Schaller, N.; Arnold, P.; Antoniadis, H.; Pachkov, M.; van Nimwegen, E.; Christofori, G. Klf4 is a transcriptional regulator of genes critical for EMT, including Jnk1 (Mapk8). PLoS ONE 2013, 8, e57329. [Google Scholar] [CrossRef]
- Chen, J.; Zhao, J.; Ma, R.; Lin, H.; Liang, X.; Cai, X. Prognostic significance of E-cadherin expression in hepatocellular carcinoma: A meta-analysis. PLoS ONE 2014, 9, e103952. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, T.; Makino, R.; Mitamura, K. Frequent down-regulation of E-cadherin by genetic and epigenetic changes in the malignant progression of hepatocellular carcinomas. Clin. Cancer Res. 2001, 7, 594–599. [Google Scholar]
- Wei, Y.; Van Nhieu, J.T.; Prigent, S.; Srivatanakul, P.; Tiollais, P.; Buendia, M.A. Altered expression of E-cadherin in hepatocellular carcinoma: Correlations with genetic alterations, beta-catenin expression, and clinical features. Hepatology 2002, 36, 692–701. [Google Scholar] [CrossRef]
- Soyama, A.; Eguchi, S.; Takatsuki, M.; Kawashita, Y.; Hidaka, M.; Tokai, H.; Nagayoshi, S.; Mochizuki, S.; Matsumoto, S.; Hamasaki, K.; et al. Significance of the serum level of soluble E-cadherin in patients with HCC. Hepatogastroenterology 2008, 55, 1390–1393. [Google Scholar]
- Orsulic, S.; Huber, O.; Aberle, H.; Arnold, S.; Kemler, R. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. J. Cell Sci. 1999, 112, 1237–1245. [Google Scholar]
- Yamada, S.; Pokutta, S.; Drees, F.; Weis, W.I.; Nelson, W.J. Deconstructing the cadherin-catenin-actin complex. Cell 2005, 123, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.; Lisby, A.; Ma, C.; Lo, N.; Ehmer, U.; Hayer, K.E.; Furth, E.E.; Viatour, P. Promotion of growth factor signaling as a critical function of beta-catenin during HCC progression. Nat. Commun. 2019, 10, 1909. [Google Scholar] [CrossRef]
- Vicente-Duenas, C.; Romero-Camarero, I.; Cobaleda, C.; Sanchez-Garcia, I. Function of oncogenes in cancer development: A changing paradigm. EMBO J. 2013, 32, 1502–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Hsu, H.S.; Chen, Y.W.; Tsai, T.H.; How, C.K.; Wang, C.Y.; Hung, S.C.; Chang, Y.L.; Tsai, M.L.; Lee, Y.Y.; et al. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS ONE 2008, 3, e2637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.M.; Liu, S.; Lu, H.; Zhang, H.; Zhang, P.J.; Gimotty, P.A.; Guerra, M.; Guo, W.; Xu, X. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene 2012, 31, 4898–4911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, P.; Qiu, J.; Li, B.; Hong, J.; Lu, C.; Wang, L.; Wang, J.; Hu, Y.; Jia, W.; Yuan, Y. Role of Sox2 and Oct4 in predicting survival of hepatocellular carcinoma patients after hepatectomy. Clin. Biochem. 2011, 44, 582–589. [Google Scholar] [CrossRef]
- Zheng, Y.W.; Nie, Y.Z.; Taniguchi, H. Cellular reprogramming and hepatocellular carcinoma development. World J. Gastroenterol. 2013, 19, 8850–8860. [Google Scholar] [CrossRef]
- Sun, C.; Sun, L.; Li, Y.; Kang, X.; Zhang, S.; Liu, Y. Sox2 expression predicts poor survival of hepatocellular carcinoma patients and it promotes liver cancer cell invasion by activating Slug. Med. Oncol. 2013, 30, 503. [Google Scholar] [CrossRef]
- Li, Q.; Gao, Y.; Jia, Z.; Mishra, L.; Guo, K.; Li, Z.; Le, X.; Wei, D.; Huang, S.; Xie, K. Dysregulated Kruppel-like factor 4 and vitamin D receptor signaling contribute to progression of hepatocellular carcinoma. Gastroenterology 2012, 143, 799–810 e792. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.S.; Chu, H.C.; Yen, Y.C.; Lewis, B.C.; Chen, Y.W. Kruppel-like factor 4, a tumor suppressor in hepatocellular carcinoma cells reverts epithelial mesenchymal transition by suppressing slug expression. PLoS ONE 2012, 7, e43593. [Google Scholar] [CrossRef]
- Pang, L.; Xu, L.; Yuan, C.; Li, X.; Zhang, X.; Wang, W.; Guo, X.; Ouyang, Y.; Qiao, L.; Wang, Z.; et al. Activation of EGFR-KLF4 positive feedback loop results in acquired resistance to sorafenib in hepatocellular carcinoma. Mol. Carcinog 2019, 58, 2118–2126. [Google Scholar] [CrossRef]
- Hsu, H.T.; Wu, P.R.; Chen, C.J.; Hsu, L.S.; Yeh, C.M.; Hsing, M.T.; Chiang, Y.S.; Lai, M.T.; Yeh, K.T. High cytoplasmic expression of Kruppel-like factor 4 is an independent prognostic factor of better survival in hepatocellular carcinoma. Int. J. Mol. Sci. 2014, 15, 9894–9906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, X.; Li, Y.W.; Jin, J.J.; Zhou, Y.; Ren, Z.G.; Qiu, S.J.; Zhang, B.H. The clinical and prognostic implications of pluripotent stem cell gene expression in hepatocellular carcinoma. Oncol. Lett. 2013, 5, 1155–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prange, W.; Breuhahn, K.; Fischer, F.; Zilkens, C.; Pietsch, T.; Petmecky, K.; Eilers, R.; Dienes, H.P.; Schirmacher, P. Beta-catenin accumulation in the progression of human hepatocarcinogenesis correlates with loss of E-cadherin and accumulation of p53, but not with expression of conventional WNT-1 target genes. J. Pathol. 2003, 201, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, L.; Weis, W.I. Structure and biochemistry of cadherins and catenins. Cold Spring Harb. Perspect Biol. 2009, 1, a003053. [Google Scholar] [CrossRef] [PubMed]
- De La Coste, A.; Romagnolo, B.; Billuart, P.; Renard, C.A.; Buendia, M.A.; Soubrane, O.; Fabre, M.; Chelly, J.; Beldjord, C.; Kahn, A.; et al. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc. Natl. Acad. Sci. USA 1998, 95, 8847–8851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurent-Puig, P.; Legoix, P.; Bluteau, O.; Belghiti, J.; Franco, D.; Binot, F.; Monges, G.; Thomas, G.; Bioulac-Sage, P.; Zucman-Rossi, J. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology 2001, 120, 1763–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Utikal, J.; Polo, J.M.; Stadtfeld, M.; Maherali, N.; Kulalert, W.; Walsh, R.M.; Khalil, A.; Rheinwald, J.G.; Hochedlinger, K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 2009, 460, 1145–1148. [Google Scholar] [CrossRef]
- Carette, J.E.; Pruszak, J.; Varadarajan, M.; Blomen, V.A.; Gokhale, S.; Camargo, F.D.; Wernig, M.; Jaenisch, R.; Brummelkamp, T.R. Generation of iPSCs from cultured human malignant cells. Blood 2010, 115, 4039–4042. [Google Scholar] [CrossRef] [Green Version]
- Miyoshi, N.; Ishii, H.; Nagai, K.; Hoshino, H.; Mimori, K.; Tanaka, F.; Nagano, H.; Sekimoto, M.; Doki, Y.; Mori, M. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc. Natl. Acad. Sci. USA 2010, 107, 40–45. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Mejia, V.; Fraga, M.F.; Menendez, P. iPSCs from cancer cells: Challenges and opportunities. Trends Mol. Med. 2012, 18, 245–247. [Google Scholar] [CrossRef]
- Van Keymeulen, A.; Lee, M.Y.; Ousset, M.; Brohee, S.; Rorive, S.; Giraddi, R.R.; Wuidart, A.; Bouvencourt, G.; Dubois, C.; Salmon, I.; et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature 2015, 525, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Murayama, Y.; Hashimoto, K.; Nakamura, Y.; Lin, C.S.; Yokoyama, K.K.; Saito, S. Role of tumor suppressor genes in the cancer-associated reprogramming of human induced pluripotent stem cells. Stem. Cell Res. Ther. 2014, 5, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Jeong, J.; Park, S.; Jin, Y.W.; Lee, S.S.; Lee, S.B.; Choi, D. Establishment of Hepatocellular Cancer Induced Pluripotent Stem Cells Using a Reprogramming Technique. Gut. Liver 2017, 11, 261–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krizhanovsky, V.; Lowe, S.W. Stem cells: The promises and perils of p53. Nature 2009, 460, 1085–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scaffidi, P.; Misteli, T. In vitro generation of human cells with cancer stem cell properties. Nat. Cell Biol. 2011, 13, 1051–1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Firtina Karagonlar, Z.; Akbari, S.; Karabicici, M.; Sahin, E.; Tercan Avci, S.; Ersoy, N.; Eren Ates, K.; Balli, T.; Karacicek, B.; Kaplan, K.N.; et al. A Novel Function for KLF4 in Modulating the De-Differentiation of EpCAM−/CD133− nonStem Cells into EpCAM+/CD133+ Liver Cancer Stem Cells in HCC Cell Line HuH7. Cells 2020, 9, 1198. https://doi.org/10.3390/cells9051198
Firtina Karagonlar Z, Akbari S, Karabicici M, Sahin E, Tercan Avci S, Ersoy N, Eren Ates K, Balli T, Karacicek B, Kaplan KN, et al. A Novel Function for KLF4 in Modulating the De-Differentiation of EpCAM−/CD133− nonStem Cells into EpCAM+/CD133+ Liver Cancer Stem Cells in HCC Cell Line HuH7. Cells. 2020; 9(5):1198. https://doi.org/10.3390/cells9051198
Chicago/Turabian StyleFirtina Karagonlar, Zeynep, Soheil Akbari, Mustafa Karabicici, Eren Sahin, Sanem Tercan Avci, Nevin Ersoy, Kıvılcım Eren Ates, Tugsan Balli, Bilge Karacicek, Kubra Nur Kaplan, and et al. 2020. "A Novel Function for KLF4 in Modulating the De-Differentiation of EpCAM−/CD133− nonStem Cells into EpCAM+/CD133+ Liver Cancer Stem Cells in HCC Cell Line HuH7" Cells 9, no. 5: 1198. https://doi.org/10.3390/cells9051198
APA StyleFirtina Karagonlar, Z., Akbari, S., Karabicici, M., Sahin, E., Tercan Avci, S., Ersoy, N., Eren Ates, K., Balli, T., Karacicek, B., Kaplan, K. N., Celiker, C., Atabey, N., & Erdal, E. (2020). A Novel Function for KLF4 in Modulating the De-Differentiation of EpCAM−/CD133− nonStem Cells into EpCAM+/CD133+ Liver Cancer Stem Cells in HCC Cell Line HuH7. Cells, 9(5), 1198. https://doi.org/10.3390/cells9051198




