Nuclear Envelope Proteins Modulating the Heterochromatin Formation and Functions in Fission Yeast
Abstract
:1. Introduction
2. Nuclear Membrane Proteins and Heterochromatin Formation in Fission Yeast
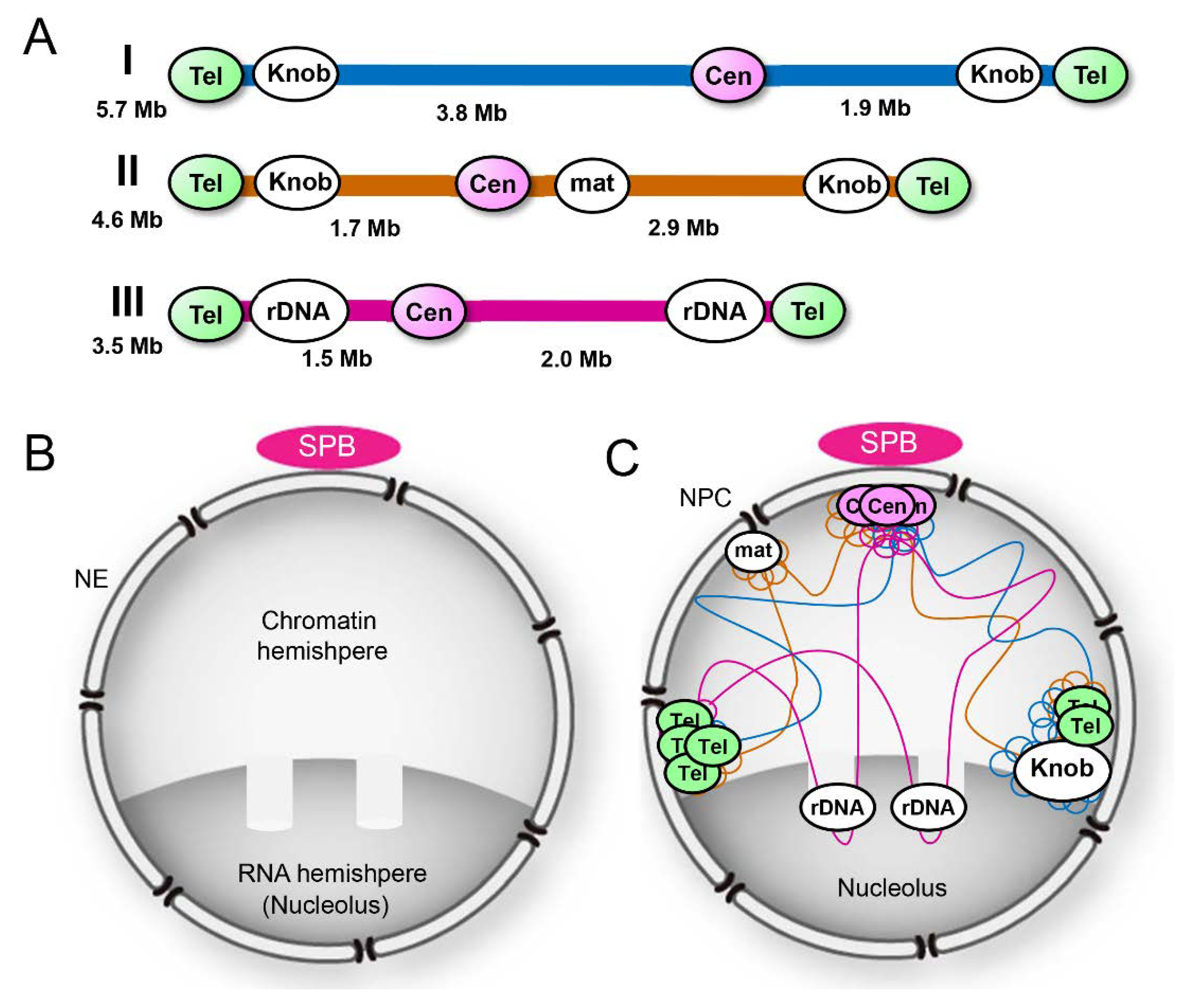
2.1. Organization of Chromosome in the Nucleus
2.2. Heterochromatin as Transcriptionally Silent Regions
2.3. “Knob” Regions
3. Mechanisms for Heterochromatin Formation in Fission Yeast
3.1. RNAi-Mediated Silencing Machinery
3.2. HDAC-Mediated Silencing Machinery
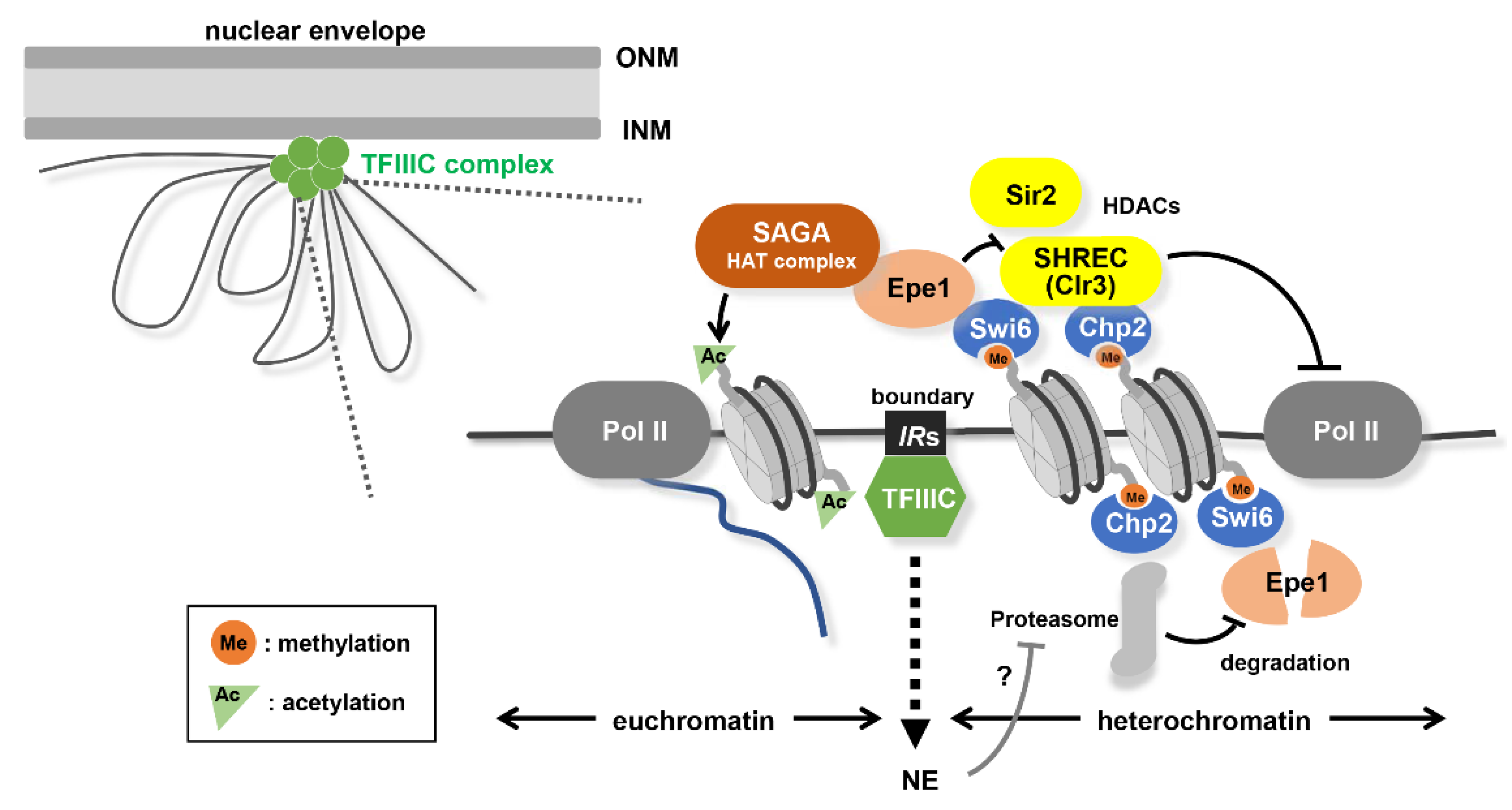
3.3. Boundary Elements Between Heterochromatin and Euchromatin
3.4. DAPI-Dense “Knob” Region
4. Proteins Attaching Heterochromatic Regions to the NE
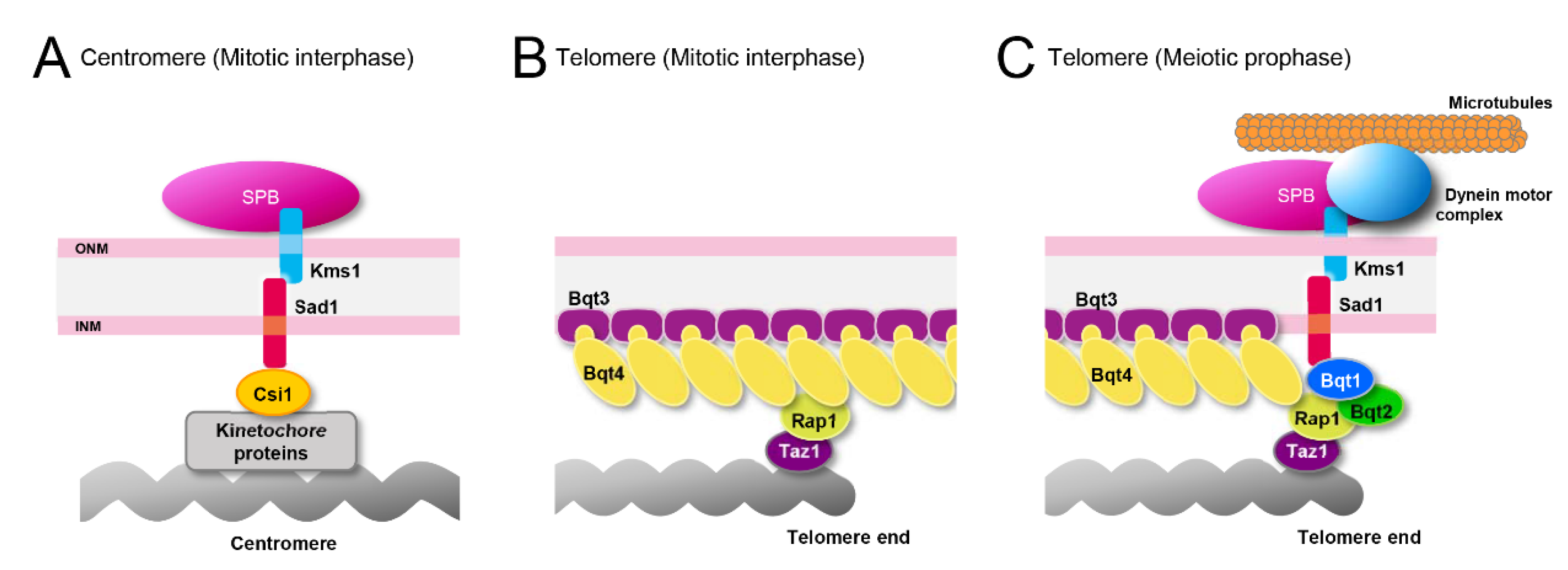
4.1. Proteins Attaching Heterochromatic Regions to the NE in Mitosis
4.2. Proteins Attaching Heterochromatic Regions to the NE in Meiosis
5. NE Proteins Modulating Heterochromatin Formation
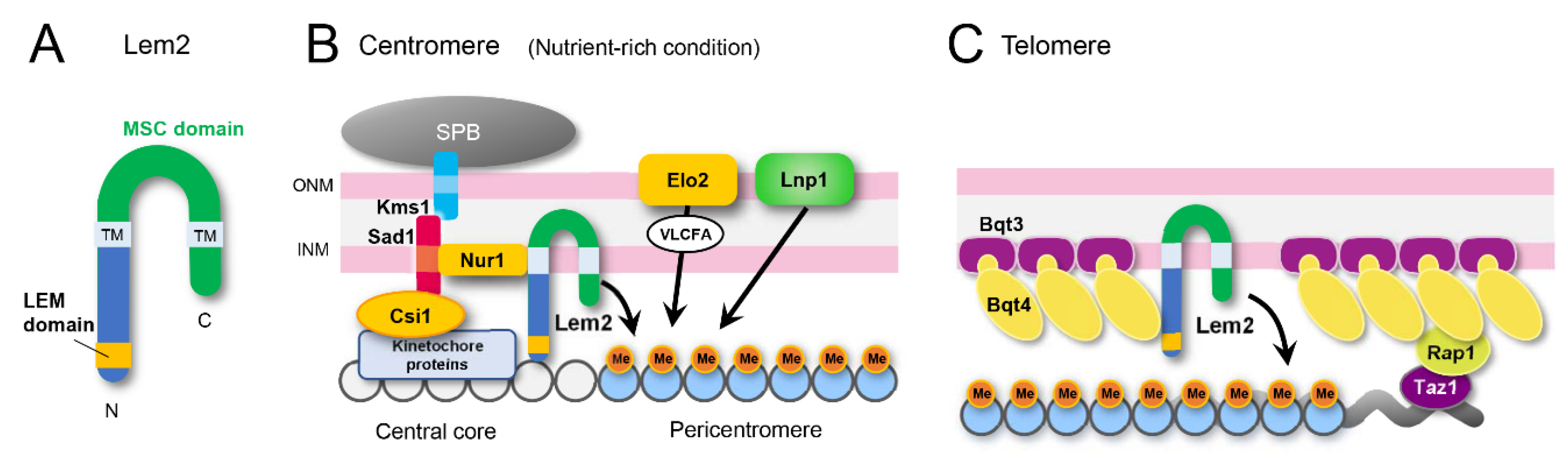
5.1. Lem2 Functions in Heterochromatin Formation
5.1.1. Lem2 is a Conserved Protein
5.1.2. Lem2 on Centromeric Heterochromatin
5.1.3. Lem2 on Telomeric Heterochromatin
5.1.4. Lem2 on LTR Sequences
5.1.5. Molecular Domains of Lem2 for Heterochromatin Functions
5.2. Regulation of Lem2 Localization
5.3. Membrane Protein Network Regulating Lem2 Functions
5.3.1. Lem2 Functions through Lnp1
5.3.2. Lem2 Functions through Bqt4
5.3.3. Lem2 Functions through the ESCRT-III Complex
6. Nucleoporins and Heterochromatin
6.1. Nucleoporins Modulate Gene Silencing
6.2. Nucleoporin Amo1 Sequesters Heterochromatin to the NE
7. Perspectives
Funding
Conflicts of Interest
References
- Briand, N.; Collas, P. Lamina-associated domains: Peripheral matters and internal affairs. Genome Biol. 2020, 21, 85. [Google Scholar] [CrossRef] [Green Version]
- Iwamoto, M.; Hiraoka, Y.; Haraguchi, T. Uniquely designed nuclear structures of lower eukaryotes. Curr Opin. Cell Biol. 2016, 40, 66–73. [Google Scholar] [CrossRef] [Green Version]
- Mans, B.J.; Anantharaman, V.; Aravind, L.; Koonin, E.V. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 2004, 3, 1612–1637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worman, H.J.; Yuan, J.; Blobel, G.; Georgatos, S.D. A lamin B receptor in the nuclear envelope. Proc. Natl. Acad. Sci. USA 1988, 85, 8531–8534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirano, Y.; Hizume, K.; Kimura, H.; Takeyasu, K.; Haraguchi, T.; Hiraoka, Y. Lamin B receptor recognizes specific modifications of histone H4 in heterochromatin formation. J. Biol. Chem. 2012, 287, 42654–42663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solovei, I.; Kreysing, M.; Lanctot, C.; Kosem, S.; Peichl, L.; Cremer, T.; Guck, J.; Joffe, B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 2009, 137, 356–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solovei, I.; Wang, A.S.; Thanisch, K.; Schmidt, C.S.; Krebs, S.; Zwerger, M.; Cohen, T.V.; Devys, D.; Foisner, R.; Peichl, L.; et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 2013, 152, 584–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dechat, T.; Vlcek, S.; Foisner, R. Review: Lamina-associated polypeptide 2 isoforms and related proteins in cell cycle-dependent nuclear structure dynamics. J. Struct Biol. 2000, 129, 335–345. [Google Scholar] [CrossRef]
- Gruenbaum, Y.; Margalit, A.; Goldman, R.D.; Shumaker, D.K.; Wilson, K.L. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 2005, 6, 21–31. [Google Scholar] [CrossRef]
- Lee, K.K.; Wilson, K.L. All in the family: Evidence for four new LEM-domain proteins Lem2 (NET-25), Lem3, Lem4 and Lem5 in the human genome. Symp. Soc. Exp. Biol. 2004, 329–339. [Google Scholar]
- Brachner, A.; Foisner, R. Evolvement of LEM proteins as chromatin tethers at the nuclear periphery. Biochem. Soc. Trans. 2011, 39, 1735–1741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, N.; Krohne, G. LEM-Domain proteins: New insights into lamin-interacting proteins. Int Rev. Cytol. 2007, 261, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Fukuda, Y.; Osakada, H.; Mori, C.; Hiraoka, Y.; Haraguchi, T. Identification of the evolutionarily conserved nuclear envelope proteins Lem2 and MicLem2 in Tetrahymena thermophila. Gene X 2019, 1, 100006. [Google Scholar] [CrossRef] [PubMed]
- Korfali, N.; Wilkie, G.S.; Swanson, S.K.; Srsen, V.; de Las Heras, J.; Batrakou, D.G.; Malik, P.; Zuleger, N.; Kerr, A.R.; Florens, L.; et al. The nuclear envelope proteome differs notably between tissues. Nucleus 2012, 3, 552–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bui, K.H.; von Appen, A.; DiGuilio, A.L.; Ori, A.; Sparks, L.; Mackmull, M.T.; Bock, T.; Hagen, W.; Andres-Pons, A.; Glavy, J.S.; et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell 2013, 155, 1233–1243. [Google Scholar] [CrossRef] [Green Version]
- Kosinski, J.; Mosalaganti, S.; von Appen, A.; Teimer, R.; DiGuilio, A.L.; Wan, W.; Bui, K.H.; Hagen, W.J.; Briggs, J.A.; Glavy, J.S.; et al. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 2016, 352, 363–365. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.H.; Stuwe, T.; Schilbach, S.; Rundlet, E.J.; Perriches, T.; Mobbs, G.; Fan, Y.; Thierbach, K.; Huber, F.M.; Collins, L.N.; et al. Architecture of the symmetric core of the nuclear pore. Science 2016, 352, aaf1015. [Google Scholar] [CrossRef] [Green Version]
- Von Appen, A.; Kosinski, J.; Sparks, L.; Ori, A.; DiGuilio, A.L.; Vollmer, B.; Mackmull, M.T.; Banterle, N.; Parca, L.; Kastritis, P.; et al. In situ structural analysis of the human nuclear pore complex. Nature 2015, 526, 140–143. [Google Scholar] [CrossRef]
- Alber, F.; Dokudovskaya, S.; Veenhoff, L.M.; Zhang, W.; Kipper, J.; Devos, D.; Suprapto, A.; Karni-Schmidt, O.; Williams, R.; Chait, B.T.; et al. The molecular architecture of the nuclear pore complex. Nature 2007, 450, 695–701. [Google Scholar] [CrossRef]
- Eisenhardt, N.; Redolfi, J.; Antonin, W. Interaction of Nup53 with Ndc1 and Nup155 is required for nuclear pore complex assembly. J. Cell Sci. 2014, 127, 908–921. [Google Scholar] [CrossRef] [Green Version]
- Onischenko, E.; Stanton, L.H.; Madrid, A.S.; Kieselbach, T.; Weis, K. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J. Cell Biol. 2009, 185, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Amlacher, S.; Sarges, P.; Flemming, D.; van Noort, V.; Kunze, R.; Devos, D.P.; Arumugam, M.; Bork, P.; Hurt, E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 2011, 146, 277–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asakawa, H.; Hiraoka, Y.; Haraguchi, T. A method of correlative light and electron microscopy for yeast cells. Micron 2014, 61, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Cronshaw, J.M.; Krutchinsky, A.N.; Zhang, W.; Chait, B.T.; Matunis, M.J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002, 158, 915–927. [Google Scholar] [CrossRef] [Green Version]
- DeGrasse, J.A.; DuBois, K.N.; Devos, D.; Siegel, T.N.; Sali, A.; Field, M.C.; Rout, M.P.; Chait, B.T. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell Proteomics 2009, 8, 2119–2130. [Google Scholar] [CrossRef] [Green Version]
- Iwamoto, M.; Osakada, H.; Mori, C.; Fukuda, Y.; Nagao, K.; Obuse, C.; Hiraoka, Y.; Haraguchi, T. Compositionally distinct nuclear pore complexes of functionally distinct dimorphic nuclei in the ciliate Tetrahymena. J. Cell Sci. 2017, 130, 1822–1834. [Google Scholar] [CrossRef] [Green Version]
- Obado, S.O.; Brillantes, M.; Uryu, K.; Zhang, W.; Ketaren, N.E.; Chait, B.T.; Field, M.C.; Rout, M.P. Interactome Mapping Reveals the Evolutionary History of the Nuclear Pore Complex. PLoS Biol. 2016, 14, e1002365. [Google Scholar] [CrossRef] [Green Version]
- Rout, M.P.; Aitchison, J.D.; Suprapto, A.; Hjertaas, K.; Zhao, Y.; Chait, B.T. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J. Cell Biol. 2000, 148, 635–651. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Fukao, Y.; Iwamoto, M.; Haraguchi, T.; Hara-Nishimura, I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant. Cell 2010, 22, 4084–4097. [Google Scholar] [CrossRef] [Green Version]
- Knockenhauer, K.E.; Schwartz, T.U. The Nuclear Pore Complex as a Flexible and Dynamic Gate. Cell 2016, 164, 1162–1171. [Google Scholar] [CrossRef] [Green Version]
- Holla, S.; Dhakshnamoorthy, J.; Folco, H.D.; Balachandran, V.; Xiao, H.; Sun, L.L.; Wheeler, D.; Zofall, M.; Grewal, S.I.S. Positioning Heterochromatin at the Nuclear Periphery Suppresses Histone Turnover to Promote Epigenetic Inheritance. Cell 2020, 180, 150–164 e115. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, N.; Paulo, J.A.; Tatarakis, A.; Wang, X.; Edwards, A.L.; Bhanu, N.V.; Garcia, B.A.; Haas, W.; Gygi, S.P.; Moazed, D. Native Chromatin Proteomics Reveals a Role for Specific Nucleoporins in Heterochromatin Organization and Maintenance. Mol. Cell 2020, 77, 51–66 e58. [Google Scholar] [CrossRef] [PubMed]
- Barrales, R.R.; Forn, M.; Georgescu, P.R.; Sarkadi, Z.; Braun, S. Control of heterochromatin localization and silencing by the nuclear membrane protein Lem2. Genes Dev. 2016, 30, 133–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emmerth, S.; Schober, H.; Gaidatzis, D.; Roloff, T.; Jacobeit, K.; Buhler, M. Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Dev. Cell 2010, 18, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Tange, Y.; Chikashige, Y.; Takahata, S.; Kawakami, K.; Higashi, M.; Mori, C.; Kojidani, T.; Hirano, Y.; Asakawa, H.; Murakami, Y.; et al. Inner nuclear membrane protein Lem2 augments heterochromatin formation in response to nutritional conditions. Genes Cells 2016, 21, 812–832. [Google Scholar] [CrossRef]
- Wood, V.; Gwilliam, R.; Rajandream, M.A.; Lyne, M.; Lyne, R.; Stewart, A.; Sgouros, J.; Peat, N.; Hayles, J.; Baker, S.; et al. The genome sequence of Schizosaccharomyces pombe. Nature 2002, 415, 871–880. [Google Scholar] [CrossRef] [Green Version]
- Chikashige, Y.; Ding, D.Q.; Funabiki, H.; Haraguchi, T.; Mashiko, S.; Yanagida, M.; Hiraoka, Y. Telomere-led premeiotic chromosome movement in fission yeast. Science 1994, 264, 270–273. [Google Scholar] [CrossRef]
- Funabiki, H.; Hagan, I.; Uzawa, S.; Yanagida, M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 1993, 121, 961–976. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, A.; Asakawa, H.; Haraguchi, T.; Hiraoka, Y. Spatial organization of the Schizosaccharomyces pombe genome within the nucleus. Yeast 2017, 34, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Kanbe, T. Mitosis in the fission yeast Schizosaccharomyces pombe as revealed by freeze-substitution electron microscopy. J. Cell Sci. 1986, 80, 253–268. [Google Scholar]
- Toda, T.; Yamamoto, M.; Yanagida, M. Sequential alterations in the nuclear chromatin region during mitosis of the fission yeast Schizosaccharomyces pombe: Video fluorescence microscopy of synchronously growing wild-type and cold-sensitive cdc mutants by using a DNA-binding fluorescent probe. J. Cell Sci. 1981, 52, 271–287. [Google Scholar] [PubMed]
- Uzawa, S.; Yanagida, M. Visualization of centromeric and nucleolar DNA in fission yeast by fluorescence in situ hybridization. J. Cell Sci. 1992, 101 Pt 2, 267–275. [Google Scholar]
- Yanagida, M.; Hiraoka, Y. [Dynamic structures of DNA, chromatin and chromosomes studied by video fluorescence microscopy]. Tanpakushitsu Kakusan Koso 1984, 29, 329–343. [Google Scholar] [PubMed]
- Fedorova, E.; Zink, D. Nuclear architecture and gene regulation. Biochim. Biophys. Acta 2008, 1783, 2174–2184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allshire, R.C.; Ekwall, K. Epigenetic Regulation of Chromatin States in Schizosaccharomyces pombe. Cold Spring Harb Perspect Biol. 2015, 7, a018770. [Google Scholar] [CrossRef] [Green Version]
- Grewal, S.I.; Jia, S. Heterochromatin revisited. Nat. Rev. Genet. 2007, 8, 35–46. [Google Scholar] [CrossRef]
- Martienssen, R.; Moazed, D. RNAi and heterochromatin assembly. Cold Spring Harb. Perspect. Biol. 2015, 7, a019323. [Google Scholar] [CrossRef]
- Matsuda, A.; Chikashige, Y.; Ding, D.Q.; Ohtsuki, C.; Mori, C.; Asakawa, H.; Kimura, H.; Haraguchi, T.; Hiraoka, Y. Highly condensed chromatins are formed adjacent to subtelomeric and decondensed silent chromatin in fission yeast. Nat. Commun. 2015, 6, 7753. [Google Scholar] [CrossRef] [Green Version]
- Egel, R.; Beach, D.H.; Klar, A.J. Genes required for initiation and resolution steps of mating-type switching in fission yeast. Proc. Natl. Acad. Sci. USA 1984, 81, 3481–3485. [Google Scholar] [CrossRef] [Green Version]
- Klar, A.J. The developmental fate of fission yeast cells is determined by the pattern of inheritance of parental and grandparental DNA strands. EMBO J. 1990, 9, 1407–1415. [Google Scholar] [CrossRef]
- Klar, A.J.; Bonaduce, M.J. swi6, a gene required for mating-type switching, prohibits meiotic recombination in the mat2-mat3 “cold spot” of fission yeast. Genetics 1991, 129, 1033–1042. [Google Scholar] [PubMed]
- Lorentz, A.; Ostermann, K.; Fleck, O.; Schmidt, H. Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene 1994, 143, 139–143. [Google Scholar] [CrossRef]
- Folco, H.D.; Chalamcharla, V.R.; Sugiyama, T.; Thillainadesan, G.; Zofall, M.; Balachandran, V.; Dhakshnamoorthy, J.; Mizuguchi, T.; Grewal, S.I. Untimely expression of gametogenic genes in vegetative cells causes uniparental disomy. Nature 2017, 543, 126–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roche, B.; Arcangioli, B.; Martienssen, R.A. RNA interference is essential for cellular quiescence. Science 2016, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef]
- Mello, C.C.; Conte, D., Jr. Revealing the world of RNA interference. Nature 2004, 431, 338–342. [Google Scholar] [CrossRef]
- Volpe, T.A.; Kidner, C.; Hall, I.M.; Teng, G.; Grewal, S.I.; Martienssen, R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 2002, 297, 1833–1837. [Google Scholar] [CrossRef] [Green Version]
- Grewal, S.I. RNAi-dependent formation of heterochromatin and its diverse functions. Curr. Opin. Genet. Dev. 2010, 20, 134–141. [Google Scholar] [CrossRef] [Green Version]
- Cam, H.P.; Sugiyama, T.; Chen, E.S.; Chen, X.; FitzGerald, P.C.; Grewal, S.I. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005, 37, 809–819. [Google Scholar] [CrossRef]
- Djupedal, I.; Portoso, M.; Spahr, H.; Bonilla, C.; Gustafsson, C.M.; Allshire, R.C.; Ekwall, K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005, 19, 2301–2306. [Google Scholar] [CrossRef] [Green Version]
- Kato, H.; Goto, D.B.; Martienssen, R.A.; Urano, T.; Furukawa, K.; Murakami, Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 2005, 309, 467–469. [Google Scholar] [CrossRef] [PubMed]
- Verdel, A.; Jia, S.; Gerber, S.; Sugiyama, T.; Gygi, S.; Grewal, S.I.; Moazed, D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 2004, 303, 672–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iida, T.; Nakayama, J.; Moazed, D. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol. Cell 2008, 31, 178–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motamedi, M.R.; Verdel, A.; Colmenares, S.U.; Gerber, S.A.; Gygi, S.P.; Moazed, D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 2004, 119, 789–802. [Google Scholar] [CrossRef]
- Bayne, E.H.; White, S.A.; Kagansky, A.; Bijos, D.A.; Sanchez-Pulido, L.; Hoe, K.L.; Kim, D.U.; Park, H.O.; Ponting, C.P.; Rappsilber, J.; et al. Stc1: A critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 2010, 140, 666–677. [Google Scholar] [CrossRef] [Green Version]
- Nakayama, J.; Rice, J.C.; Strahl, B.D.; Allis, C.D.; Grewal, S.I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 2001, 292, 110–113. [Google Scholar] [CrossRef] [Green Version]
- Rea, S.; Eisenhaber, F.; O’Carroll, D.; Strahl, B.D.; Sun, Z.W.; Schmid, M.; Opravil, S.; Mechtler, K.; Ponting, C.P.; Allis, C.D.; et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000, 406, 593–599. [Google Scholar] [CrossRef]
- Goto, D.B.; Nakayama, J. RNA and epigenetic silencing: Insight from fission yeast. Dev. Growth Differ. 2012, 54, 129–141. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Mosch, K.; Fischle, W.; Grewal, S.I. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 2008, 15, 381–388. [Google Scholar] [CrossRef]
- Conte, D., Jr.; Mello, C.C. Primal RNAs: The end of the beginning? Cell 2010, 140, 452–454. [Google Scholar] [CrossRef] [Green Version]
- Bühler, M.; Spies, N.; Bartel, D.P.; Moazed, D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat. Struct Mol. Biol. 2008, 15, 1015–1023. [Google Scholar] [CrossRef] [PubMed]
- Chalamcharla, V.R.; Folco, H.D.; Dhakshnamoorthy, J.; Grewal, S.I. Conserved factor Dhp1/Rat1/Xrn2 triggers premature transcription termination and nucleates heterochromatin to promote gene silencing. Proc. Natl. Acad. Sci. USA 2015, 112, 15548–15555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marina, D.B.; Shankar, S.; Natarajan, P.; Finn, K.J.; Madhani, H.D. A conserved ncRNA-binding protein recruits silencing factors to heterochromatin through an RNAi-independent mechanism. Genes Dev. 2013, 27, 1851–1856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parsa, J.Y.; Boudoukha, S.; Burke, J.; Homer, C.; Madhani, H.D. Polymerase pausing induced by sequence-specific RNA-binding protein drives heterochromatin assembly. Genes Dev. 2018, 32, 953–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes-Turcu, F.E.; Zhang, K.; Zofall, M.; Chen, E.; Grewal, S.I. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat. Struct. Mol. Biol. 2011, 18, 1132–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tucker, J.F.; Ohle, C.; Schermann, G.; Bendrin, K.; Zhang, W.; Fischer, T.; Zhang, K. A Novel Epigenetic Silencing Pathway Involving the Highly Conserved 5’-3’ Exoribonuclease Dhp1/Rat1/Xrn2 in Schizosaccharomyces pombe. PLoS Genet. 2016, 12, e1005873. [Google Scholar] [CrossRef] [Green Version]
- Vo, T.V.; Dhakshnamoorthy, J.; Larkin, M.; Zofall, M.; Thillainadesan, G.; Balachandran, V.; Holla, S.; Wheeler, D.; Grewal, S.I.S. CPF Recruitment to Non-canonical Transcription Termination Sites Triggers Heterochromatin Assembly and Gene Silencing. Cell Rep. 2019, 28, 267–281 e265. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, T.; Cam, H.P.; Sugiyama, R.; Noma, K.; Zofall, M.; Kobayashi, R.; Grewal, S.I. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 2007, 128, 491–504. [Google Scholar] [CrossRef] [Green Version]
- Nicolas, E.; Yamada, T.; Cam, H.P.; Fitzgerald, P.C.; Kobayashi, R.; Grewal, S.I. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat. Struct. Mol. Biol. 2007, 14, 372–380. [Google Scholar] [CrossRef]
- Sanulli, S.; Trnka, M.J.; Dharmarajan, V.; Tibble, R.W.; Pascal, B.D.; Burlingame, A.L.; Griffin, P.R.; Gross, J.D.; Narlikar, G.J. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 2019, 575, 390–394. [Google Scholar] [CrossRef]
- Aguilar-Arnal, L.; Marsellach, F.X.; Azorin, F. The fission yeast homologue of CENP-B, Abp1, regulates directionality of mating-type switching. EMBO J. 2008, 27, 1029–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, S.; Noma, K.; Grewal, S.I. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 2004, 304, 1971–1976. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Choi, E.S.; Shin, J.A.; Jang, Y.K.; Park, S.D. Regulation of Swi6/HP1-dependent heterochromatin assembly by cooperation of components of the mitogen-activated protein kinase pathway and a histone deacetylase Clr6. J. Biol. Chem. 2004, 279, 42850–42859. [Google Scholar] [CrossRef] [Green Version]
- Buscaino, A.; White, S.A.; Houston, D.R.; Lejeune, E.; Simmer, F.; de Lima Alves, F.; Diyora, P.T.; Urano, T.; Bayne, E.H.; Rappsilber, J.; et al. Raf1 Is a DCAF for the Rik1 DDB1-like protein and has separable roles in siRNA generation and chromatin modification. PLoS Genet. 2012, 8, e1002499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, S.; Xiong, Y. CRL4s: The CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 2009, 34, 562–570. [Google Scholar] [CrossRef] [Green Version]
- Oya, E.; Nakagawa, R.; Yoshimura, Y.; Tanaka, M.; Nishibuchi, G.; Machida, S.; Shirai, A.; Ekwall, K.; Kurumizaka, H.; Tagami, H.; et al. H3K14 ubiquitylation promotes H3K9 methylation for heterochromatin assembly. EMBO Rep. 2019, 20, e48111. [Google Scholar] [CrossRef]
- Noma, K.; Allis, C.D.; Grewal, S.I. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 2001, 293, 1150–1155. [Google Scholar] [CrossRef]
- Thon, G.; Bjerling, P.; Bunner, C.M.; Verhein-Hansen, J. Expression-state boundaries in the mating-type region of fission yeast. Genetics 2002, 161, 611–622. [Google Scholar]
- Noma, K.; Cam, H.P.; Maraia, R.J.; Grewal, S.I. A role for TFIIIC transcription factor complex in genome organization. Cell 2006, 125, 859–872. [Google Scholar] [CrossRef] [Green Version]
- Ayoub, N.; Noma, K.; Isaac, S.; Kahan, T.; Grewal, S.I.; Cohen, A. A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol. Cell Biol. 2003, 23, 4356–4370. [Google Scholar] [CrossRef] [Green Version]
- Zofall, M.; Grewal, S.I. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol. Cell 2006, 22, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Trewick, S.C.; Minc, E.; Antonelli, R.; Urano, T.; Allshire, R.C. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J. 2007, 26, 4670–4682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukada, Y.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006, 439, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Isaac, S.; Walfridsson, J.; Zohar, T.; Lazar, D.; Kahan, T.; Ekwall, K.; Cohen, A. Interaction of Epe1 with the heterochromatin assembly pathway in Schizosaccharomyces pombe. Genetics 2007, 175, 1549–1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, S.; Garcia, J.F.; Rowley, M.; Rougemaille, M.; Shankar, S.; Madhani, H.D. The Cul4-Ddb1(Cdt)(2) ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell 2011, 144, 41–54. [Google Scholar] [CrossRef] [Green Version]
- Audergon, P.N.; Catania, S.; Kagansky, A.; Tong, P.; Shukla, M.; Pidoux, A.L.; Allshire, R.C. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science 2015, 348, 132–135. [Google Scholar] [CrossRef] [Green Version]
- Ragunathan, K.; Jih, G.; Moazed, D. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science 2015, 348, 1258699. [Google Scholar] [CrossRef] [Green Version]
- Shimada, A.; Dohke, K.; Sadaie, M.; Shinmyozu, K.; Nakayama, J.; Urano, T.; Murakami, Y. Phosphorylation of Swi6/HP1 regulates transcriptional gene silencing at heterochromatin. Genes Dev. 2009, 23, 18–23. [Google Scholar] [CrossRef] [Green Version]
- Bao, K.; Shan, C.M.; Moresco, J.; Yates, J., 3rd; Jia, S. Anti-silencing factor Epe1 associates with SAGA to regulate transcription within heterochromatin. Genes Dev. 2019, 33, 116–126. [Google Scholar] [CrossRef] [Green Version]
- Garcia, J.F.; Al-Sady, B.; Madhani, H.D. Intrinsic Toxicity of Unchecked Heterochromatin Spread Is Suppressed by Redundant Chromatin Boundary Functions in Schizosacchromyces pombe. G3 (Bethesda) 2015, 5, 1453–1461. [Google Scholar] [CrossRef] [Green Version]
- Buchanan, L.; Durand-Dubief, M.; Roguev, A.; Sakalar, C.; Wilhelm, B.; Stralfors, A.; Shevchenko, A.; Aasland, R.; Shevchenko, A.; Ekwall, K.; et al. The Schizosaccharomyces pombe JmjC-protein, Msc1, prevents H2A.Z localization in centromeric and subtelomeric chromatin domains. PLoS Genet. 2009, 5, e1000726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tashiro, S.; Handa, T.; Matsuda, A.; Ban, T.; Takigawa, T.; Miyasato, K.; Ishii, K.; Kugou, K.; Ohta, K.; Hiraoka, Y.; et al. Shugoshin forms a specialized chromatin domain at subtelomeres that regulates transcription and replication timing. Nat. Commun. 2016, 7, 10393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murawska, M.; Schauer, T.; Matsuda, A.; Wilson, M.D.; Pysik, T.; Wojcik, F.; Muir, T.W.; Hiraoka, Y.; Straub, T.; Ladurner, A.G. The Chaperone FACT and Histone H2B Ubiquitination Maintain S. pombe Genome Architecture through Genic and Subtelomeric Functions. Mol. Cell 2020, 77, 501–513 e507. [Google Scholar] [CrossRef] [PubMed]
- Mata, J.; Lyne, R.; Burns, G.; Bahler, J. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 2002, 32, 143–147. [Google Scholar] [CrossRef]
- Ishii, K.; Ogiyama, Y.; Chikashige, Y.; Soejima, S.; Masuda, F.; Kakuma, T.; Hiraoka, Y.; Takahashi, K. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 2008, 321, 1088–1091. [Google Scholar] [CrossRef]
- Gallardo, P.; Barrales, R.R.; Daga, R.R.; Salas-Pino, S. Nuclear Mechanics in the Fission Yeast. Cells 2019, 8, 1285. [Google Scholar] [CrossRef] [Green Version]
- Hagan, I.; Yanagida, M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 1995, 129, 1033–1047. [Google Scholar] [CrossRef] [Green Version]
- Hou, H.; Kallgren, S.P.; Jia, S. Csi1 illuminates the mechanism and function of Rabl configuration. Nucleus 2013, 4, 176–181. [Google Scholar] [CrossRef] [Green Version]
- Hou, H.; Zhou, Z.; Wang, Y.; Wang, J.; Kallgren, S.P.; Kurchuk, T.; Miller, E.A.; Chang, F.; Jia, S. Csi1 links centromeres to the nuclear envelope for centromere clustering. J. Cell Biol. 2012, 199, 735–744. [Google Scholar] [CrossRef] [Green Version]
- Crisp, M.; Liu, Q.; Roux, K.; Rattner, J.B.; Shanahan, C.; Burke, B.; Stahl, P.D.; Hodzic, D. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol. 2006, 172, 41–53. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.I.; Birendra, K.C.; Roux, K.J. Making the LINC: SUN and KASH protein interactions. Biol. Chem. 2015, 396, 295–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starr, D.A. KASH and SUN proteins. Curr. Biol. 2011, 21, R414–R415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tapley, E.C.; Starr, D.A. Connecting the nucleus to the cytoskeleton by SUN-KASH bridges across the nuclear envelope. Curr. Opin. Cell Biol. 2013, 25, 57–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Alvarez, A.; Cooper, J.P. The functionally elusive RabI chromosome configuration directly regulates nuclear membrane remodeling at mitotic onset. Cell Cycle 2017, 16, 1392–1396. [Google Scholar] [CrossRef]
- Chikashige, Y.; Yamane, M.; Okamasa, K.; Tsutsumi, C.; Kojidani, T.; Sato, M.; Haraguchi, T.; Hiraoka, Y. Membrane proteins Bqt3 and -4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J. Cell Biol. 2009, 187, 413–427. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, Y.; Saito, A.; Sazer, S. Fission yeast Lem2 and Man1 perform fundamental functions of the animal cell nuclear lamina. Nucleus 2012, 3, 60–76. [Google Scholar] [CrossRef] [Green Version]
- Steglich, B.; Stralfors, A.; Khorosjutina, O.; Persson, J.; Smialowska, A.; Javerzat, J.P.; Ekwall, K. The Fun30 chromatin remodeler Fft3 controls nuclear organization and chromatin structure of insulators and subtelomeres in fission yeast. PLoS Genet. 2015, 11, e1005101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chikashige, Y.; Tsutsumi, C.; Yamane, M.; Okamasa, K.; Haraguchi, T.; Hiraoka, Y. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 2006, 125, 59–69. [Google Scholar] [CrossRef] [Green Version]
- Chikashige, Y.; Haraguchi, T.; Hiraoka, Y. Another way to move chromosomes. Chromosoma 2007, 116, 497–505. [Google Scholar] [CrossRef]
- Hiraoka, Y.; Dernburg, A.F. The SUN rises on meiotic chromosome dynamics. Dev. Cell 2009, 17, 598–605. [Google Scholar] [CrossRef] [Green Version]
- Brachner, A.; Reipert, S.; Foisner, R.; Gotzmann, J. LEM2 is a novel MAN1-related inner nuclear membrane protein associated with A-type lamins. J. Cell Sci. 2005, 118, 5797–5810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batsios, P.; Graf, R.; Koonce, M.P.; Larochelle, D.A.; Meyer, I. Nuclear envelope organization in Dictyostelium discoideum. Int. J. Dev. Biol. 2019, 63, 509–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batsios, P.; Ren, X.; Baumann, O.; Larochelle, D.A.; Graf, R. Src1 is a Protein of the Inner Nuclear Membrane Interacting with the Dictyostelium Lamin NE81. Cells 2016, 5, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banday, S.; Farooq, Z.; Rashid, R.; Abdullah, E.; Altaf, M. Role of Inner Nuclear Membrane Protein Complex Lem2-Nur1 in Heterochromatic Gene Silencing. J. Biol. Chem. 2016, 291, 20021–20029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pidoux, A.L.; Allshire, R.C. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res. 2004, 12, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Chen, E.S.; Yanagida, M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 2000, 288, 2215–2219. [Google Scholar] [CrossRef]
- Chikashige, Y.; Kinoshita, N.; Nakaseko, Y.; Matsumoto, T.; Murakami, S.; Niwa, O.; Yanagida, M. Composite motifs and repeat symmetry in S. pombe centromeres: Direct analysis by integration of NotI restriction sites. Cell 1989, 57, 739–751. [Google Scholar] [CrossRef]
- Hirano, Y.; Kinugasa, Y.; Asakawa, H.; Chikashige, Y.; Obuse, C.; Haraguchi, T.; Hiraoka, Y. Lem2 is retained at the nuclear envelope through its interaction with Bqt4 in fission yeast. Genes Cells 2018, 23, 122–135. [Google Scholar] [CrossRef] [Green Version]
- Ebrahimi, H.; Masuda, H.; Jain, D.; Cooper, J.P. Distinct ‘safe zones’ at the nuclear envelope ensure robust replication of heterochromatic chromosome regions. eLife 2018, 7. [Google Scholar] [CrossRef] [Green Version]
- Finnegan, D.J. Retrotransposons. Curr. Biol. 2012, 22, R432–R437. [Google Scholar] [CrossRef] [Green Version]
- Cam, H.P.; Noma, K.; Ebina, H.; Levin, H.L.; Grewal, S.I. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature 2008, 451, 431–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaratiegui, M.; Vaughn, M.W.; Irvine, D.V.; Goto, D.; Watt, S.; Bahler, J.; Arcangioli, B.; Martienssen, R.A. CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR. Nature 2011, 469, 112–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, C.; Inoue, H.; Sun, W.; Takeshita, Y.; Huang, Y.; Xu, Y.; Kanoh, J.; Chen, Y. The Inner Nuclear Membrane Protein Bqt4 in Fission Yeast Contains a DNA-Binding Domain Essential for Telomere Association with the Nuclear Envelope. Structure 2019, 27, 335–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, C.; Inoue, H.; Sun, W.; Takeshita, Y.; Huang, Y.; Xu, Y.; Kanoh, J.; Chen, Y. Structural insights into chromosome attachment to the nuclear envelope by an inner nuclear membrane protein Bqt4 in fission yeast. Nucleic Acids Res. 2019, 47, 1573–1584. [Google Scholar] [CrossRef]
- Pieper, G.H.; Sprenger, S.; Teis, D.; Oliferenko, S. ESCRT-III/Vps4 Controls Heterochromatin-Nuclear Envelope Attachments. Dev. Cell 2020. [Google Scholar] [CrossRef]
- Hiraoka, Y.; Maekawa, H.; Asakawa, H.; Chikashige, Y.; Kojidani, T.; Osakada, H.; Matsuda, A.; Haraguchi, T. Inner nuclear membrane protein Ima1 is dispensable for intranuclear positioning of centromeres. Genes Cells 2011, 16, 1000–1011. [Google Scholar] [CrossRef]
- Chen, S.; Desai, T.; McNew, J.A.; Gerard, P.; Novick, P.J.; Ferro-Novick, S. Lunapark stabilizes nascent three-way junctions in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2015, 112, 418–423. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Novick, P.; Ferro-Novick, S. ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat. Cell Biol. 2012, 14, 707–716. [Google Scholar] [CrossRef]
- Wang, S.; Powers, R.E.; Gold, V.A.; Rapoport, T.A. The ER morphology-regulating lunapark protein induces the formation of stacked bilayer discs. Life Sci. Alliance 2018, 1, e201700014. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Tukachinsky, H.; Romano, F.B.; Rapoport, T.A. Cooperation of the ER-shaping proteins atlastin, lunapark, and reticulons to generate a tubular membrane network. eLife 2016, 5. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, T.; Huo, H.; Ye, Y.; Liu, Y. Lunapark Is a Component of a Ubiquitin Ligase Complex Localized to the Endoplasmic Reticulum Three-way Junctions. J. Biol. Chem. 2016, 291, 18252–18262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirano, Y.; Kinugasa, Y.; Osakada, H.; Shindo, T.; Kubota, Y.; Shibata, S.; Haraguchi, T.; Hiraoka, Y. Lem2 and Lnp1 maintain the membrane boundary between the nuclear envelope and endoplasmic reticulum. Commun. Biol. 2020, 3, 276. [Google Scholar] [CrossRef] [PubMed]
- Kume, K.; Cantwell, H.; Burrell, A.; Nurse, P. Nuclear membrane protein Lem2 regulates nuclear size through membrane flow. Nat. Commun. 2019, 10, 1871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinugasa, Y.; Hirano, Y.; Sawai, M.; Ohno, Y.; Shindo, T.; Asakawa, H.; Chikashige, Y.; Shibata, S.; Kihara, A.; Haraguchi, T.; et al. The very-long-chain fatty acid elongase Elo2 rescues lethal defects associated with loss of the nuclear barrier function in fission yeast cells. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imgrund, S.; Hartmann, D.; Farwanah, H.; Eckhardt, M.; Sandhoff, R.; Degen, J.; Gieselmann, V.; Sandhoff, K.; Willecke, K. Adult ceramide synthase 2 (CERS2)-deficient mice exhibit myelin sheath defects, cerebellar degeneration, and hepatocarcinomas. J. Biol. Chem. 2009, 284, 33549–33560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kihara, A. Synthesis and degradation pathways, functions, and pathology of ceramides and epidermal acylceramides. Prog. Lipid Res. 2016, 63, 50–69. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, Y.; Mitsutake, S.; Tsuji, K.; Kihara, A.; Igarashi, Y. Ceramide biosynthesis in keratinocyte and its role in skin function. Biochimie 2009, 91, 784–790. [Google Scholar] [CrossRef]
- Gu, M.; LaJoie, D.; Chen, O.S.; von Appen, A.; Ladinsky, M.S.; Redd, M.J.; Nikolova, L.; Bjorkman, P.J.; Sundquist, W.I.; Ullman, K.S.; et al. LEM2 recruits CHMP7 for ESCRT-mediated nuclear envelope closure in fission yeast and human cells. Proc. Natl. Acad. Sci. USA 2017, 114, E2166–E2175. [Google Scholar] [CrossRef] [Green Version]
- Olmos, Y.; Hodgson, L.; Mantell, J.; Verkade, P.; Carlton, J.G. ESCRT-III controls nuclear envelope reformation. Nature 2015, 522, 236–239. [Google Scholar] [CrossRef] [Green Version]
- Vietri, M.; Schink, K.O.; Campsteijn, C.; Wegner, C.S.; Schultz, S.W.; Christ, L.; Thoresen, S.B.; Brech, A.; Raiborg, C.; Stenmark, H. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 2015, 522, 231–235. [Google Scholar] [CrossRef]
- von Appen, A.; LaJoie, D.; Johnson, I.E.; Trnka, M.J.; Pick, S.M.; Burlingame, A.L.; Ullman, K.S.; Frost, A. LEM2 phase separation promotes ESCRT-mediated nuclear envelope reformation. Nature 2020, 582, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Willan, J.; Cleasby, A.J.; Flores-Rodriguez, N.; Stefani, F.; Rinaldo, C.; Pisciottani, A.; Grant, E.; Woodman, P.; Bryant, H.E.; Ciani, B. ESCRT-III is necessary for the integrity of the nuclear envelope in micronuclei but is aberrant at ruptured micronuclear envelopes generating damage. Oncogenesis 2019, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.J. Inner nuclear membrane protein Lem2 facilitates Rad3-mediated checkpoint signaling under replication stress induced by nucleotide depletion in fission yeast. Cell Signal. 2016, 28, 235–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.J.; Fernandez-Martinez, J.; Nudelman, I.; Shi, Y.; Zhang, W.; Raveh, B.; Herricks, T.; Slaughter, B.D.; Hogan, J.A.; Upla, P.; et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature 2018, 555, 475–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosalaganti, S.; Kosinski, J.; Albert, S.; Schaffer, M.; Strenkert, D.; Salome, P.A.; Merchant, S.S.; Plitzko, J.M.; Baumeister, W.; Engel, B.D.; et al. In situ architecture of the algal nuclear pore complex. Nat. Commun. 2018, 9, 2361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asakawa, H.; Kojidani, T.; Yang, H.J.; Ohtsuki, C.; Osakada, H.; Matsuda, A.; Iwamoto, M.; Chikashige, Y.; Nagao, K.; Obuse, C.; et al. Asymmetrical localization of Nup107-160 subcomplex components within the nuclear pore complex in fission yeast. PLoS Genet. 2019, 15, e1008061. [Google Scholar] [CrossRef] [Green Version]
- Woolcock, K.J.; Stunnenberg, R.; Gaidatzis, D.; Hotz, H.R.; Emmerth, S.; Barraud, P.; Buhler, M. RNAi keeps Atf1-bound stress response genes in check at nuclear pores. Genes Dev. 2012, 26, 683–692. [Google Scholar] [CrossRef] [Green Version]
- Thon, G.; Hansen, K.R.; Altes, S.P.; Sidhu, D.; Singh, G.; Verhein-Hansen, J.; Bonaduce, M.J.; Klar, A.J. The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics 2005, 171, 1583–1595. [Google Scholar] [CrossRef] [Green Version]
- Gozalo, A.; Duke, A.; Lan, Y.; Pascual-Garcia, P.; Talamas, J.A.; Nguyen, S.C.; Shah, P.P.; Jain, R.; Joyce, E.F.; Capelson, M. Core Components of the Nuclear Pore Bind Distinct States of Chromatin and Contribute to Polycomb Repression. Mol. Cell 2020, 77, 67–81 e67. [Google Scholar] [CrossRef]
- Pardo, M.; Nurse, P. The nuclear rim protein Amo1 is required for proper microtubule cytoskeleton organisation in fission yeast. J. Cell Sci. 2005, 118, 1705–1714. [Google Scholar] [CrossRef] [Green Version]
- Lejeune, E.; Bortfeld, M.; White, S.A.; Pidoux, A.L.; Ekwall, K.; Allshire, R.C.; Ladurner, A.G. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr. Biol. 2007, 17, 1219–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motamedi, M.R.; Hong, E.J.; Li, X.; Gerber, S.; Denison, C.; Gygi, S.; Moazed, D. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol. Cell 2008, 32, 778–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frey, S.; Richter, R.P.; Gorlich, D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 2006, 314, 815–817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.J.; Iwamoto, M.; Hiraoka, Y.; Haraguchi, T. Function of nuclear membrane proteins in shaping the nuclear envelope integrity during closed mitosis. J. Biochem. 2017, 161, 471–477. [Google Scholar] [CrossRef] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hirano, Y.; Asakawa, H.; Sakuno, T.; Haraguchi, T.; Hiraoka, Y. Nuclear Envelope Proteins Modulating the Heterochromatin Formation and Functions in Fission Yeast. Cells 2020, 9, 1908. https://doi.org/10.3390/cells9081908
Hirano Y, Asakawa H, Sakuno T, Haraguchi T, Hiraoka Y. Nuclear Envelope Proteins Modulating the Heterochromatin Formation and Functions in Fission Yeast. Cells. 2020; 9(8):1908. https://doi.org/10.3390/cells9081908
Chicago/Turabian StyleHirano, Yasuhiro, Haruhiko Asakawa, Takeshi Sakuno, Tokuko Haraguchi, and Yasushi Hiraoka. 2020. "Nuclear Envelope Proteins Modulating the Heterochromatin Formation and Functions in Fission Yeast" Cells 9, no. 8: 1908. https://doi.org/10.3390/cells9081908
APA StyleHirano, Y., Asakawa, H., Sakuno, T., Haraguchi, T., & Hiraoka, Y. (2020). Nuclear Envelope Proteins Modulating the Heterochromatin Formation and Functions in Fission Yeast. Cells, 9(8), 1908. https://doi.org/10.3390/cells9081908





