Regulation and Functions of ROP GTPases in Plant–Microbe Interactions
Abstract
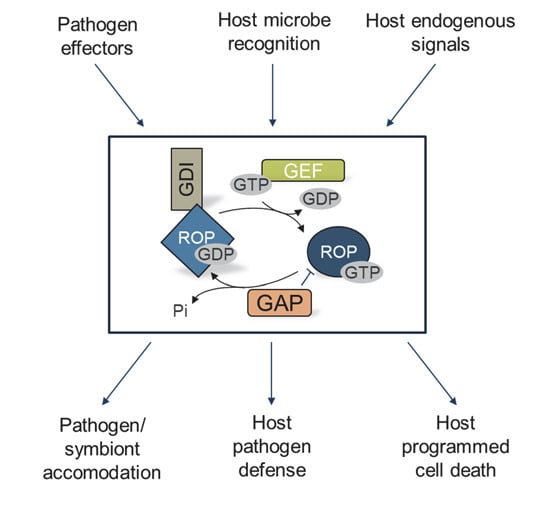
:1. Introduction
2. Rice OsRAC1 Acts in Plant Immunity
2.1. Activation of OsRAC1 in MTI and ETI
2.2. Interactors of Activated OsRAC1
3. Barley HvRACB Acts in Cell Polarity and Susceptibility to Powdery Mildew
3.1. Activation and Inactivation of HvRACB
3.2. Scaffolds and Executors of HvRACB Function in Susceptibility or Resistance
4. An Excursion into Mammalian Rho Signaling in Bacterial Pathogen Entry and Immunity
5. ROPs Involved in Further Plant-Microbe Interactions
5.1. The Role of Arabidopsis ROP6 in Response to Powdery Mildew
5.2. ROP GTPases Involved in Symbiosis
6. Concluding Remarks
Funding
Conflicts of Interest
References
- Berken, A.; Wittinghofer, A. Structure and function of Rho-type molecular switches in plants. Plant Physiol. Biochem. 2008, 46, 380–393. [Google Scholar] [CrossRef]
- Takai, Y.; Sasaki, T.; Matozaki, T. Small GTP-Binding Proteins. Physiol. Rev. 2001, 81, 153–208. [Google Scholar] [CrossRef]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, T.; Henmi, K.; Ono, E.; Hatakeyama, S.; Iwano, M.; Satoh, H.; Shimamoto, K. The small GTP-binding protein Rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA 1999, 96, 10922–10926. [Google Scholar] [CrossRef] [Green Version]
- Winge, P.; Brembu, T.; Kristensen, R.; Bones, A.M. Genetic structure and evolution of RAC-GTPases in Arabidopsis thaliana. Genetics 2000, 156, 1959–1971. [Google Scholar]
- Zheng, Z.L.; Yang, Z. The Rop GTPase: An emerging signaling switch in plants. Plant Mol. Biol. 2000, 44, 1–9. [Google Scholar] [CrossRef]
- Oda, Y.; Fukuda, H. Emerging roles of small GTPases in secondary cell wall development. Front. Plant Sci. 2014, 5, 428. [Google Scholar] [CrossRef] [Green Version]
- Fehér, A.; Lajkó, D.B. Signals fly when kinases meet Rho-of-plants (ROP) small G-proteins. Plant Sci. 2015, 237, 93–107. [Google Scholar] [CrossRef] [Green Version]
- Yalovsky, S. Protein lipid modifications and the regulation of ROP GTPase function. J. Exp. Bot. 2015, 66, 1617–1624. [Google Scholar] [CrossRef] [Green Version]
- Feiguelman, G.; Fu, Y.; Yalovsky, S. ROP GTPases structure-function and signaling pathways. Plant Physiol. 2018, 176, 57–79. [Google Scholar] [CrossRef]
- Hoefle, C.; Huesmann, C.; Schultheiss, H.; Börnke, F.; Hensel, G.; Kumlehn, J.; Hückelhoven, R. A barley ROP GTPase ACTIVATING PROTEIN associates with microtubules and regulates entry of the barley powdery mildew fungus into leaf epidermal cells. Plant Cell 2011, 23, 2422–2439. [Google Scholar] [CrossRef] [Green Version]
- Boulter, E.; Garcia-Mata, R. RhoGDI: A rheostat for the Rho switch. Small GTPases 2010, 1, 65–68. [Google Scholar] [CrossRef] [Green Version]
- Dangl, J.L.; Jones, J.D.G. Defence Responses to Infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef]
- Gust, A.A.; Pruitt, R.; Nürnberger, T. Sensing Danger: Key to Activating Plant Immunity. Trends Plant Sci. 2017, 22, 779–791. [Google Scholar] [CrossRef]
- Kawano, Y.; Shimamoto, K. Early signaling network in rice PRR-mediated and R-mediated immunity. Curr. Opin. Plant Biol. 2013, 16, 496–504. [Google Scholar] [CrossRef]
- Kawano, Y.; Kaneko-Kawano, T.; Shimamoto, K. Rho family GTPase-dependent immunity in plants and animals. Front. Plant Sci. 2014, 5, 522. [Google Scholar] [CrossRef] [Green Version]
- Miki, D.; Itoh, R.; Shimamoto, K. RNA silencing of single and multiple members in a gene family in rice. Plant Physiol. 2005, 138, 1903–1913. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Shiotani, K.; Togashi, T.; Miki, D.; Aoyama, M.; Wong, H.L.; Kawasaki, T.; Shimamoto, K. Analysis of the Rac/Rop small gtpase family in rice: Expression, subcellular localization and role in disease resistance. Plant Cell Physiol. 2010, 51, 585–595. [Google Scholar] [CrossRef]
- Ono, E.; Wong, H.L.; Kawasaki, T.; Hasegawa, M.; Kodama, O.; Shimamoto, K. Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 2001, 98, 759–764. [Google Scholar] [CrossRef] [PubMed]
- Suharsono, U.; Fujisawa, Y.; Kawasaki, T.; Iwasaki, Y.; Satoh, H.; Shimamoto, K. The heterotrimeric G protein α subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 2002, 99, 13307–13312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akamatsu, A.; Wong, H.L.; Fujiwara, M.; Okuda, J.; Nishide, K.; Uno, K.; Imai, K.; Umemura, K.; Kawasaki, T.; Kawano, Y.; et al. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 2013, 13, 465–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, K.; Imai, K.; Akamatsu, A.; Mihashi, M.; Hayashi, N.; Shimamoto, K.; Kawasaki, T. SWAP70 functions as a Rac/Rop guanine nucleotide-exchange factor in rice. Plant J. 2012, 70, 389–397. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Kawasaki, T. Function of Arabidopsis SWAP70 GEF in immune response. Plant Signal. Behav. 2012, 7, 465–468. [Google Scholar] [CrossRef] [Green Version]
- Lieberherr, D.; Nguyen, P.T.; Nakashima, A.; Umemura, K.; Kawasaki, T.; Shimamoto, K. A sphingolipid elicitor-inducible mitogen-activated protein kinase is regulated by the small GTPase OsRac1 and heterotrimeric G-protein in rice. Plant Physiol. 2005, 138, 1644–1652. [Google Scholar] [CrossRef] [Green Version]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Minami, E.; Shibuya, N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef] [Green Version]
- Akamatsu, A.; Uno, K.; Kato, M.; Wong, H.L.; Shimamoto, K.; Kawano, Y. New insights into the dimerization of small GTPase Rac/ROP guanine nucleotide exchange factors in rice. Plant Signal. Behav. 2015, 10, e1044702. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Hamada, S.; Fujiwara, M.; Zhu, T.; Thao, N.P.; Wong, H.L.; Krishna, P.; Ueda, T.; Kaku, H.; Shibuya, N.; et al. The Hop/Sti1-Hsp90 Chaperone Complex Facilitates the Maturation and Transport of a PAMP Receptor in Rice Innate Immunity. Cell Host Microbe 2010, 7, 185–196. [Google Scholar] [CrossRef] [Green Version]
- Kawano, Y.; Akamatsu, A.; Hayashi, K.; Housen, Y.; Okuda, J.; Yao, A.; Nakashima, A.; Takahashi, H.; Yoshida, H.; Wong, H.L.; et al. Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe 2010, 7, 362–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawano, Y.; Chen, L.; Shimamoto, K. The function of rac small GTPase and associated proteins in rice innate immunity. Rice 2010, 3, 112–121. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Li, Y.; Ishikawa, K.; Kosami, K.; Uno, K.; Nagawa, S.; Tan, L.; Du, J.; Shimamoto, K.; Kawano, Y. Resistance protein Pit interacts with the GEF OsSPK1 to activate OsRac1 and trigger rice immunity. Proc. Natl. Acad. Sci. USA 2018, 115, E11551–E11560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawano, Y.; Fujiwara, T.; Yao, A.; Housen, Y.; Hayashi, K.; Shimamoto, K. Palmitoylation-dependent membrane localization of the rice resistance protein pit is critical for the activation of the small GTPase OsRac1. J. Biol. Chem. 2014, 289, 19079–19088. [Google Scholar] [CrossRef] [Green Version]
- Césari, S.; Kanzaki, H.; Fujiwara, T.; Bernoux, M.; Chalvon, V.; Kawano, Y.; Shimamoto, K.; Dodds, P.; Terauchi, R.; Kroj, T. The NB - LRR proteins RGA 4 and RGA 5 interact functionally and physically to confer disease resistance. EMBO J. 2014, 33, 1941–1959. [Google Scholar] [CrossRef]
- Okuyama, Y.; Kanzaki, H.; Abe, A.; Yoshida, K.; Tamiru, M.; Saitoh, H.; Fujibe, T.; Matsumura, H.; Shenton, M.; Galam, D.C.; et al. A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 2011, 66, 467–479. [Google Scholar] [CrossRef]
- Ortiz, D.; de Guillen, K.; Cesari, S.; Chalvon, V.; Gracy, J.; Padilla, A.; Kroj, T. Recognition of the magnaporthe oryzae effector AVR-pia by the decoy domain of the rice NLR immune receptor RGA5. Plant Cell 2017, 29, 156–168. [Google Scholar] [CrossRef] [Green Version]
- Thao, N.P.; Chen, L.; Nakashima, A.; Hara, S.I.; Umemura, K.; Takahashi, A.; Shirasu, K.; Kawasaki, T.; Shimamoto, K. RAR1 and HSP90 form a complex with Rac/Rop GTPase and function in innate-immune responses in rice. Plant Cell 2007, 19, 4035–4045. [Google Scholar] [CrossRef] [Green Version]
- Wong, H.L.; Pinontoan, R.; Hayashi, K.; Tabata, R.; Yaeno, T.; Hasegawa, K.; Kojima, C.; Yoshioka, H.; Iba, K.; Kawasaki, T.; et al. Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 2007, 19, 4022–4034. [Google Scholar] [CrossRef] [Green Version]
- Kosami, K.I.; Ohki, I.; Nagano, M.; Furuita, K.; Sugiki, T.; Kawano, Y.; Kawasaki, T.; Fujiwara, T.; Nakagawa, A.; Shimamoto, K.; et al. The crystal structure of the plant small GTPase OsRac1 reveals its mode of binding to NADPH oxidase. J. Biol. Chem. 2014, 289, 28569–28578. [Google Scholar] [CrossRef] [Green Version]
- Oda, T.; Hashimoto, H.; Kuwabara, N.; Akashi, S.; Hayashi, K.; Kojima, C.; Wong, H.L.; Kawasaki, T.; Shimamoto, K.; Sato, M.; et al. Structure of the N-terminal regulatory domain of a plant NADPH oxidase and its functional implications. J. Biol. Chem. 2010, 285, 1435–1445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagano, M.; Ishikawa, T.; Fujiwara, M.; Fukao, Y.; Kawano, Y.; Kawai-Yamada, M.; Shimamoto, K. Plasma membrane microdomains are essential for Rac1-RbohB/H-mediated immunity in rice. Plant Cell 2016, 28, 1966–1983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawasaki, T.; Koita, H.; Nakatsubo, T.; Hasegawa, K.; Wakabayashi, K.; Takahashi, H.; Umemura, K.; Umezawa, T.; Shimamoto, K. Cinnamoyl-CoA reductase, a key in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 230–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakashima, A.; Chen, L.; Nguyen, P.T.; Fujiwara, M.; Hann, L.W.; Kuwano, M.; Umemura, K.; Shirasu, K.; Kawasaki, T.; Shimamotoa, K. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 2008, 20, 2265–2279. [Google Scholar] [CrossRef] [Green Version]
- Morino, K.; Kimizu, M.; Fujiwara, M. Disulfide proteomics of rice cultured cells in response to OsRacl and probenazole-related immune signaling pathway in rice. Proteome Sci. 2017, 15, 1–2. [Google Scholar] [CrossRef]
- Wamaitha, M.J.; Yamamoto, R.; Wong, H.L.; Kawasaki, T.; Kawano, Y.; Shimamoto, K. OsRap2.6 transcription factor contributes to rice innate immunity through its interaction with receptor for activated Kinase-C 1 (RACK1). Rice 2012, 5, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Dong, S.; Sun, D.; Liu, W.; Gu, F.; Liu, Y.; Guo, T.; Wang, H.; Wang, J.; Chen, Z. CONSTANS-like 9 (OsCOL9) interacts with receptor for activated C-kinase 1(OsRACK1) to regulate blast resistance through salicylic acid and ethylene signaling pathways. PLoS ONE 2016, 11, e166249. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Li, J.F.; Niu, Y.; Zhang, X.C.; Woody, O.Z.; Xiong, Y.; Djonović, S.; Millet, Y.; Bush, J.; McConkey, B.J.; et al. Pathogen-secreted proteases activate a novel plant immune pathway. Nature 2015, 521, 213–216. [Google Scholar] [CrossRef]
- Kim, S.H.; Oikawa, T.; Kyozuka, J.; Wong, H.L.; Umemura, K.; Kishi-Kaboshi, M.; Takahashi, A.; Kawano, Y.; Kawasaki, T.; Shimamoto, K. The bHLH Rac immunity1 (RAI1) is activated by OsRac1 via OsMAPK3 and OsMAPK6 in rice immunity. Plant Cell Physiol. 2012, 53, 740–754. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Yao, S.; Kosami, K.; Guo, T.; Li, J.; Zhang, Y.; Fukao, Y.; Kaneko-Kawano, T.; Zhang, H.; She, Y.M.; et al. Identification of endogenous small peptides involved in rice immunity through transcriptomics- and proteomics-based screening. Plant Biotechnol. J. 2020, 18, 415–428. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Xiong, Y.; Liu, R.; Xue, H.W.; Yang, Z. The Rho-family GTPase OsRac1 controls rice grain size and yield by regulating cell division. Proc. Natl. Acad. Sci. USA 2019, 116, 16121–16126. [Google Scholar] [CrossRef] [Green Version]
- Dong, Z.; Li, W.; Liu, J.; Li, L.; Pan, S.; Liu, S.; Gao, J.; Liu, L.; Liu, X.; Wang, G.L.; et al. The Rice Phosphate Transporter Protein OsPT8 Regulates Disease Resistance and Plant Growth. Sci. Rep. 2019, 9, 2–11. [Google Scholar] [CrossRef]
- Zhou, Z.; Pang, Z.; Zhao, S.; Zhang, L.; Lv, Q.; Yin, D.; Li, D.; Liu, X.; Zhao, X.; Li, X.; et al. Importance of OsRac1 and RAI1 in signalling of nucleotide-binding site leucine-rich repeat protein-mediated resistance to rice blast disease. New Phytol. 2019, 223, 828–838. [Google Scholar] [CrossRef]
- Moeder, W.; Yoshioka, K.; Klessig, D.F. Involvement of the small GTPase rac in the defense responses of tobacco to pathogens. Mol. Plant-Microbe Interact. 2005, 18, 116–124. [Google Scholar] [CrossRef]
- Bai, P.; Park, C.H.; Shirsekar, G.; Songkumarn, P.; Bellizzi, M.; Wang, G.L. Role of lysine residues of the Magnaporthe oryzae effector AvrPiz-t in effector- and PAMP-triggered immunity. Mol. Plant Pathol. 2019, 20, 599–608. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plantĝ€” pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Wang, J.; Hu, M.; Wang, J.; Qi, J.; Han, Z.; Wang, G.; Qi, Y.; Wang, H.W.; Zhou, J.M.; Chai, J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 2019, 364. [Google Scholar] [CrossRef]
- Liu, J.; Park, C.H.; He, F.; Nagano, M.; Wang, M.; Bellizzi, M.; Zhang, K.; Zeng, X.; Liu, W.; Ning, Y.; et al. The RhoGAP SPIN6 Associates with SPL11 and OsRac1 and Negatively Regulates Programmed Cell Death and Innate Immunity in Rice. PLoS Pathog. 2015, 11, e1004629. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, J.; He, F.; Wang, Z.; Ning, Y.; Wang, G.L. OsHUB1 and OsHUB2 interact with spin6 and form homo- and hetero-dimers in rice. Plant Signal. Behav. 2015, 10, 6–10. [Google Scholar] [CrossRef] [Green Version]
- Schultheiss, H.; Dechert, C.; Kogel, K.H.; Hückelhoven, R. A small GTP-binding host protein is required for entry of powdery mildew fungus into epidermal cells of barley. Plant Physiol. 2002, 128, 1447–1454. [Google Scholar] [CrossRef] [Green Version]
- Hückelhoven, R.; Kogel, K.H. Reactive oxygen intermediates in plant-microbe interactions: Who is who in powdery mildew resistance? Planta 2003, 216, 891–902. [Google Scholar] [CrossRef]
- Schultheiss, H.; Dechert, C.; Kogel, K.H.; Hückelhoven, R. Functional analysis of barley RAC/ROP G-protein family members in susceptibility to the powdery mildew fungus. Plant J. 2003, 36, 589–601. [Google Scholar] [CrossRef]
- Opalski, K.S.; Schultheiss, H.; Kogel, K.H.; Hückelhoven, R. The receptor-like MLO protein and the RAC/ROP family G-protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp. hordei. Plant J. 2005, 41, 291–303. [Google Scholar] [CrossRef]
- Šamaj, J.; Müller, J.; Beck, M.; Böhm, N.; Menzel, D. Vesicular trafficking, cytoskeleton and signalling in root hairs and pollen tubes. Trends Plant Sci. 2006, 11, 594–600. [Google Scholar] [CrossRef]
- Lee, Y.J.; Yang, Z. Tip growth: Signaling in the apical dome. Curr. Opin. Plant Biol. 2008, 11, 662–671. [Google Scholar] [CrossRef] [Green Version]
- Humphries, J.A.; Vejlupkova, Z.; Luo, A.; Meeley, R.B.; Sylvester, A.W.; Fowler, J.E.; Smith, L.G. ROP GTPases act with the receptor-like protein PAN1 to polarize asymmetric cell division in maize. Plant Cell 2011, 23, 2273–2284. [Google Scholar] [CrossRef] [Green Version]
- Scheler, B.; Schnepf, V.; Galgenmüller, C.; Ranf, S.; Hückelhoven, R. Barley disease susceptibility factor RACB acts in epidermal cell polarity and positioning of the nucleus. J. Exp. Bot. 2016, 67, 3263–3275. [Google Scholar] [CrossRef]
- Pathuri, I.P.; Zellerhoff, N.; Schaffrath, U.; Hensel, G.; Kumlehn, J.; Kogel, K.H.; Eichmann, R.; Hückelhoven, R. Constitutively activated barley ROPs modulate epidermal cell size, defense reactions and interactions with fungal leaf pathogens. Plant Cell Rep. 2008, 27, 1877–1887. [Google Scholar] [CrossRef]
- Nottensteiner, M.; Zechmann, B.; McCollum, C.; Hückelhoven, R. A barley powdery mildew fungus non-autonomous retrotransposon encodes a peptide that supports penetration success on barley. J. Exp. Bot. 2018, 69, 3745–3758. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; McCormick, S. A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 18830–18835. [Google Scholar] [CrossRef] [Green Version]
- Duan, Q.; Kita, D.; Li, C.; Cheung, A.Y.; Wu, H.M. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. USA 2010, 107, 17821–17826. [Google Scholar] [CrossRef] [Green Version]
- Nissen, K.S.; Willats, W.G.T.; Malinovsky, F.G. Understanding CrRLK1L Function: Cell Walls and Growth Control. Trends Plant Sci. 2016, 21, 516–527. [Google Scholar] [CrossRef]
- Kessler, S.A.; Shimosato-Asano, H.; Keinath, N.F.; Wuest, S.E.; Ingram, G.; Panstruga, R.; Grossniklaus, U. Conserved molecular components for pollen tube reception and fungal invasion. Science 2010, 330, 968–971. [Google Scholar] [CrossRef]
- Schnepf, V.; Vlot, A.C.; Kugler, K.; Hückelhoven, R. Barley susceptibility factor RACB modulates transcript levels of signalling protein genes in compatible interaction with Blumeria graminis f.sp. hordei. Mol. Plant Pathol. 2018, 19, 393–404. [Google Scholar] [CrossRef] [Green Version]
- Fodor-Dunai, C.; Fricke, I.; Potocký, M.; Dorjgotov, D.; Domoki, M.; Jurca, M.E.; Ötvös, K.; Ẑárský, V.; Berken, A.; Fehér, A. The phosphomimetic mutation of an evolutionarily conserved serine residue affects the signaling properties of Rho of plants (ROPs). Plant J. 2011, 66, 669–679. [Google Scholar] [CrossRef]
- Weiß, L.; Reiner, T.; Mergner, J.; Kuster, B.; Fehér, A.; Hensel, G.; Gahrtz, M.; Kumlehn, J.; Engelhardt, S.; Hückelhoven, R. Posttranslational modification of the RHO of plants protein RACB by phosphorylation and cross-kingdom conserved ubiquitination. bioRxiv 2020. [Google Scholar] [CrossRef]
- Huesmann, C.; Reiner, T.; Hoefle, C.; Preuss, J.; Jurca, M.E.; Domoki, M.; Fehér, A.; Hückelhoven, R. Barley rop binding kinase1 is involved in microtubule organization and in basal penetration resistance to the barley powdery mildew fungus. Plant Physiol. 2012, 159, 311–320. [Google Scholar] [CrossRef] [Green Version]
- Reiner, T.; Hoefle, C.; Hückelhoven, R. A barley SKP1-like protein controls abundance of the susceptibility factor RACB and influences the interaction of barley with the barley powdery mildew fungus. Mol. Plant Pathol. 2016, 17, 184–195. [Google Scholar] [CrossRef]
- Zhao, J.; Mialki, R.K.; Wei, J.; Coon, T.A.; Zou, C.; Chen, B.B.; Mallampalli, R.K.; Zhao, Y. SCF E3 ligase F-box protein complex SCFFBXL19 regulates cell migration by mediating Rac1 ubiquitination and degradation. FASEB J. 2013, 27, 2611–2619. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Gu, Y.; Li, S.; Yang, Z. A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets. Plant Cell 2001, 13, 2841–2856. [Google Scholar] [CrossRef]
- Schultheiss, H.; Preuss, J.; Pircher, T.; Eichmann, R.; Hückelhoven, R. Barley RIC171 interacts with RACB in planta and supports entry of the powdery mildew fungus. Cell Microbiol. 2008, 10, 1815–1826. [Google Scholar] [CrossRef]
- Engelhardt, S.; Kopischke, M.; Hofer, J.; Probst, K.; McCollum, C.; Hückelhoven, R. Barley RIC157 is involved in RACB-mediated susceptibility to powdery mildew. bioRxiv 2019, 848226. [Google Scholar] [CrossRef] [Green Version]
- Hückelhoven, R.; Panstruga, R. Cell biology of the plant-powdery mildew interaction. Curr. Opin. Plant Biol. 2011, 14, 738–746. [Google Scholar] [CrossRef]
- Wu, G.; Li, H.; Yang, Z. Arabidopsis RopGAPSs are a novel family of Rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for Rop-specific GTPase stimulation. Plant Physiol. 2000, 124, 1625–1636. [Google Scholar] [CrossRef] [Green Version]
- Hoefle, C.; McCollum, C.; Hückelhoven, R. Barley ROP-Interactive Partner-a organizes into RAC1- A nd MICROTUBULE-ASSOCIATED ROP-GTPASE ACTIVATING PROTEIN 1-dependent membrane domains. BMC Plant Biol. 2020, 20, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Hoefle, C.; Hückelhoven, R. A barley Engulfment and Motility domain containing protein modulates Rho GTPase activating protein HvMAGAP1 function in the barley powdery mildew interaction. Plant Mol. Biol. 2014, 84, 469–478. [Google Scholar] [CrossRef]
- Kinchen, J.M.; Ravichandran, K.S. Journey to the grave: Signaling events regulating removal of apoptotic cells. J. Cell Sci. 2007, 120, 2143–2149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Jin, T. ELMO proteins transduce G protein-coupled receptor signal to control reorganization of actin cytoskeleton in chemotaxis of eukaryotic cells. Small GTPases 2019, 10, 271–279. [Google Scholar] [CrossRef]
- Mccollum, C.; Engelhardt, S.; Weiss, L. ROP INTERACTIVE PARTNER b interacts with RACB and supports fungal penetration into barley epidermal cells. Plant Physiol. 2020. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Kobayashi, I.; Funaki, Y.; Fujimoto, S.; Takemoto, T.; Kunoh, H. Dynamic reorganization of microfilaments and microtubules is necessary for the expression of non-host resistance in barley coleoptile cells. Plant J. 1997, 11, 525–537. [Google Scholar] [CrossRef]
- Miklis, M.; Consonni, C.; Bhat, R.A.; Lipka, V.; Schulze-Lefert, P.; Panstruga, R. Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery. Plant Physiol. 2007, 144, 1132–1143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ly, K.T.; Casanova, J.E. Mechanisms of Salmonella entry into host cells. Cell Microbiol. 2007, 9, 2103–2111. [Google Scholar] [CrossRef] [PubMed]
- Aepfelbacher, M.; Wolters, M. Acting on Actin: Rac and Rho played by Yersinia. Curr. Top. Microbiol. Immunol. 2016, 399, 201–220. [Google Scholar] [CrossRef]
- Bohn, E.; Sonnabend, M.; Klein, K.; Autenrieth, I.B. Bacterial adhesion and host cell factors leading to effector protein injection by type III secretion system. Int. J. Med. Microbiol. 2019, 309, 344–350. [Google Scholar] [CrossRef]
- McGhie, E.J.; Hayward, R.D.; Koronakis, V. Cooperation between actin-binding proteins of invasive Salmonella: SipA potentiates SipC nucleation and bundling of actin. EMBO J. 2001, 20, 2131–2139. [Google Scholar] [CrossRef] [Green Version]
- Zhou, D.; Galán, J. Salmonella entry into host cells: The work in concert of type III secreted effector proteins. Microbes Infect. 2001, 3, 1293–1298. [Google Scholar] [CrossRef]
- Patel, J.C.; Galán, J.E. Differential activation and function of Rho GTPases during Salmonella-host cell interactions. J. Cell Biol. 2006, 175, 453–463. [Google Scholar] [CrossRef] [Green Version]
- Hardt, W.D.; Chen, L.M.; Schuebel, K.E.; Bustelo, X.R.; Galán, J.E.S. typhimurium Encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 1998, 93, 815–826. [Google Scholar] [CrossRef] [Green Version]
- Buchwald, G.; Friebel, A.; Galán, J.E.; Hardt, W.D.; Wittinghofer, A.; Scheffzek, K. Structural basis for the reversible activation of a Rho protein by the bacterial toxin SopE. EMBO J. 2002, 21, 3286–3295. [Google Scholar] [CrossRef]
- Williams, C.; Galyov, E.E.; Bagby, S. Solution structure, backbone dynamics, and interaction with Cdc42 of Salmonella guanine nucleotide exchange factor SopE2. Biochemistry 2004, 43, 11998–12008. [Google Scholar] [CrossRef]
- Truong, D.; Boddy, K.C.; Canadien, V.; Brabant, D.; Fairn, G.D.; D’Costa, V.M.; Coyaud, E.; Raught, B.; Pérez-Sala, D.; Park, W.S.; et al. Salmonella exploits host Rho GTPase signalling pathways through the phosphatase activity of SopB. Cell Microbiol. 2018, 20, 1–12. [Google Scholar] [CrossRef]
- Von Pawel-Rammingen, U.; Telepnev, M.V.; Schmidt, G.; Aktories, K.; Wolf-Watz, H.; Rosqvist, R. GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: A mechanism for disruption of actin microfilament structure. Mol. Microbiol. 2000, 36, 737–748. [Google Scholar] [CrossRef]
- Roppenser, B.; Röder, A.; Hentschke, M.; Ruckdeschel, K.; Aepfelbacher, M. Yersinia enterocolitica differentially modulates RhoG activity in host cells. J. Cell Sci. 2009, 122, 696–705. [Google Scholar] [CrossRef] [Green Version]
- Aepfelbacher, M.; Trasak, C.; Wilharm, G.; Wiedemann, A.; Trülzsch, K.; Krauss, K.; Gierschik, P.; Heesemann, J. Characterization of YopT effects on Rho GTPases in Yersinia enterocolitica-infected cells. J. Biol. Chem. 2003, 278, 33217–33223. [Google Scholar] [CrossRef] [Green Version]
- Van der Biezen, E.A.; Jones, J.D.G. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 1998, 23, 454–456. [Google Scholar] [CrossRef]
- Boyer, L.; Magoc, L.; Dejardin, S.; Cappillino, M.; Hinault, C.; Charriere, G.M.; Ip, W.K.E.; Fracchia, S.; Hennessy, E.; Erturk-hasdemir, D.; et al. Pathogen-derived effectors trigger protective immunity via activation of the Rac2 enzyme and the IMD or Rip kinase signaling pathway. Immunity 2011, 35, 536–549. [Google Scholar] [CrossRef] [Green Version]
- Boyer, L.; Lemichez, E. Switching Rho GTPase activation into effective antibacterial defenses requires the caspase-1/IL-1beta signaling axis. Small GTPases 2015, 6, 186–188. [Google Scholar] [CrossRef] [Green Version]
- Bruno, V.M.; Hannemann, S.; Lara-Tejero, M.; Flavell, R.A.; Kleinstein, S.H.; Galán, J.E. Salmonella typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS Pathog. 2009, 5, e1000538. [Google Scholar] [CrossRef] [Green Version]
- Keestra, A.M.; Winter, M.G.; Auburger, J.J.; Fräßle, S.P.; Xavier, M.N.; Winter, S.E.; Kim, A.; Poon, V.; Ravesloot, M.M.; Waldenmaier, J.F.T.; et al. Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature 2013, 496, 233–237. [Google Scholar] [CrossRef] [Green Version]
- Keestra-Gounder, A.M.; Tsolis, R.M. NOD1 and NOD2: Beyond Peptidoglycan Sensing. Trends Immunol. 2017, 38, 758–767. [Google Scholar] [CrossRef]
- Fukazawa, A.; Alonso, C.; Kurachi, K.; Gupta, S.; Lesser, C.F.; McCormick, B.A.; Reinecker, H.C. GEF-H1 mediated control of NOD1 dependent NF-κB activation by Shigella effectors. PLoS Pathog. 2008, 4, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Perona, R.; Montaner, S.; Saniger, L.; Sánchez-Pérez, I.; Bravo, R.; Lacal, J.C. Activation of the nuclear factor-κB by Rho, CDC42, and Rac-1 proteins. Genes Dev. 1997, 11, 463–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legrand-Poels, S.; Kustermans, G.; Bex, F.; Kremmer, E.; Kufer, T.A.; Piette, J. Modulation of Nod2-dependent NF-κB signaling by the actin cytoskeleton. J. Cell Sci. 2007, 120, 1299–1310. [Google Scholar] [CrossRef] [Green Version]
- Craddock, C.; Lavagi, I.; Yang, Z. New Insights into Rho signaling from plant ROP/Rac GTPases. Trends Cell Biol. 2012, 22, 492–501. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Wen, M.; Nagawa, S.; Fu, Y.; Chen, J.; Wu, M.; Perrot-Rechenmann, C.; Friml, J.; Jones, A.M.; Yang, Z. Cell surface- and Rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 2010, 143, 99–110. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Grandont, L.; Li, H.; Hauschild, R.; Paque, S.; Abuzeineh, A.; Rakusová, H.; Benkova, E.; Perrot-Rechenmann, C.; Friml, J. Inhibition of cell expansion by rapid ABP1-mediated auxin effect on microtubules. Nature 2014, 516, 90–93. [Google Scholar] [CrossRef]
- Venus, Y.; Oelmüller, R. Arabidopsis ROP1 and ROP6 influence germination time, root morphology, the formation of F-actin bundles, and symbiotic fungal interactions. Mol. Plant 2013, 6, 872–886. [Google Scholar] [CrossRef] [Green Version]
- Basu, D.; Le, J.; Zakharova, T.; Mallery, E.L.; Szymanski, D.B. A SPIKE1 signaling complex controls actin-dependent cell morphogenesis through the heteromeric WAVE and ARP2/3 complexes. Proc. Natl. Acad. Sci. USA 2008, 105, 4044–4049. [Google Scholar] [CrossRef] [Green Version]
- Ren, H.; Dang, X.; Yang, Y.; Huang, D.; Liu, M.; Gao, X.; Lin, D. SPIKE1 activates ROP GTPase to modulate petal growth and shape. Plant Physiol. 2016, 172, 358–371. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Shi, Y.; Liu, G.; Guo, Y.; Yang, Y. Activation of ROP6 GTPase by phosphatidylglycerol in arabidopsis. Front. Plant Sci. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Poraty-Gavra, L.; Zimmermann, P.; Haigis, S.; Bednarek, P.; Hazak, O.; Stelmakh, O.R.; Sadot, E.; Schulze-Lefert, P.; Gruissem, W.; Yalovsky, S. The Arabidopsis Rho of plants GTPase AtROP6 functions in developmental and pathogen response pathways. Plant Physiol. 2013, 161, 1172–1188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huesmann, C.; Hoefle, C.; Hückelhoven, R. Ropgaps of arabidopsis limit susceptibility to powdery mildew. Plant Signal. Behav. 2011, 6, 1691–1694. [Google Scholar] [CrossRef] [Green Version]
- Reiner, T.; Hoefle, C.; Huesmann, C.; Ménesi, D.; Fehér, A.; Hückelhoven, R. The Arabidopsis ROP-activated receptor-like cytoplasmic kinase RLCK VI_A3 is involved in control of basal resistance to powdery mildew and trichome branching. Plant Cell Rep. 2014, 34, 457–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durr, J.; Reyt, G.; Spaepen, S.; Hilton, S.; Meehan, C.; Qi, W.; Kamiya, T.; Flis, P.; Dickinson, H.G.; Feher, A.; et al. Two Receptor-Like Kinases Required For Arabidopsis Endodermal Root Organisation Shape The Rhizosphere Microbiome. bioRxiv 2019, 816330. [Google Scholar] [CrossRef]
- Zhai, L.; Sun, C.; Feng, Y.; Li, D.; Chai, X.; Wang, L.; Sun, Q.; Zhang, G.; Li, Y.; Wu, T.; et al. AtROP6 is involved in reactive oxygen species signaling in response to iron-deficiency stress in Arabidopsis thaliana. FEBS Lett. 2018, 592, 3446–3459. [Google Scholar] [CrossRef] [Green Version]
- Oldroyd, G.E.D.; Murray, J.D.; Poole, P.S.; Downie, J.A. The Rules of Engagement in the Legume-Rhizobial Symbiosis. Annu. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef] [PubMed]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Jurkiewicz, A.; Fukai, E.; Quistgaard, E.M.H.; Albrektsen, A.S.; James, E.K.; Thirup, S.; Stougaard, J. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007, 26, 3923–3935. [Google Scholar] [CrossRef] [Green Version]
- Bek, A.S.; Sauer, J.; Thygesen, M.B.; Duus, J.; Petersen, B.O.; Thirup, S.; James, E.; Jensen, K.J.; Stougaard, J.; Radutoiu, S. Improved characterization of nod factors and genetically based variation in lysm receptor domains identify amino acids expendable for nod factor recognition in Lotus spp. Mol. Plant-Microbe Interact. 2010, 23, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Gough, C.; Cottret, L.; Lefebvre, B.; Bono, J.J. Evolutionary history of plant LysM receptor proteins related to root endosymbiosis. Front. Plant Sci. 2018, 9, 923. [Google Scholar] [CrossRef]
- Zipfel, C.; Oldroyd, G.E.D. Plant signalling in symbiosis and immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef]
- Ke, D.; Fang, Q.; Chen, C.; Zhu, H.; Chen, T.; Chang, X.; Yuan, S.; Kang, H.; Ma, L.; Hong, Z.; et al. The small GTPase ROP6 interacts with NFR5 and is involved in nodule formation in Lotus japonicus. Plant Physiol. 2012, 159, 131–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, D.; Li, X.; Han, Y.; Cheng, L.; Yuan, H.; Wang, L. ROP6 is involved in root hair deformation induced by Nod factors in Lotus japonicus. Plant Physiol. Biochem. 2016, 108, 488–498. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, M.; Duan, L.; Yu, H.; Chang, X.; Li, L.; Kang, H.; Feng, Y.; Zhu, H.; Hong, Z.; et al. Lotus japonicus clathrin heavy chain1 is associated with rho-like GTPase ROP6 and involved in nodule formation. Plant Physiol. 2015, 167, 1497–1510. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Chen, A.M.; Luo, L.; Sun, J.; Cao, L.P.; Yu, G.Q.; Zhu, J.B.; Wang, Y.Z. Characterization and Expression Analysis of Medicago truncatula ROP GTPase Family during the Early Stage of Symbiosis. J. Integr. Plant Biol. 2010, 52, 639–652. [Google Scholar] [CrossRef]
- Kiirika, L.M.; Bergmann, H.F.; Schikowsky, C.; Wimmer, D.; Korte, J.; Schmitz, U.; Niehaus, K.; Colditz, F. Silencing of the Rac1 GTPase MtRoP9 in medicago truncatula stimulates early mycorrhizal and oomycete root colonizations but negatively affects rhizobial infection. Plant Physiol. 2012, 159, 501–516. [Google Scholar] [CrossRef] [Green Version]
- Kiirika, L.M.; Schmitz, U.; Colditz, F. The alternative Medicago truncatula defense proteome of ROS—defective transgenic roots during early microbial infection. Front. Plant Sci. 2014, 5, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Lei, M.J.; Wang, Q.; Li, X.; Chen, A.; Luo, L.; Xie, Y.; Li, G.; Luo, D.; Mysore, K.S.; Wen, J.; et al. The small Gtpase ROP10 of medicago truncatula is required for both tip growth of root hairs and nod factor-induced root hair deformation. Plant Cell 2015, 27, 806–822. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Lei, M.; Chen, A.; Wang, R.; Li, G.; Wang, Y. MtROP8 is involved in root hair development and the establishment of symbiotic interaction between Medicago truncatula and Sinorhizobium meliloti. Chinese Sci. Bull. 2014, 59, 4289–4297. [Google Scholar] [CrossRef]
- Riely, B.K.; He, H.; Venkateshwaran, M.; Sarma, B.; Schraiber, J.; Ané, J.M.; Cook, D.R. Identification of legume RopGEF gene families and characterization of a Medicago truncatula RopGEF mediating polar growth of root hairs. Plant J. 2011, 65, 230–243. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Engelhardt, S.; Trutzenberg, A.; Hückelhoven, R. Regulation and Functions of ROP GTPases in Plant–Microbe Interactions. Cells 2020, 9, 2016. https://doi.org/10.3390/cells9092016
Engelhardt S, Trutzenberg A, Hückelhoven R. Regulation and Functions of ROP GTPases in Plant–Microbe Interactions. Cells. 2020; 9(9):2016. https://doi.org/10.3390/cells9092016
Chicago/Turabian StyleEngelhardt, Stefan, Adriana Trutzenberg, and Ralph Hückelhoven. 2020. "Regulation and Functions of ROP GTPases in Plant–Microbe Interactions" Cells 9, no. 9: 2016. https://doi.org/10.3390/cells9092016
APA StyleEngelhardt, S., Trutzenberg, A., & Hückelhoven, R. (2020). Regulation and Functions of ROP GTPases in Plant–Microbe Interactions. Cells, 9(9), 2016. https://doi.org/10.3390/cells9092016