Epigenetic Features of Human Perinatal Stem Cells Redefine Their Stemness Potential
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture of Human Cells
2.2. RNA Extraction and Reverse Transcription
2.3. Real Time Quantitative PCR (qPCR)
2.4. MTT (3[4,5-Dimethylthiazol-2yl]-2,5-Diphenyl Tetrazolium Bromide) Assay
2.5. Flow Cytometry
2.6. Immunofluorescent Analysis
2.7. Pyrosequencing
2.8. Absolute Telomere Length Quantification
2.9. Statistical Analysis
3. Results
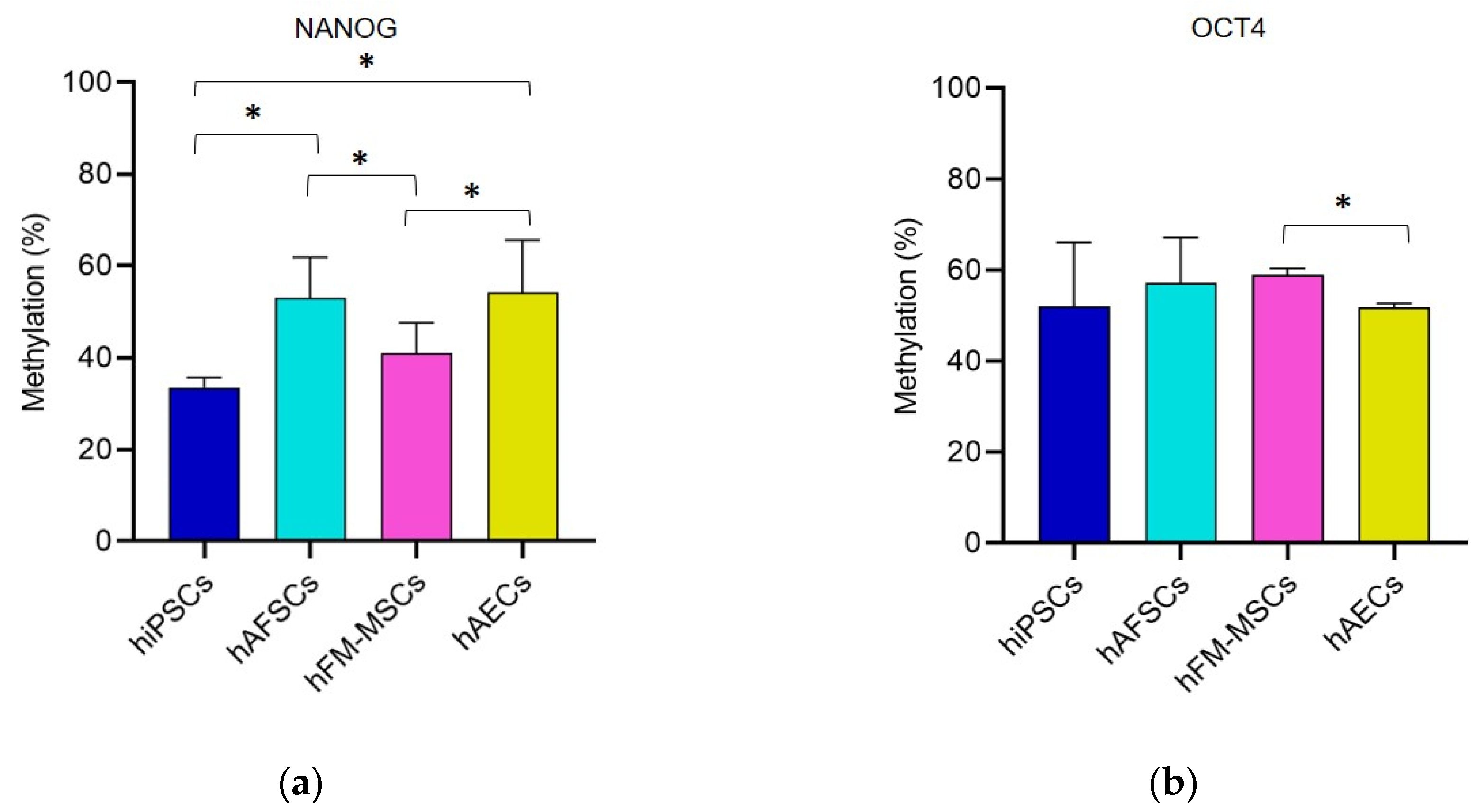
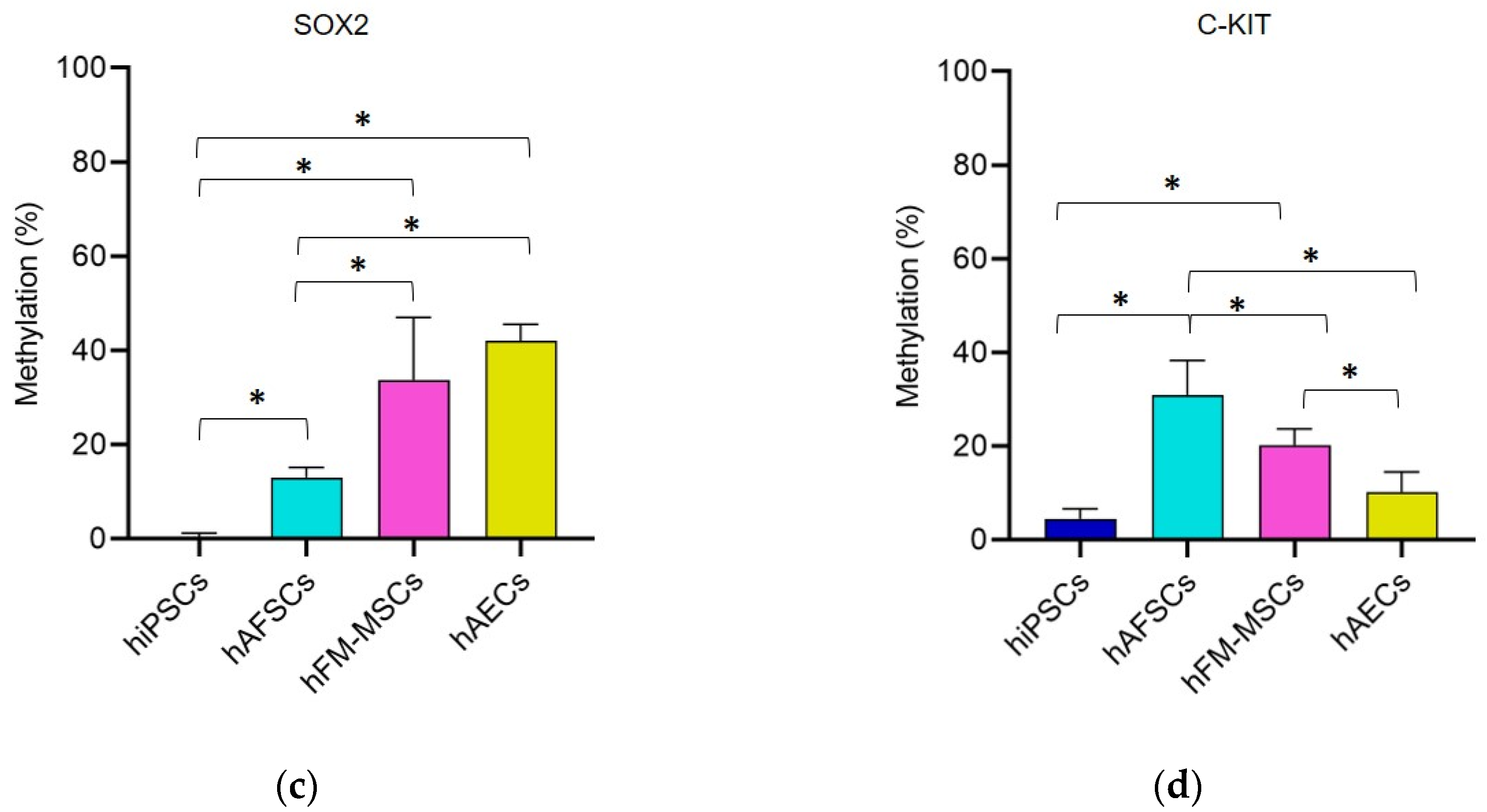
3.1. Promoter DNA Methylation Profile of Stemness Gene in Perinatal SCs
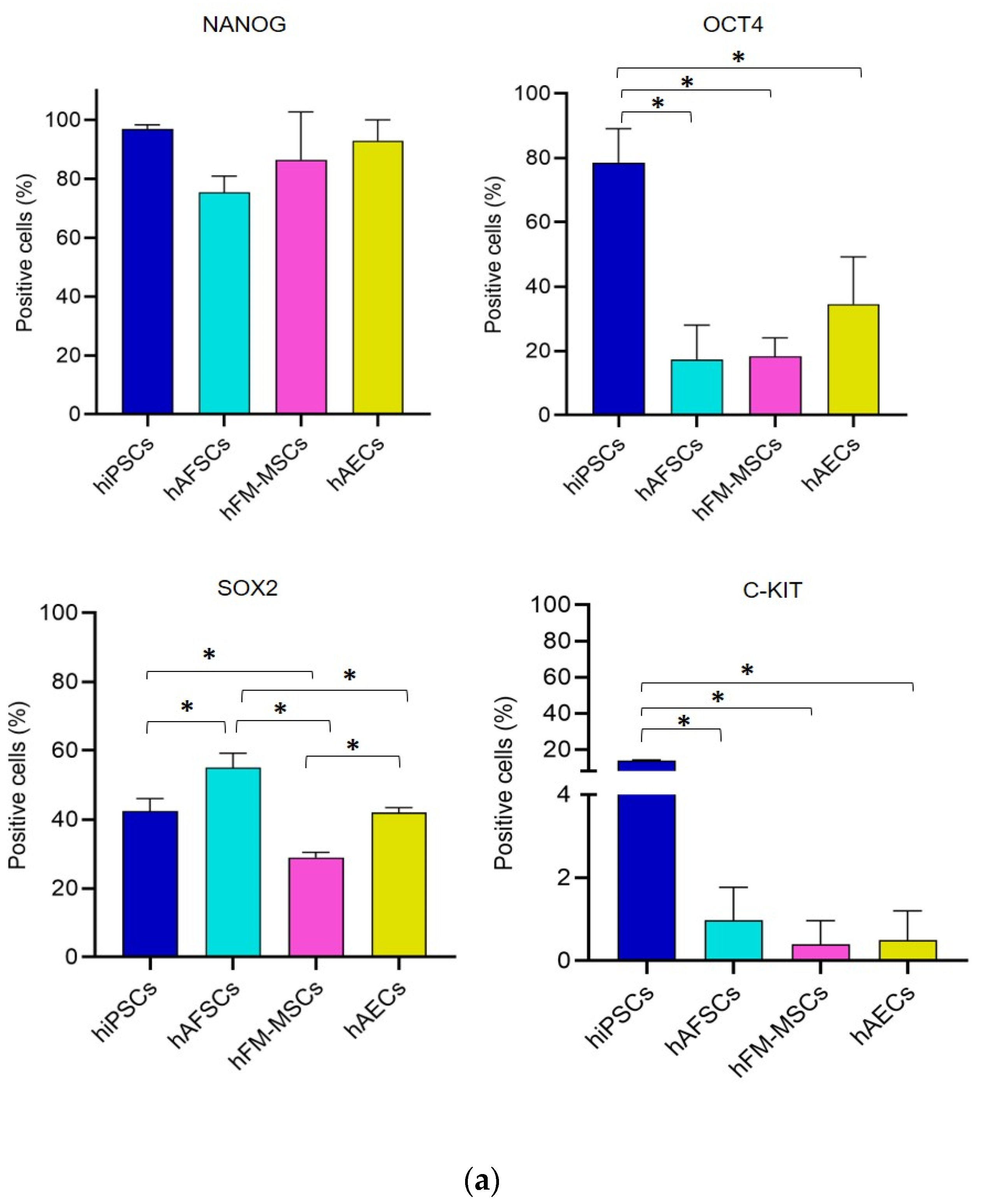
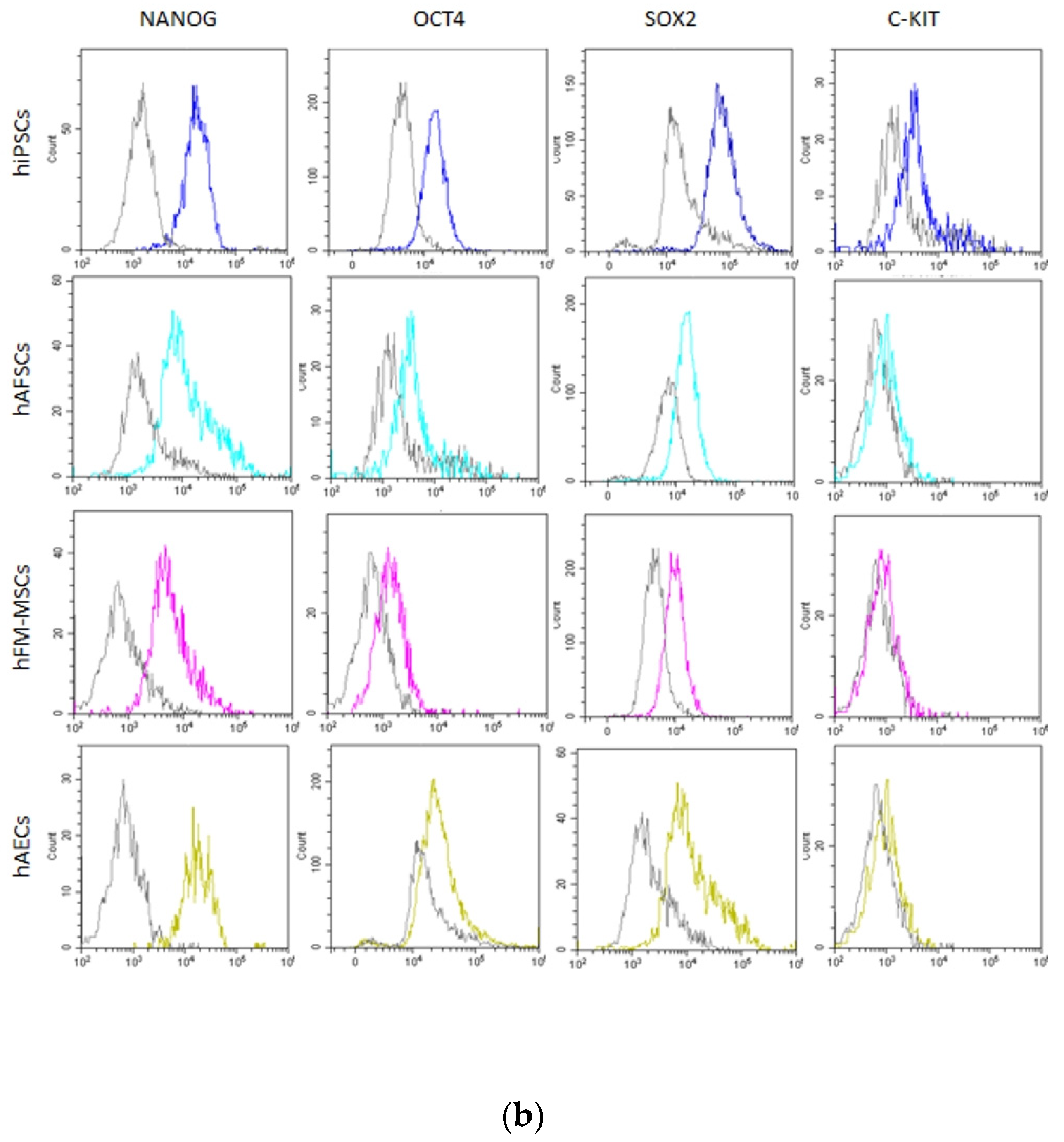
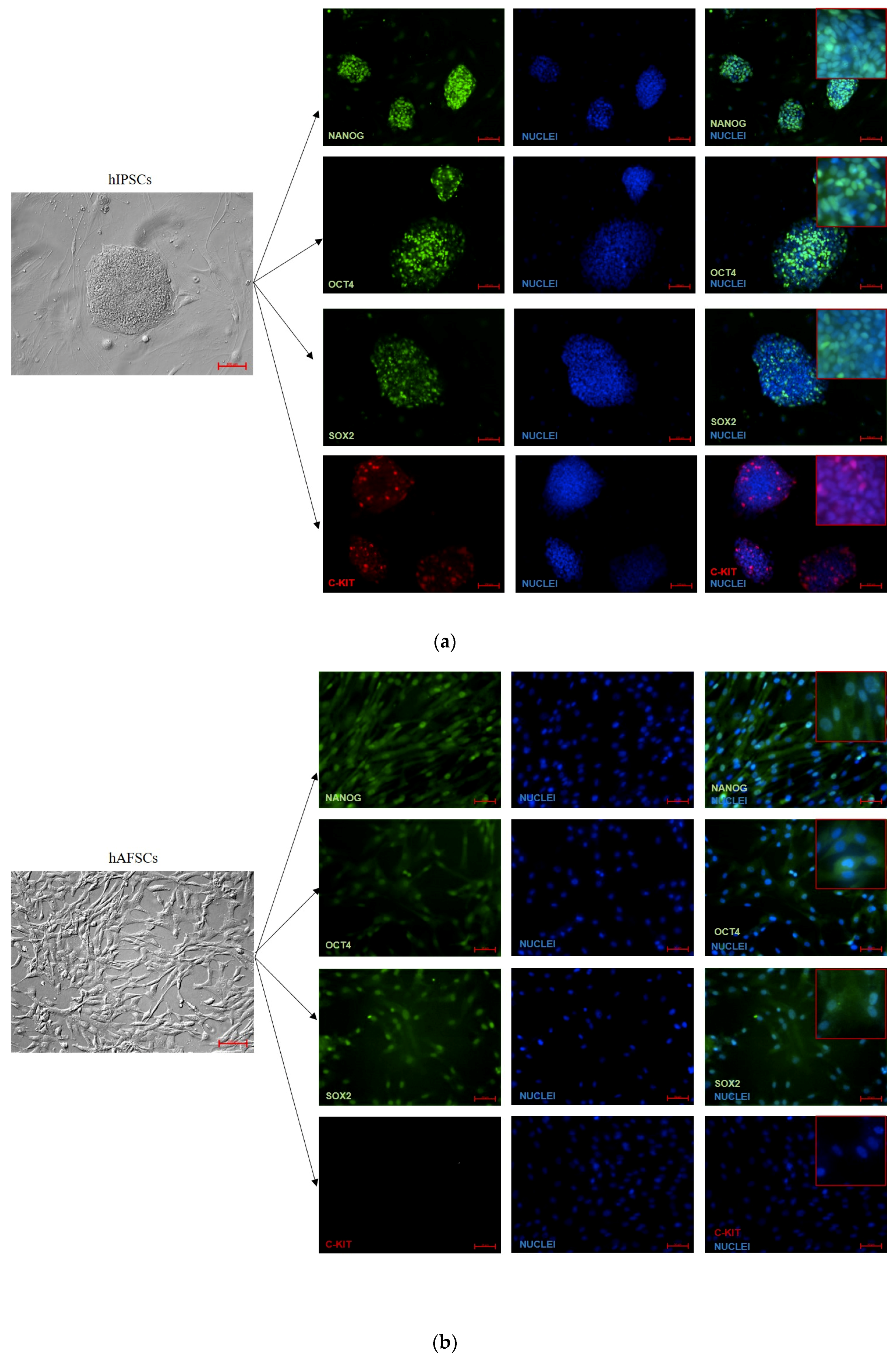
3.2. Analysis of Pluripotency Gene and Protein Expression in hiPSCs and Human Perinatal SCs
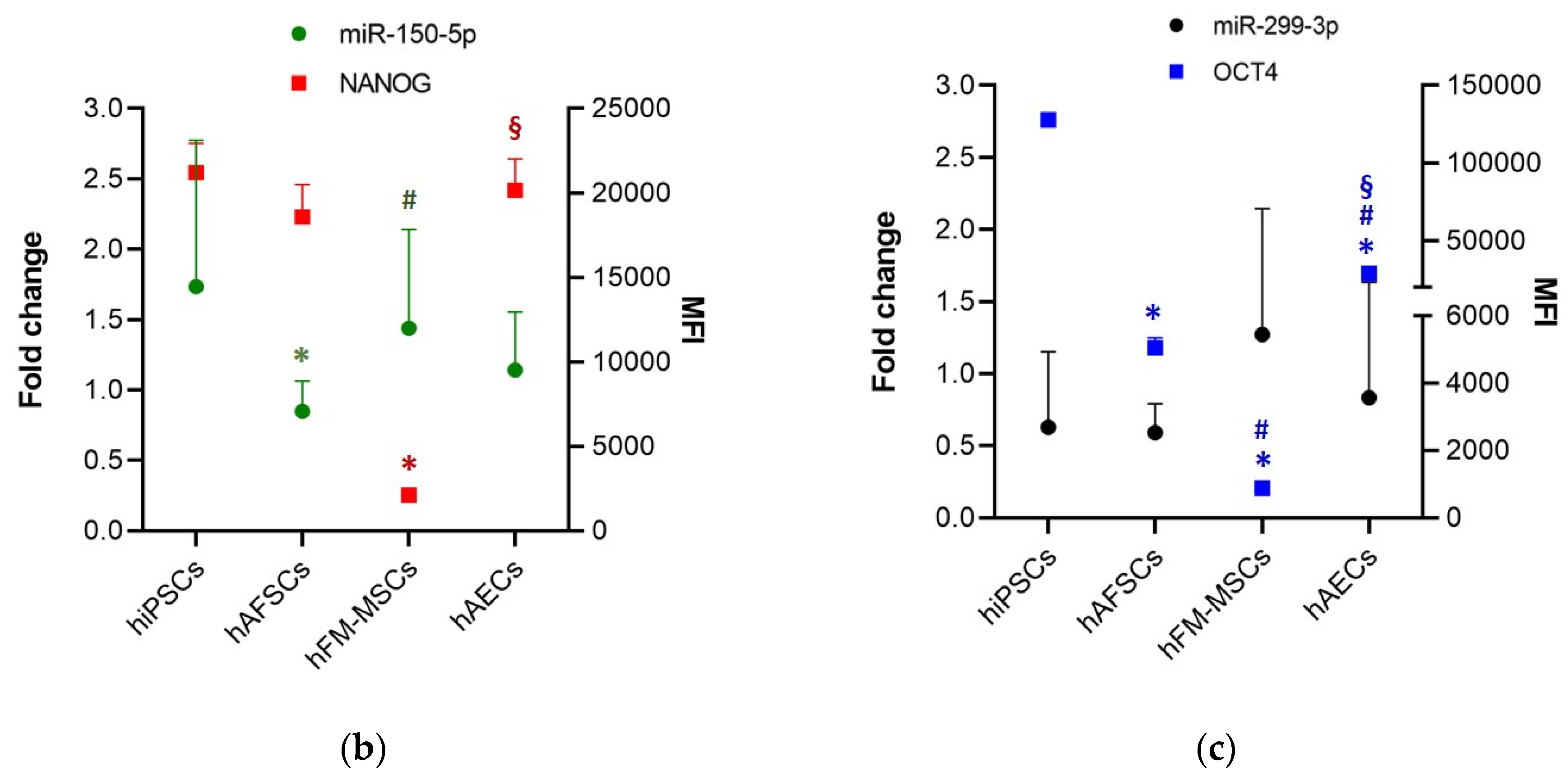
3.3. Perinatal SCs Expressed miRNAs That Are Involved in the Regulation of the “Stemness” Gene
3.4. Telomere Length Quantification in hiPSCs and Perinatal SCs
3.5. Metabolic Activity in hiPSCs and Perinatal SCs
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bieback, K. Critical Parameters for the Isolation of Mesenchymal Stem Cells from Umbilical Cord Blood. Stem Cells 2004, 22, 625–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prentice, D.A. Adult Stem Cells: Successful Standard for Regenerative Medicine. Circ. Res. 2019, 124, 837–839. [Google Scholar] [CrossRef] [PubMed]
- Ilancheran, S.; Moodley, Y.; Manuelpillai, U. Human Fetal Membranes: A Source of Stem Cells for Tissue Regeneration and Repair? Placenta 2009, 30, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Gaggi, G.; Izzicupo, P.; di Credico, A.; Sancilio, S.; di Baldassarre, A.; Ghinassi, B. Spare Parts from Discarded Materials: Fetal Annexes in Regenerative Medicine. Int. J. Mol. Sci. 2019, 20, 1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, G.; Zhang, Y. Embryonic stem cell and induced pluripotent stem cell: An epigenetic perspective. Cell Res. 2013, 23, 49–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Baldassarre, A.; Cimetta, E.; Bollini, S.; Gaggi, G.; Ghinassi, B. Human-Induced Pluripotent Stem Cell Technology and Cardiomyocyte Generation: Progress and Clinical Applications. Cells 2018, 7, 48. [Google Scholar] [CrossRef] [Green Version]
- Ilic, D.; Polak, J.M. Stem cells in regenerative medicine: Introduction. Br. Med. Bull. 2011, 98, 117–126. [Google Scholar] [CrossRef] [Green Version]
- Kmiecik, G.; Niklińska, W.; Kuć, P.; Pancewicz-Wojtkiewicz, J.; Fil, D.; Karwowska, A.; Karczewski, J.; Mackiewicz, Z. Fetal membranes as a source of stem cells. Adv. Med. Sci. 2013, 58, 185–195. [Google Scholar] [CrossRef] [Green Version]
- Okere, B.; Alviano, F.; Costa, R.; Quaglino, D.; Ricci, F.; Dominici, M.; Paolucci, P.; Bonsi, L.; Iughetti, L. In vitro differentiation of human amniotic epithelial cells into insulin-producing 3D spheroids. Int. J. Immunopathol. Pharmacol. 2015, 28, 390–402. [Google Scholar] [CrossRef] [Green Version]
- Di Baldassarre, A.; D’Amico, M.A.; Izzicupo, P.; Gaggi, G.; Guarnieri, S.; Mariggiò, M.A.; Antonucci, I.; Corneo, B.; Sirabella, D.; Stuppia, L.; et al. Cardiomyocytes Derived from Human CardiopoieticAmniotic Fluids. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Rodrigues, M.; Antonucci, I.; Elabd, S.; Kancherla, S.; Marchisio, M.; Blattner, C.; Stuppia, L. p53 Is Active in Human Amniotic Fluid Stem Cells. Stem Cells Dev. 2018, 27, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Amabile, G.; Welner, R.S.; Nombela-Arrieta, C.; D’Alise, A.M.; Di Ruscio, A.; Ebralidze, A.K.; Kraytsberg, Y.; Ye, M.; Kocher, O.; Neuberg, D.S.; et al. In vivo generation of transplantable human hematopoietic cells from induced pluripotent stem cells. Blood 2013, 121, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Antonucci, I.; Di Pietro, R.; Alfonsi, M.; Centurione, M.A.; Centurione, L.; Sancilio, S.; Pelagatti, F.; D’amico, M.A.; Di Baldassarre, A.; Piattelli, A.; et al. Human Second Trimester Amniotic Fluid Cells are Able to Create Embryoid Body-Like Structures in Vitro and to Show Typical Expression Profiles of Embryonic and Primordial Germ Cells. Cell Transplant. 2014, 23, 1501–1515. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, A.; Rossi, M.; Alviano, F.; Poggi, P.; Zannini, C.; Marchionni, C.; Ricci, F.; Tazzari, P.L.; Taglioli, V.; Calder, P.C.; et al. Restored in vivo-like membrane lipidomics positively influence in vitro features of cultured mesenchymal stromal/stem cells derived from human placenta. Stem Cell Res. Ther. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Mauro, M.; Izzicupo, P.; Santarelli, F.; Falone, S.; Pennelli, A.; Amicarelli, F.; Calafiore, A.M.; Di Baldassarre, A.; Gallina, S. ACE and AGTR1 Polymorphisms and Left Ventricular Hypertrophy in Endurance Athletes. Med. Sci. Sports Exerc. 2010, 42, 915–921. [Google Scholar] [CrossRef] [PubMed]
- Chen, C. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Mannelli, M.; Ferruzzi, P.; Luciani, P.; Crescioli, C.; Buci, L.; Corona, G.; Serio, M.; Peri, A. Cushing’s Syndrome in a Patient with Bilateral Macronodular Adrenal Hyperplasia Responding to Cisapride: An in Vivo and in Vitro Study. J. Clin. Endocrinol. Metab. 2003, 88, 4616–4622. [Google Scholar] [CrossRef] [Green Version]
- Aasen, T.; Raya, A.; Barrero, M.J.; Garreta, E.; Consiglio, A.; Gonzalez, F.; Vassena, R.; Bilić, J.; Pekarik, V.; Tiscornia, G.; et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 2008, 26, 1276–1284. [Google Scholar] [CrossRef]
- Jiang, Y.; Jakovcevski, M.; Bharadwaj, R.; Connor, C.; Schroeder, F.A.; Lin, C.L.; Straubhaar, J.; Martin, G.; Akbarian, S. Setdb1 Histone Methyltransferase Regulates Mood-Related Behaviors and Expression of the NMDA Receptor Subunit NR2B. J. Neurosci. 2010, 30, 7152–7167. [Google Scholar] [CrossRef] [Green Version]
- Ghinassi, B.; Ferro, L.; Masiello, F.; Tirelli, V.; Sanchez, M.; Migliaccio, G.; Whitsett, C.; Kachala, S.; Riviere, I.; Sadelain, M.; et al. Recovery and Biodistribution of Ex Vivo Expanded Human Erythroblasts Injected into NOD/SCID/IL2R γ null mice. Stem Cells Int. 2011, 1–13. [Google Scholar] [CrossRef] [Green Version]
- D’amico, M.A.; Ghinassi, B.; Izzicupo, P.; di Ruscio, A.; di Baldassarre, A. IL-6 Activates PI3K and PKCζ Signaling and Determines Cardiac Differentiation in Rat Embryonic H9c2 Cells: IL-6 AND CARDIAC DIFFERENTIATION OF H9c2 CELLS. J. Cell. Physiol. 2016, 231, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Ghinassi, B.; D’Addazio, G.; Di Baldassarre, A.; Femminella, B.; Di Vincenzo, G.; Piattelli, M.; Gaggi, G.; Sinjari, B. Immunohistochemical Results of Soft Tissues around a New Implant Healing-Abutment Surface: A Human Study. J. Clin. Med. 2020, 9, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashyap, V.; Rezende, N.C.; Scotland, K.B.; Shaffer, S.M.; Persson, J.L.; Gudas, L.J.; Mongan, N.P. Regulation of Stem Cell Pluripotency and Differentiation Involves a Mutual Regulatory Circuit of the Nanog, OCT4, and SOX2 Pluripotency Transcription Factors With Polycomb Repressive Complexes and Stem Cell microRNAs. Stem Cells Dev. 2009, 18, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Fraser, L.; Taylor, A.H.; Forrester, L.M. SCF/KIT Inhibition Has a Cumulative but Reversible Effect on the Self-Renewal of Embryonic Stem Cells and on the Survival of Differentiating Cells. Cell. Reprogramming 2013, 15, 259–268. [Google Scholar] [CrossRef]
- Migliaccio, A.R.; Martelli, F.; Verrucci, M.; Sanchez, M.; Valeri, M.; Migliaccio, G.; Vannucchi, A.M.; Zigariello, M.; Di Baldassarre, A.; Ghinassi, B.; et al. Gata1 expression driven by the alternative HS2 enhancer in the spleen rescues the hematopoietic failure induced by the hypomorphic Gata1low mutation. Blood 2009, 114, 2107–2120. [Google Scholar] [CrossRef] [Green Version]
- Martelli, F.; Ghinassi, B.; Lorenzini, R.; Vannucchi, A.M.; Rana, R.A.; Nishikawa, M.; Partamian, S.; Migliaccio, G.; Migliaccio, A.R. Thrombopoietin Inhibits Murine Mast Cell Differentiation. Stem Cells 2008, 26, 912–919. [Google Scholar] [CrossRef] [Green Version]
- Nishino, K.; Toyoda, M.; Yamazaki-Inoue, M.; Fukawatase, Y.; Chikazawa, E.; Sakaguchi, H.; Akutsu, H.; Umezawa, A. DNA Methylation Dynamics in Human Induced Pluripotent Stem Cells over Time. PLoS Genet. 2011, 7, e1002085. [Google Scholar] [CrossRef] [Green Version]
- Tesarova, L.; Simara, P.; Stejskal, S.; Koutna, I. The Aberrant DNA Methylation Profile of Human Induced Pluripotent Stem Cells Is Connected to the Reprogramming Process and Is Normalized During In Vitro Culture. PLoS ONE 2016, 11, e0157974. [Google Scholar] [CrossRef]
- Göhring, A.R.; Reuter, S.; Clement, J.H.; Cheng, X.; Theobald, J.; Wölfl, S.; Mrowka, R. Human microRNA-299-3p decreases invasive behavior of cancer cells by downregulation of Oct4 expression and causes apoptosis. PLoS ONE 2017, 12, e0174912. [Google Scholar] [CrossRef] [Green Version]
- Xu, N.; Papagiannakopoulos, T.; Pan, G.; Thomson, J.A.; Kosik, K.S. MicroRNA-145 Regulates OCT4, SOX2, and KLF4 and Represses Pluripotency in Human Embryonic Stem Cells. Cell 2009, 137, 647–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, D.; Zhou, P.; Wang, Y.; Zhang, Y.; Zhang, R.; Zhang, L.; Chen, S.; Fu, W.; Ruan, B.; Xu, H.; et al. miR-150 Suppresses the Proliferation and Tumorigenicity of Leukemia Stem Cells by Targeting the Nanog Signaling Pathway. Front. Pharmacol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yue, Y.; Wang, R.; Gong, B.; Duan, Z. MicroRNA-145 inhibits tumorigenesis and invasion of cervical cancer stem cells. Int. J. Oncol. 2017, 50, 853–862. [Google Scholar] [CrossRef] [Green Version]
- Marion, R.M.; Strati, K.; Li, H.; Tejera, A.; Schoeftner, S.; Ortega, S.; Serrano, M.; Blasco, M.A. Telomeres Acquire Embryonic Stem Cell Characteristics in Induced Pluripotent Stem Cells. Cell Stem Cell 2009, 4, 141–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shyh-Chang, N.; Daley, G.Q.; Cantley, L.C. Stem cell metabolism in tissue development and aging. Development 2013, 140, 2535–2547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, I.M.; Biswas, A.; Verfaillie, C.; Khurana, S. Energy Producing Metabolic Pathways in Functional Regulation of the Hematopoietic Stem Cells: HSC PROLIFERATION, AGEING. IUBMB Life 2018, 70, 612–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volarevic, V.; Markovic, B.S.; Gazdic, M.; Volarevic, A.; Jovicic, N.; Arsenijevic, N.; Armstrong, L.; Djonov, V.; Lako, M.; Stojkovic, M. Ethical and Safety Issues of Stem Cell-Based Therapy. Int. J. Med. Sci. 2018, 15, 36–45. [Google Scholar] [CrossRef] [Green Version]
- Toda, A.; Okabe, M.; Yoshida, T.; Nikaido, T. The Potential of Amniotic Membrane/Amnion-Derived Cells for Regeneration of Various Tissues. J. Pharmacol. Sci. 2007, 105, 215–228. [Google Scholar] [CrossRef] [Green Version]
- Merrill, B.J. Wnt Pathway Regulation of Embryonic Stem Cell Self-Renewal. Cold Spring Harb. Perspect. Biol. 2012, 4, a007971. [Google Scholar] [CrossRef] [Green Version]
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef] [Green Version]
- Kopp, J.L.; Ormsbee, B.D.; Desler, M.; Rizzino, A. Small Increases in the Level of Sox2 Trigger the Differentiation of Mouse Embryonic Stem Cells. Stem Cells 2008, 26, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Chambers, I.; Silva, J.; Colby, D.; Nichols, J.; Nijmeijer, B.; Robertson, M.; Vrana, J.; Jones, K.; Grotewold, L.; Smith, A. Nanog safeguards pluripotency and mediates germline development. Nature 2007, 450, 1230–1234. [Google Scholar] [CrossRef] [PubMed]
- Perumal, D.; Pillai, S.; Nguyen, J.; Schaal, C.; Coppola, D.; Chellappan, S.P. Nicotinic acetylcholine receptors induce c-Kit ligand/Stem Cell Factor and promote stemness in an ARRB1/ β-arrestin-1 dependent manner in NSCLC. Oncotarget 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Pfeifer, G.P. Defining Driver DNA Methylation Changes in Human Cancer. Int. J. Mol. Sci. 2018, 19, 1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilic, J.; Belmonte, J.C.I. Concise Review: Induced Pluripotent Stem Cells Versus Embryonic Stem Cells: Close Enough or Yet Too Far Apart? Stem Cells 2012, 30, 33–41. [Google Scholar] [CrossRef]
- Suelves, M.; Carrió, E.; Núñez-Álvarez, Y.; Peinado, M.A. DNA methylation dynamics in cellular commitment and differentiation. Brief. Funct. Genom. 2016, 15, 443–453. [Google Scholar] [CrossRef] [PubMed]
- van Schaijik, B.; Davis, P.F.; Wickremesekera, A.C.; Tan, S.T.; Itinteang, T. Subcellular localisation of the stem cell markers OCT4, SOX2, NANOG, KLF4 and c-MYC in cancer: A review. J. Clin. Pathol. 2018, 71, 88–91. [Google Scholar] [CrossRef]
- Jeong, C.-H.; Cho, Y.-Y.; Kim, M.-O.; Kim, S.-H.; Cho, E.-J.; Lee, S.-Y.; Jeon, Y.-J.; Yeong Lee, K.; Yao, K.; Keum, Y.-S.; et al. Phosphorylation of Sox2 Cooperates in Reprogramming to Pluripotent Stem Cells. Stem Cells 2010, 28, 2141–2150. [Google Scholar] [CrossRef]
- Huang, J.; Wang, F.; Okuka, M.; Liu, N.; Ji, G.; Ye, X.; Zuo, B.; Li, M.; Liang, P.; Ge, W.W.; et al. Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res. 2011, 21, 779–792. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhang, K.; Liu, Y.; Fu, Y.; Gao, S.; Gong, P.; Wang, H.; Zhou, Z.; Zeng, M.; Wu, Z.; et al. Telomere heterogeneity linked to metabolism and pluripotency state revealed by simultaneous analysis of telomere length and RNA-seq in the same human embryonic stem cell. BMC Biol. 2017, 15. [Google Scholar] [CrossRef] [Green Version]
- Motedayyen, H.; Esmaeil, N.; Tajik, N.; Khadem, F.; Ghotloo, S.; Khani, B.; Rezaei, A. Method and key points for isolation of human amniotic epithelial cells with high yield, viability and purity. BMC Res. Notes 2017, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falone, S.; Mirabilio, A.; Passerini, A.; Izzicupo, P.; Cacchio, M.; Gallina, S.; Baldassarre, A.D.; Amicarelli, F. Aerobic Performance and Antioxidant Protection in Runners. Int. J. Sports Med. 2009, 30, 782–788. [Google Scholar] [CrossRef] [PubMed]





| Cell Type | Medium | Supplements | Provider |
|---|---|---|---|
| hiPSCs | DMEM F/12 | 20%KO | All the media and supplements were purchased by Thermo Fisher Scientific (Waltham, MA, USA) |
| 5 ng/mL bFGF | |||
| 1%MEM non essential amino acid | |||
| hAFSCs | IMDM | 20% FBS | |
| 5 ng/mL bFGF | |||
| hFM-MSCs | DMEM | 10% FBS | |
| hAECs | DMEM | 10% FBS | |
| 10 ng/mL EGF |
| miRNA | Sequence (5′–3′) |
|---|---|
| miR_299-3p_RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAGCGG |
| miR_145-5p_RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGGGAT |
| miR_150-5p_RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCACTGG |
| miR_U6_RT | GAACGCTTCACGAATTTGCGTGTCAT |
| Gene | Sequence (5′–3′) |
|---|---|
| NANOG-FW | CCAGACCCAGAACATCCAGTC |
| NANOG-RW | CACTGGCAGGAGAATTTGGC |
| Endo-OCT4-FW [18] | GGGTTTTTGGGATTAAGTTCTTCA |
| Endo-OCT4-RW [18] | GCCCCCACCCTTTGTGTT |
| Endo-SOX2-FW [18] | CAAAAATGGCCATGCAGGTT |
| Endo-SOX2-RW [18] | AGTTGGGATCGAACAAAAGCTATT |
| c-kit-FW | CCACACCCTGTTCACTCCTT |
| c-kit-RW | TTCTGGGAAACTCCCATTTGTG |
| 18S-FW [19] | CATGGCCGTTCTTAGTTGGT |
| 18S-RW [19] | CGCTGAGCCAGTCAGTGTAG |
| miR-299-3p_FW | CGTGGAGTATGTGGGATGGTAAA |
| mir-150-5p_FW | GCATGTCTCCCAACCCTTGTA |
| miR-145-5p_FW | GAAGGTCCAGTTTTCCCAGGA |
| miR_universal_RW | GTGCAGGGTCCGAGGT |
| miR_U6_FW | CTCGCTTCGGCAGCACA |
| miR_U6_RW | AACGCTTCACGAATTTGCGT |
| Gene Promoter | FW Primer | RW Primer | Sequence Primer (5′–3′) | No. of CpGs Assayed | Amplicon Size (bp) |
|---|---|---|---|---|---|
| NANOG | [Bio]TGTATTTTTAGTAGAGAGGGGGTTT | ACCCAACAACAAATACTTCTAAATTCACC | ATTCACCACCTTTCCAACTT | 6 | 237 |
| OCT4 | [Bio]ATGGGGGAATTTTTTATATTTTAGAGTT | CACCACCATTAAACAAACATCC | AAAAAATTAAATAATCCCTT | 10 | 373 |
| SOX2 | AGTAAGGAAGGTTTTGAGGATAGA | [Bio]ATATCATTATTCTCCCCCTCATCCACAA | AGGTTTGGGTTTTTTAAT | 6 | 187 |
| C-KIT | GGAGGGGGGAAAAAGTGTATGAAAATTTG | [Bio]TTCTACTCAATTTCTCCACCTACTT | AAATTTGGGTTTTTAGAGTAA | 8 | 179 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaggi, G.; Di Credico, A.; Izzicupo, P.; Antonucci, I.; Crescioli, C.; Di Giacomo, V.; Di Ruscio, A.; Amabile, G.; Alviano, F.; Di Baldassarre, A.; et al. Epigenetic Features of Human Perinatal Stem Cells Redefine Their Stemness Potential. Cells 2020, 9, 1304. https://doi.org/10.3390/cells9051304
Gaggi G, Di Credico A, Izzicupo P, Antonucci I, Crescioli C, Di Giacomo V, Di Ruscio A, Amabile G, Alviano F, Di Baldassarre A, et al. Epigenetic Features of Human Perinatal Stem Cells Redefine Their Stemness Potential. Cells. 2020; 9(5):1304. https://doi.org/10.3390/cells9051304
Chicago/Turabian StyleGaggi, Giulia, Andrea Di Credico, Pascal Izzicupo, Ivana Antonucci, Clara Crescioli, Viviana Di Giacomo, Annalisa Di Ruscio, Giovanni Amabile, Francesco Alviano, Angela Di Baldassarre, and et al. 2020. "Epigenetic Features of Human Perinatal Stem Cells Redefine Their Stemness Potential" Cells 9, no. 5: 1304. https://doi.org/10.3390/cells9051304
APA StyleGaggi, G., Di Credico, A., Izzicupo, P., Antonucci, I., Crescioli, C., Di Giacomo, V., Di Ruscio, A., Amabile, G., Alviano, F., Di Baldassarre, A., & Ghinassi, B. (2020). Epigenetic Features of Human Perinatal Stem Cells Redefine Their Stemness Potential. Cells, 9(5), 1304. https://doi.org/10.3390/cells9051304











