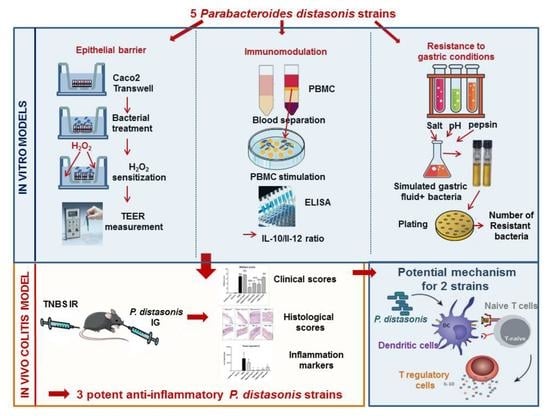
In Vitro Characterization of Gut Microbiota-Derived Commensal Strains: Selection of Parabacteroides distasonis Strains Alleviating TNBS-Induced Colitis in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
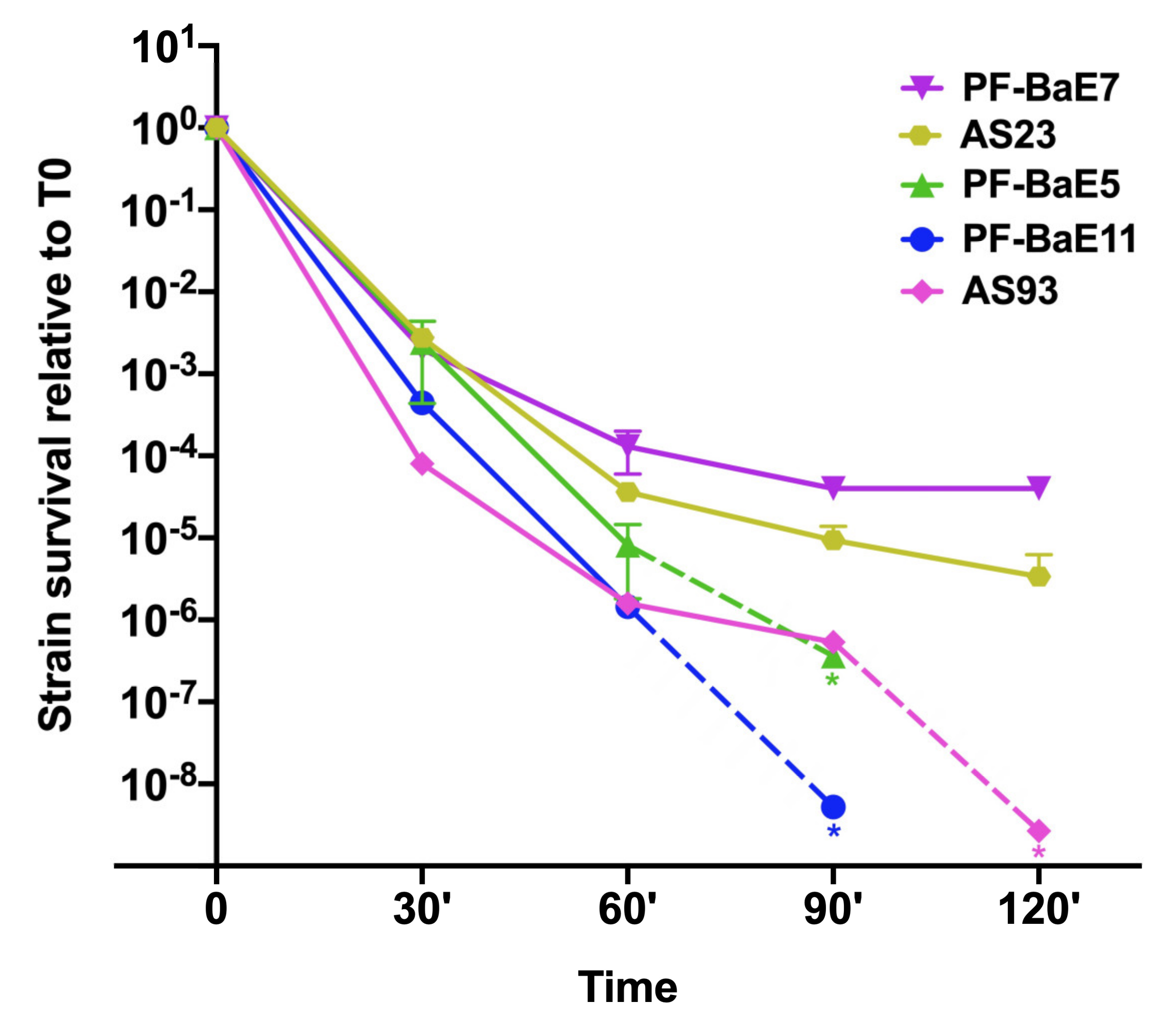
2.2. Resistance to Gastric Conditions
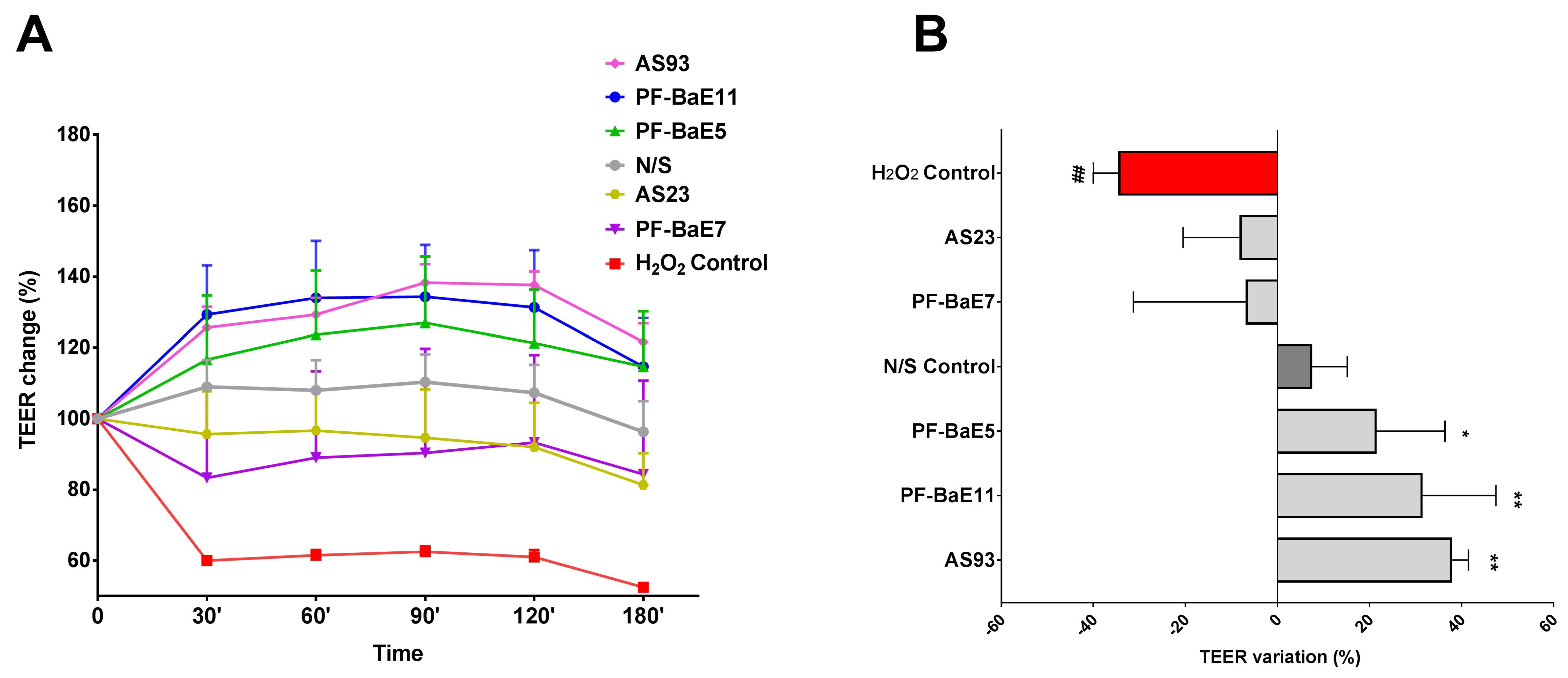
2.3. The In Vitro Epithelial Barrier Model
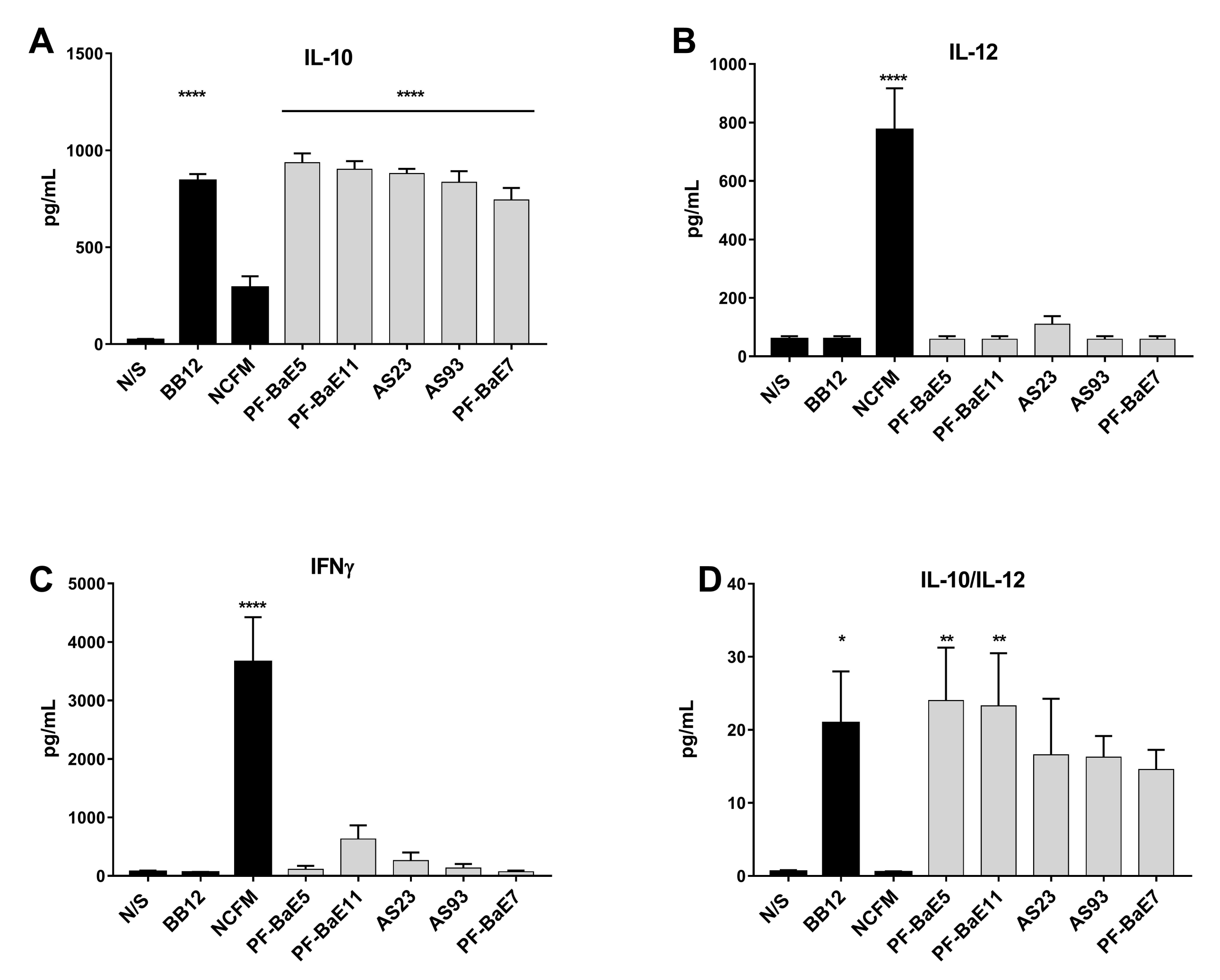
2.4. In Vitro Immunomodulation Assay
2.5. Murine Model of 2,4,6-Trinitrobenzenesulfonic Acid (TNBS)-Induced Colitis
2.6. Quantification of Fecal Lipocalin 2 (Lcn-2)
2.7. Real-Time Quantitative PCR (qRT-PCR)
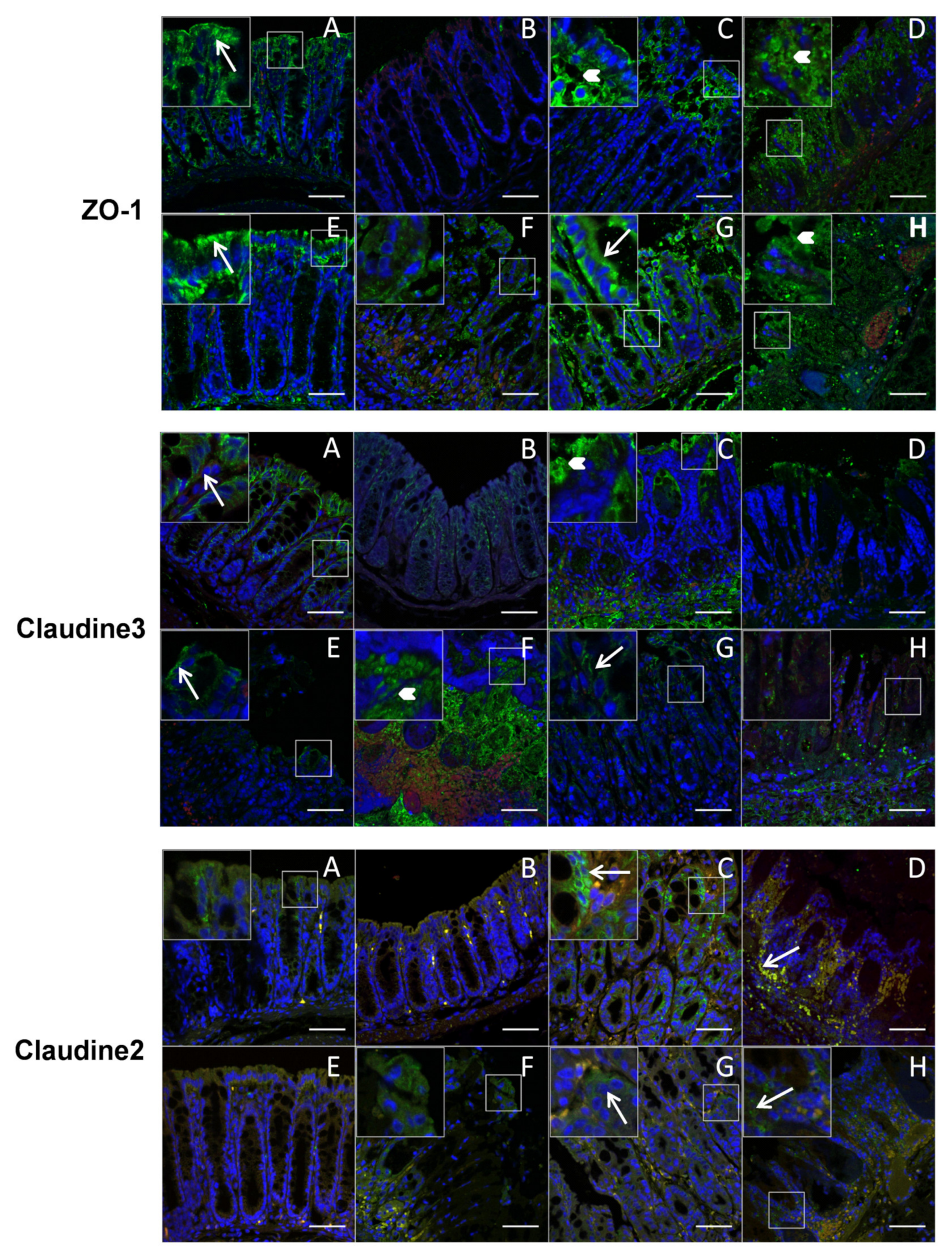
2.8. Immunofluorescence Staining and Confocal Analysis
2.9. DNA Extraction from Stool and P. distasonis Quantification by qPCR
2.10. Preparation of Bacteria-Primed Bone Marrow Derived Dendritic Cells (BMDCs) and Co-Culture with Naive CD4+ T Cells
2.11. Statistical Analysis
3. Results
3.1. P. distasonis Strains Displayed Different Ability to Restore the H2O2-Induced Disruption of the Epithelial Barrier
3.2. P. distasonis PF-BaE5 and PF-BaE11 Exhibit the Best Anti-Inflammatory In Vitro Profile
3.3. Strains Survive Differently to Gastric Condition
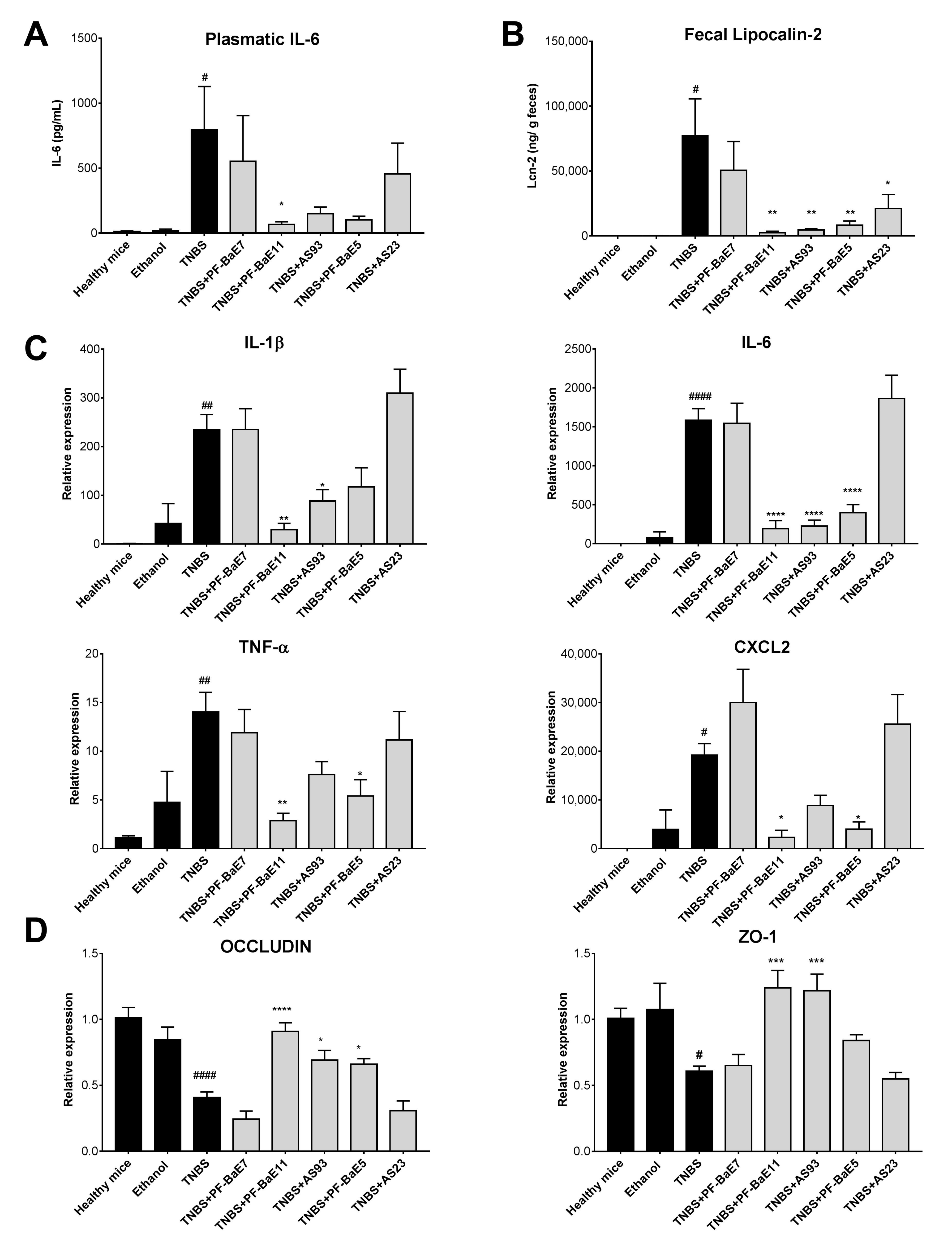
3.4. P. distasonis PF-BaE5, PF-BaE11 and AS93 Were the Most Potent Strains to Counteract Inflammation in a Murine Model of TNBS-Induced Colitis
3.5. P. distasonis PF-BaE5 and PF-BaE11 Led to Immature Bone Marrow Dendritic Cells (BMDCs) In Vitro and Regulatory T Lymphocytes When Co-Cultured with Naïve CD4+ T Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and fungi of the human gut microbiome: Correlations with diet and bacterial residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbaugh, P.J.; Gordon, J.I. The core gut microbiome, energy balance and obesity. J. Physiol. 2009, 587, 4153–4158. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slack, E.; Hapfelmeier, S.; Stecher, B.; Velykoredko, Y.; Stoel, M.; Lawson, M.A.E.; Geuking, M.B.; Beutler, B.; Tedder, T.F.; Hardt, W.-D.; et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science 2009, 325, 617–620. [Google Scholar] [CrossRef] [Green Version]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef]
- Frank, D.N.; Amand, A.L.S.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [Green Version]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [Green Version]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef] [Green Version]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Gevers, D.; Siljander, H.; Vatanen, T.; Hyötyläinen, T.; Hämäläinen, A.-M.; Peet, A.; Tillmann, V.; Pöhö, P.; Mattila, I.; et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 2015, 17, 260–273. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Sartor, R.B. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: Antibiotics, probiotics, and prebiotics. Gastroenterology 2004, 126, 1620–1633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sellon, R.K.; Tonkonogy, S.; Schultz, M.; Dieleman, L.A.; Grenther, W.; Balish, E.; Rennick, D.M.; Sartor, R.B. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immun. 1998, 66, 5224–5231. [Google Scholar] [CrossRef] [Green Version]
- Buttó, L.F.; Haller, D. Dysbiosis in intestinal inflammation: Cause or consequence. Int. J. Med. Microbiol. 2016, 306, 302–309. [Google Scholar] [CrossRef]
- Schaubeck, M.; Clavel, T.; Calasan, J.; Lagkouvardos, I.; Haange, S.B.; Jehmlich, N.; Basic, M.; Dupont, A.; Hornef, M.; von Bergen, M.; et al. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut 2016, 65, 225–237. [Google Scholar] [CrossRef] [Green Version]
- Baumgart, M.; Dogan, B.; Rishniw, M.; Weitzman, G.; Bosworth, B.; Yantiss, R.; Orsi, R.H.; Wiedmann, M.; McDonough, P.; Kim, S.G.; et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007, 1, 403–418. [Google Scholar] [CrossRef] [Green Version]
- Darfeuille-Michaud, A. Adherent-invasive Escherichia coli: A putative new E. coli pathotype associated with Crohn’s disease. Int. J. Med. Microbiol. 2002, 292, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Pascal, V.; Pozuelo, M.; Borruel, N.; Casellas, F.; Campos, D.; Santiago, A.; Martinez, X.; Varela, E.; Sarrabayrouse, G.; Machiels, K.; et al. A microbial signature for Crohn’s disease. Gut 2017, 66, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.-P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Nishida, A.; Fujimoto, T.; Fujii, M.; Shioya, M.; Imaeda, H.; Inatomi, O.; Bamba, S.; Andoh, A.; Sugimoto, M. Reduced Abundance of Butyrate-Producing Bacteria Species in the Fecal Microbial Community in Crohn’s Disease. DIG 2016, 93, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Siles, M.; Martinez-Medina, M.; Abellà, C.; Busquets, D.; Sabat-Mir, M.; Duncan, S.H.; Aldeguer, X.; Flint, H.J.; Garcia-Gil, L.J. Mucosa-associated Faecalibacterium prausnitzii phylotype richness is reduced in patients with inflammatory bowel disease. Appl. Environ. Microbiol. 2015, 81, 7582–7592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokol, H.; Seksik, P.; Furet, J.P.; Firmesse, O.; Nion-Larmurier, I.; Beaugerie, L.; Cosnes, J.; Corthier, G.; Marteau, P.; Doré, J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 2009, 15, 1183–1189. [Google Scholar] [CrossRef]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef]
- Dave, M.; Purohit, T.; Razonable, R.; Loftus, E.V. Opportunistic infections due to inflammatory bowel disease therapy. Inflamm. Bowel Dis. 2014, 20, 196–212. [Google Scholar] [CrossRef]
- Chan, H.C.; Ng, S.C. Emerging biologics in inflammatory bowel disease. J. Gastroenterol. 2017, 52, 141–150. [Google Scholar] [CrossRef]
- Lewis, J.D.; Chen, E.Z.; Baldassano, R.N.; Otley, A.R.; Griffiths, A.M.; Lee, D.; Bittinger, K.; Bailey, A.; Friedman, E.S.; Hoffmann, C.; et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe 2017, 22, 247. [Google Scholar] [CrossRef]
- Ribaldone, D.G.; Caviglia, G.P.; Abdulle, A.; Pellicano, R.; Ditto, M.C.; Morino, M.; Fusaro, E.; Saracco, G.M.; Bugianesi, E.; Astegiano, M. Adalimumab Therapy Improves Intestinal Dysbiosis in Crohn’s Disease. J. Clin. Med. 2019, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, H.; Fu, L.; Wang, J. Protocol for Fecal Microbiota Transplantation in Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis. Biomed. Res. Int. 2018, 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Hage, R.; Hernandez-Sanabria, E.; Van de Wiele, T. Emerging Trends in “Smart Probiotics”: Functional Consideration for the Development of Novel Health and Industrial Applications. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [Green Version]
- Parvez, S.; Malik, K.A.; Kang, S.; Kim, H.-Y. Probiotics and their fermented food products are beneficial for health. J. Appl. Microbiol. 2006, 100, 1171–1185. [Google Scholar] [CrossRef]
- Foligne, B.; Zoumpopoulou, G.; Dewulf, J.; Ben Younes, A.; Chareyre, F.; Sirard, J.-C.; Pot, B.; Grangette, C. A key role of dendritic cells in probiotic functionality. PLoS ONE 2007, 2, e313. [Google Scholar] [CrossRef]
- Macho Fernandez, E.; Fernandez, E.M.; Valenti, V.; Rockel, C.; Hermann, C.; Pot, B.; Boneca, I.G.; Grangette, C. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut 2011, 60, 1050–1059. [Google Scholar] [CrossRef]
- Alard, J.; Peucelle, V.; Boutillier, D.; Breton, J.; Kuylle, S.; Pot, B.; Holowacz, S.; Grangette, C. New probiotic strains for inflammatory bowel disease management identified by combining in vitro and in vivo approaches. Benef. Microbes 2018, 9, 317–331. [Google Scholar] [CrossRef]
- Zaylaa, M.; Al Kassaa, I.; Alard, J.; Peucelle, V.; Boutillier, D.; Desramaut, J.; Dabboussi, F.; Pot, B.; Grangette, C. Probiotics in IBD: Combining in vitro and in vivo models for selecting strains with both anti-inflammatory potential as well as a capacity to restore the gut epithelial barrier. J. Funct. Foods 2018, 47, 304–315. [Google Scholar] [CrossRef]
- Ghouri, Y.A.; Richards, D.M.; Rahimi, E.F.; Krill, J.T.; Jelinek, K.A.; DuPont, A.W. Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease. Clin. Exp. Gastroenterol. 2014, 7, 473–487. [Google Scholar] [CrossRef] [Green Version]
- Saez-Lara, M.J.; Gomez-Llorente, C.; Plaza-Diaz, J.; Gil, A. The role of probiotic lactic acid bacteria and bifidobacteria in the prevention and treatment of inflammatory bowel disease and other related diseases: A systematic review of randomized human clinical trials. Biomed. Res. Int. 2015, 2015, 505878. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Langella, P. Emerging Health Concepts in the Probiotics Field: Streamlining the Definitions. Front. Microbiol. 2019, 10, 1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shetty, S.A.; Zuffa, S.; Bui, T.P.N.; Aalvink, S.; Smidt, H.; De Vos, W.M. Reclassification of Eubacterium hallii as Anaerobutyricum hallii gen. nov., comb. nov., and description of Anaerobutyricum soehngenii sp. nov., a butyrate and propionate-producing bacterium from infant faeces. Int. J. Syst. Evol. Microbiol. 2018, 68, 3741–3746. [Google Scholar] [CrossRef]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Bäckhed, F.; Mithieux, G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Udayappan, S.; Manneras-Holm, L.; Chaplin-Scott, A.; Belzer, C.; Herrema, H.; Dallinga-Thie, G.M.; Duncan, S.H.; Stroes, E.S.G.; Groen, A.K.; Flint, H.J.; et al. Oral treatment with Eubacterium hallii improves insulin sensitivity in db/db mice. NPJ Biofilms Microbiomes 2016, 2, 16009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.-R.; Lin, C.-S.; Chang, C.-J.; Lin, T.-L.; Martel, J.; Ko, Y.-F.; Ojcius, D.M.; Lu, C.-C.; Young, J.D.; Lai, H.-C. Gut commensal Parabacteroides goldsteinii plays a predominant role in the anti-obesity effects of polysaccharides isolated from Hirsutella sinensis. Gut 2019, 68, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.-J.; Lin, T.-L.; Tsai, Y.-L.; Wu, T.-R.; Lai, W.-F.; Lu, C.-C.; Lai, H.-C. Next generation probiotics in disease amelioration. J. Food Drug Anal. 2019, 27, 615–622. [Google Scholar] [CrossRef]
- Foligne, B.; Nutten, S.; Grangette, C.; Dennin, V.; Goudercourt, D.; Poiret, S.; Dewulf, J.; Brassart, D.; Mercenier, A.; Pot, B. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J. Gastroenterol. 2007, 13, 236–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burns, P.; Alard, J.; Hrdỳ, J.; Boutillier, D.; Páez, R.; Reinheimer, J.; Pot, B.; Vinderola, G.; Grangette, C. Spray-drying process preserves the protective capacity of a breast milk-derived Bifidobacterium lactis strain on acute and chronic colitis in mice. Sci. Rep. 2017, 7, 43211. [Google Scholar] [CrossRef] [PubMed]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in vitro digestion method suitable for food—An international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chantret, I.; Rodolosse, A.; Barbat, A.; Dussaulx, E.; Brot-Laroche, E.; Zweibaum, A.; Rousset, M. Differential expression of sucrase-isomaltase in clones isolated from early and late passages of the cell line Caco-2: Evidence for glucose-dependent negative regulation. J. Cell. Sci. 1994, 107 Pt 1, 213–225. [Google Scholar]
- Foligné, B.; Nutten, S.; Steidler, L.; Dennin, V.; Goudercourt, D.; Mercenier, A.; Pot, B. Recommendations for improved use of the murine TNBS-induced colitis model in evaluating anti-inflammatory properties of lactic acid bacteria: Technical and microbiological aspects. Dig. Dis. Sci. 2006, 51, 390–400. [Google Scholar] [CrossRef]
- Wallace, J.L.; MacNaughton, W.K.; Morris, G.P.; Beck, P.L. Inhibition of leukotriene synthesis markedly accelerates healing in a rat model of inflammatory bowel disease. Gastroenterology 1989, 96, 29–36. [Google Scholar] [CrossRef]
- Ameho, C.K.; Adjei, A.A.; Harrison, E.K.; Takeshita, K.; Morioka, T.; Arakaki, Y.; Ito, E.; Suzuki, I.; Kulkarni, A.D.; Kawajiri, A.; et al. Prophylactic effect of dietary glutamine supplementation on interleukin 8 and tumour necrosis factor alpha production in trinitrobenzene sulphonic acid induced colitis. Gut 1997, 41, 487–493. [Google Scholar] [CrossRef] [Green Version]
- Chassaing, B.; Srinivasan, G.; Delgado, M.A.; Young, A.N.; Gewirtz, A.T.; Vijay-Kumar, M. Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS ONE 2012, 7, e44328. [Google Scholar] [CrossRef] [Green Version]
- Dziarski, R.; Park, S.Y.; Kashyap, D.R.; Dowd, S.E.; Gupta, D. Pglyrp-Regulated Gut Microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii Enhance and Alistipes finegoldii Attenuates Colitis in Mice. PLoS ONE 2016, 11, e0146162. [Google Scholar] [CrossRef]
- Majer, M.; Macháček, T.; Súkeníková, L.; Hrdý, J.; Horák, P. The peripheral immune response of mice infected with a neuropathogenic schistosome. Parasite Immunol. 2020, e12710. [Google Scholar] [CrossRef]
- Hrdý, J.; Alard, J.; Couturier-Maillard, A.; Boulard, O.; Boutillier, D.; Delacre, M.; Lapadatescu, C.; Cesaro, A.; Blanc, P.; Pot, B.; et al. Lactobacillus reuteri 5454 and Bifidobacterium animalis ssp. lactis 5764 improve colitis while differentially impacting dendritic cells maturation and antimicrobial responses. Sci. Rep. 2020, 10, 5345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hrdý, J.; Zanvit, P.; Novotná, O.; Kocourková, I.; Zižka, J.; Prokešová, L. Cytokine expression in cord blood cells of children of healthy and allergic mothers. Folia Microbiol. 2010, 55, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Seth, A.; Yan, F.; Polk, D.B.; Rao, R.K. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 294, G1060–G1069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luettig, J.; Rosenthal, R.; Barmeyer, C.; Schulzke, J.D. Claudin-2 as a mediator of leaky gut barrier during intestinal inflammation. Tissue Barriers 2015, 3, e977176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeissig, S.; Bürgel, N.; Günzel, D.; Richter, J.; Mankertz, J.; Wahnschaffe, U.; Kroesen, A.J.; Zeitz, M.; Fromm, M.; Schulzke, J.-D. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn’s disease. Gut 2007, 56, 61–72. [Google Scholar] [CrossRef]
- Faith, J.J.; Ahern, P.P.; Ridaura, V.K.; Cheng, J.; Gordon, J.I. Identifying Gut Microbe-Host Phenotype Relationships Using Combinatorial Communities in Gnotobiotic Mice. Sci. Transl. Med. 2014, 6, 220ra11. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, R.; Sorrell, M.F.; Batra, S.K.; Dhawan, P.; Singh, A.B. Gut permeability and mucosal inflammation: Bad, good or context dependent. Mucosal Immunol. 2017, 10, 307–317. [Google Scholar] [CrossRef] [Green Version]
- Walker, A.W.; Sanderson, J.D.; Churcher, C.; Parkes, G.C.; Hudspith, B.N.; Rayment, N.; Brostoff, J.; Parkhill, J.; Dougan, G.; Petrovska, L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011, 11, 7. [Google Scholar] [CrossRef] [Green Version]
- Papa, E.; Docktor, M.; Smillie, C.; Weber, S.; Preheim, S.P.; Gevers, D.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Ingram, J.; et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS ONE 2012, 7, e39242. [Google Scholar] [CrossRef] [Green Version]
- Glocker, E.-O.; Kotlarz, D.; Klein, C.; Shah, N.; Grimbacher, B. IL-10 and IL-10 receptor defects in humans. Ann. N. Y. Acad. Sci. 2011, 1246, 102–107. [Google Scholar] [CrossRef]
- Mühl, H. Pro-Inflammatory Signaling by IL-10 and IL-22: Bad Habit Stirred Up by Interferons? Front. Immunol. 2013, 4, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, W.; Rutz, S.; Crellin, N.K.; Valdez, P.A.; Hymowitz, S.G. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol. 2011, 29, 71–109. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.-Z.; et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell 2018, 174, 1388–1405.e21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viennois, E.; Gewirtz, A.T.; Chassaing, B. Chronic Inflammatory Diseases: Are We Ready for Microbiota-based Dietary Intervention? Cell. Mol. Gastroenterol. Hepatol. 2019, 8, 61–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liew, F.Y.; Girard, J.-P.; Turnquist, H.R. Interleukin-33 in health and disease. Nat. Rev. Immunol. 2016, 16, 676–689. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, Y.; Wang, Q.; Li, C.; Deng, H.; Si, C.; Xiong, H. Interleukin-35 in immune-related diseases: Protection or destruction. Immunology 2019, 157, 13–20. [Google Scholar] [CrossRef] [Green Version]
- Xiang, X.G.; Xie, Q. IL-35: A potential therapeutic target for controlling hepatitis B virus infection. J. Dig. Dis. 2015, 16, 1–6. [Google Scholar] [CrossRef]
- Choi, J.; Leung, P.S.C.; Bowlus, C.; Gershwin, M.E. IL-35 and Autoimmunity: A Comprehensive Perspective. Clin. Rev. Allerg. Immunol. 2015, 49, 327–332. [Google Scholar] [CrossRef]
- Verdù, E.F.; Bercik, P.; Cukrowska, B.; Farre-Castany, M.A.; Bouzourene, H.; Saraga, E.; Blum, A.L.; Corthésy-Theulaz, I.; Tlaskalova-Hogenova, H.; Michetti, P. Oral administration of antigens from intestinal flora anaerobic bacteria reduces the severity of experimental acute colitis in BALB/c mice. Clin. Exp. Immunol. 2000, 120, 46–50. [Google Scholar] [CrossRef]
- Kverka, M.; Zakostelska, Z.; Klimesova, K.; Sokol, D.; Hudcovic, T.; Hrncir, T.; Rossmann, P.; Mrazek, J.; Kopecny, J.; Verdu, E.F.; et al. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin. Exp. Immunol. 2011, 163, 250–259. [Google Scholar] [CrossRef]
- Pfalzer, A.C.; Nesbeth, P.-D.C.; Parnell, L.D.; Iyer, L.K.; Liu, Z.; Kane, A.V.; Chen, C.-Y.O.; Tai, A.K.; Bowman, T.A.; Obin, M.S.; et al. Diet- and Genetically-Induced Obesity Differentially Affect the Fecal Microbiome and Metabolome in Apc1638N Mice. PLoS ONE 2015, 10, e0135758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, G.Y.; Kane, A.; Lee, K.; Xu, Q.; Wu, X.; Roper, J.; Mason, J.B.; Crott, J.W. Parabacteroides distasonis attenuates toll-like receptor 4 signaling and Akt activation and blocks colon tumor formation in high-fat diet-fed azoxymethane-treated mice. Int. J. Cancer 2018, 143, 1797–1805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cekanaviciute, E.; Yoo, B.B.; Runia, T.F.; Debelius, J.W.; Singh, S.; Nelson, C.A.; Kanner, R.; Bencosme, Y.; Lee, Y.K.; Hauser, S.L.; et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. USA 2017, 114, 10713–10718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geuking, M.B.; Cahenzli, J.; Lawson, M.A.E.; Ng, D.C.K.; Slack, E.; Hapfelmeier, S.; McCoy, K.D.; Macpherson, A.J. Intestinal Bacterial Colonization Induces Mutualistic Regulatory T Cell Responses. Immunity 2011, 34, 794–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, S.; Busetti, A.; Fotiadou, P.; Vincy Jose, N.; Reid, S.; Georgieva, M.; Brown, S.; Dunbar, H.; Beurket-Ascencio, G.; Delday, M.I.; et al. In vitro Characterization of Gut Microbiota-Derived Bacterial Strains With Neuroprotective Properties. Front. Cell. Neurosci. 2019, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, F.; Kumar, A.; Davenport, K.W.; Kelliher, J.M.; Ezeji, J.C.; Good, C.E.; Jacobs, M.R.; Conger, M.; West, G.; Fiocchi, C.; et al. Complete Genome Sequence of a Parabacteroides distasonis Strain (CavFT hAR46) Isolated from a Gut Wall-Cavitating Microlesion in a Patient with Severe Crohn’s Disease. Microbiol. Resour. Announc. 2019, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fouhy, F.; Watkins, C.; Hill, C.J.; O’Shea, C.-A.; Nagle, B.; Dempsey, E.M.; O’Toole, P.W.; Ross, R.P.; Ryan, C.A.; Stanton, C. Perinatal factors affect the gut microbiota up to four years after birth. Nat. Commun. 2019, 10, 1517. [Google Scholar] [CrossRef] [Green Version]


| Strains Designation | Species | Growth Medium | Origin |
|---|---|---|---|
| AS93 | Parabacteroides distasonis | BHIS | Healthy adult feces |
| AS23 | Parabacteroides distasonis | BHIS | Healthy adult feces |
| PF-BaE5 | Parabacteroides distasonis | BHIS | Newborn fecal samples |
| PF-BaE7 | Parabacteroides distasonis | BHIS | Newborn fecal samples |
| PF-BaE11 | Parabacteroides distasonis | BHIS | Newborn fecal samples |
| Strains | Improvement of Epithelial Barrier | IL-10 Induction | Anti-Inflammatory Profile (Il-10/IL-12) | Tolerance to Gastric Conditions (120 min) | In Vivo Protective Capacity |
|---|---|---|---|---|---|
| P. distasonis AS23 | ± | +++ | + | ++ | − |
| P. distasonis AS93 | +++ | +++ | + | ± | ++ |
| P. distasonis PF-BaE5 | ++ | +++ | ++ | ± | +++ |
| P. distasonis PF-BaE7 | ± | +++ | + | ++ | − |
| P. distasonis PF-BaE11 | +++ | +++ | ++ | ± | +++ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuffaro, B.; Assohoun, A.L.W.; Boutillier, D.; Súkeníková, L.; Desramaut, J.; Boudebbouze, S.; Salomé-Desnoulez, S.; Hrdý, J.; Waligora-Dupriet, A.-J.; Maguin, E.; et al. In Vitro Characterization of Gut Microbiota-Derived Commensal Strains: Selection of Parabacteroides distasonis Strains Alleviating TNBS-Induced Colitis in Mice. Cells 2020, 9, 2104. https://doi.org/10.3390/cells9092104
Cuffaro B, Assohoun ALW, Boutillier D, Súkeníková L, Desramaut J, Boudebbouze S, Salomé-Desnoulez S, Hrdý J, Waligora-Dupriet A-J, Maguin E, et al. In Vitro Characterization of Gut Microbiota-Derived Commensal Strains: Selection of Parabacteroides distasonis Strains Alleviating TNBS-Induced Colitis in Mice. Cells. 2020; 9(9):2104. https://doi.org/10.3390/cells9092104
Chicago/Turabian StyleCuffaro, Bernardo, Aka L. W. Assohoun, Denise Boutillier, Lenka Súkeníková, Jérémy Desramaut, Samira Boudebbouze, Sophie Salomé-Desnoulez, Jiří Hrdý, Anne-Judith Waligora-Dupriet, Emmanuelle Maguin, and et al. 2020. "In Vitro Characterization of Gut Microbiota-Derived Commensal Strains: Selection of Parabacteroides distasonis Strains Alleviating TNBS-Induced Colitis in Mice" Cells 9, no. 9: 2104. https://doi.org/10.3390/cells9092104