How Relevant Are Bone Marrow-Derived Mast Cells (BMMCs) as Models for Tissue Mast Cells? A Comparative Transcriptome Analysis of BMMCs and Peritoneal Mast Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Generation of Bone Marrow-Derived MCs and LPS Stimulation
2.3. Peritoneal Cell Extraction and MACS Sorting of Peritoneal Mast Cells
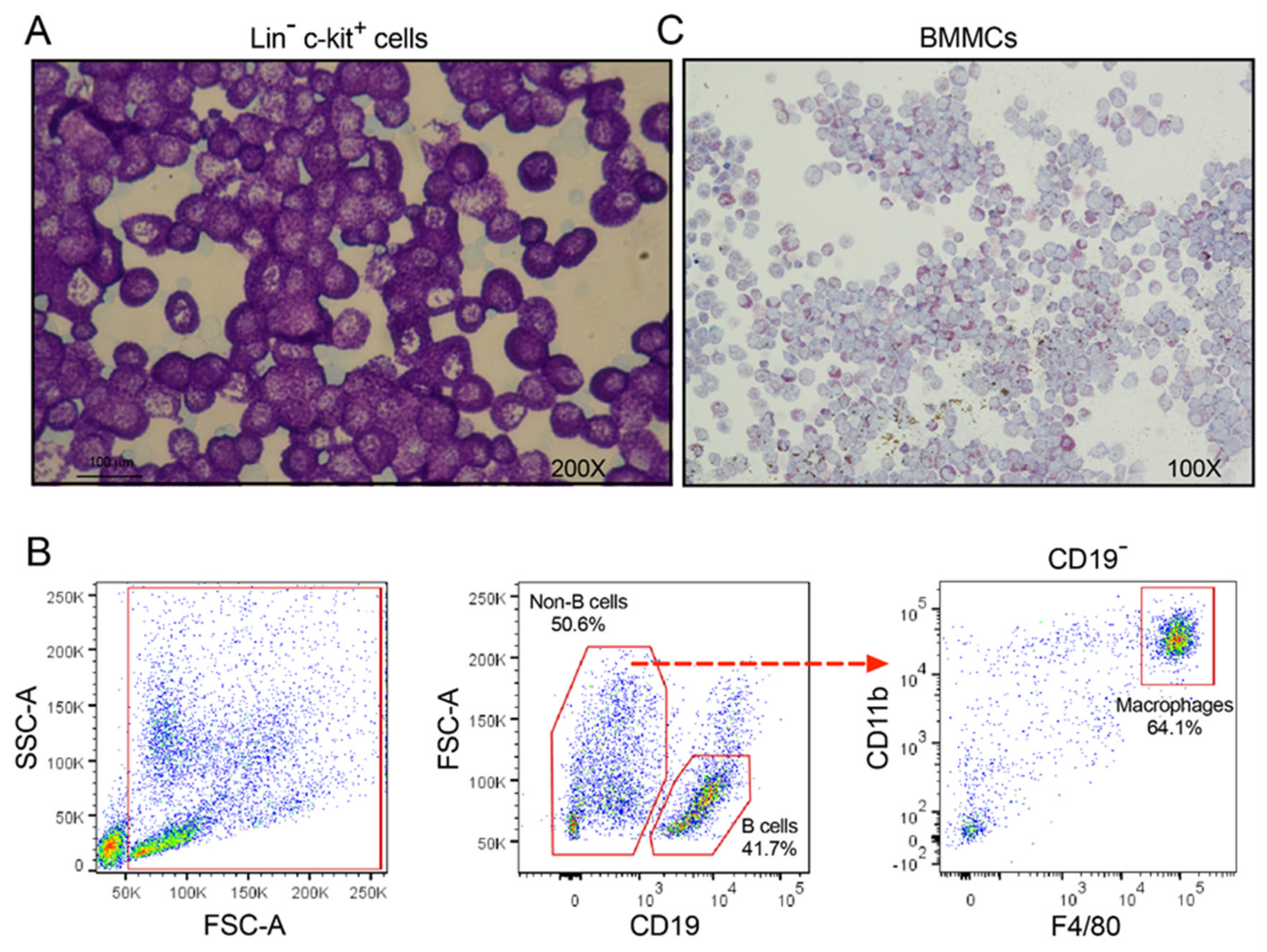
2.4. FACS Sorting of Peritoneal Macrophages and B Cells
2.5. Image Analysis
2.6. RNA Isolation from Purified Cell Fractions
2.7. Analysis of the Transcriptome by the Thermo Fisher Ampliseq PCR Based Method
3. Results
3.1. Preparation of RNA from Purified Peritoneal Cell Fractions
3.2. A Comparative Analysis of Transcript Levels in Peritoneal MCs and BMMCs
3.3. Transcripts That Were High in Peritoneal Mast Cells but Low or Almost Absent in BMMCs
3.4. Transcripts That Were High in Both Peritoneal Cells and BMMCs, but Low in Macrophages and/or B Cells
3.5. Transcripts That Were High in BMMCs but Low or Almost Absent in Peritoneal MCs
3.6. Transcripts That Were Highly Upregulated in BMMCs after Induction by Lipopolysaccharide
4. Discussion
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| MC | mast cell |
| Mcpt | mast cell protease |
References
- Galli, S.J.; Starkl, P.; Marichal, T.; Tsai, M. Mast cells and IgE in defense against venoms: Possible “good side” of allergy? Allergol. Int. 2016, 65, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Hellman, L.T.; Akula, S.; Thorpe, M.; Fu, Z. Tracing the Origins of IgE, Mast Cells, and Allergies by Studies of Wild Animals. Front. Immunol. 2017, 8, 1749. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, M.C.; de Andrade, L.R.; Du Bocage Santos-Pinto, C.; Straus, A.H.; Takahashi, H.K.; Allodi, S.; Pavao, M.S. Colocalization of heparin and histamine in the intracellular granules of test cells from the invertebrate Styela plicata (Chordata-Tunicata). J. Struct. Biol. 2002, 137, 313–321. [Google Scholar] [CrossRef]
- Wong, G.W.; Zhuo, L.; Kimata, K.; Lam, B.K.; Satoh, N.; Stevens, R.L. Ancient origin of mast cells. Biochem. Biophys. Res. Commun. 2014, 451, 314–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akula, S.; Paivandy, A.; Fu, Z.; Thorpe, M.; Pejler, G.; Hellman, L. Quantitative In-Depth Analysis of the Mouse Mast Cell Transcriptome Reveals Organ-Specific Mast Cell Heterogeneity. Cells 2020, 9, 211. [Google Scholar] [CrossRef] [Green Version]
- Abe, T.; Swieter, M.; Imai, T.; Hollander, N.D.; Befus, A.D. Mast cell heterogeneity: Two-dimensional gel electrophoretic analyses of rat peritoneal and intestinal mucosal mast cells. Eur. J. Immunol. 1990, 20, 1941–1947. [Google Scholar] [CrossRef]
- Reynolds, D.S.; Stevens, R.L.; Lane, W.S.; Carr, M.H.; Austen, K.F.; Serafin, W.E. Different mouse mast cell populations express various combinations of at least six distinct mast cell serine proteases. Proc. Natl. Acad. Sci. USA 1990, 87, 3230–3234. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.Y.; Blom, T.; Hellman, L. Cloning and structural analysis of MMCP-1, MMCP-4 and MMCP-5, three mouse mast cell-specific serine proteases. Eur. J. Immunol. 1991, 21, 1611–1621. [Google Scholar] [CrossRef]
- Lutzelschwab, C.; Pejler, G.; Aveskogh, M.; Hellman, L. Secretory granule proteases in rat mast cells. Cloning of 10 different serine proteases and a carboxypeptidase A from various rat mast cell populations. J. Exp. Med. 1997, 185, 13–29. [Google Scholar] [CrossRef] [Green Version]
- Lutzelschwab, C.; Huang, M.R.; Kullberg, M.C.; Aveskogh, M.; Hellman, L. Characterization of mouse mast cell protease-8, the first member of a novel subfamily of mouse mast cell serine proteases, distinct from both the classical chymases and tryptases. Eur. J. Immunol. 1998, 28, 1022–1033. [Google Scholar] [CrossRef]
- Lutzelschwab, C.; Lunderius, C.; Enerback, L.; Hellman, L. A kinetic analysis of the expression of mast cell protease mRNA in the intestines of Nippostrongylus brasiliensis-infected rats. Eur. J. Immunol. 1998, 28, 3730–3737. [Google Scholar] [CrossRef]
- Pejler, G.; Ronnberg, E.; Waern, I.; Wernersson, S. Mast cell proteases: Multifaceted regulators of inflammatory disease. Blood 2010, 115, 4981–4990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caughey, G.H. Mast cell proteases as protective and inflammatory mediators. Adv. Exp. Med. Biol. 2011, 716, 212–234. [Google Scholar] [CrossRef] [PubMed]
- Hellman, L.; Thorpe, M. Granule proteases of hematopoietic cells, a family of versatile inflammatory mediators-an update on their cleavage specificity, in vivo substrates, and evolution. Biol. Chem. 2014, 395, 15–49. [Google Scholar] [CrossRef]
- Akula, S.; Thorpe, M.; Boinapally, V.; Hellman, L. Granule Associated Serine Proteases of Hematopoietic Cells—An Analysis of Their Appearance and Diversification during Vertebrate Evolution. PLoS ONE 2015, 10, e0143091. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, L.B.; Irani, A.M.; Roller, K.; Castells, M.C.; Schechter, N.M. Quantitation of histamine, tryptase, and chymase in dispersed human T and TC mast cells. J. Immunol. 1987, 138, 2611–2615. [Google Scholar] [PubMed]
- Metz, M.; Piliponsky, A.M.; Chen, C.C.; Lammel, V.; Abrink, M.; Pejler, G.; Tsai, M.; Galli, S.J. Mast cells can enhance resistance to snake and honeybee venoms. Science 2006, 313, 526–530. [Google Scholar] [CrossRef] [Green Version]
- Akahoshi, M.; Song, C.H.; Piliponsky, A.M.; Metz, M.; Guzzetta, A.; Abrink, M.; Schlenner, S.M.; Feyerabend, T.B.; Rodewald, H.R.; Pejler, G.; et al. Mast cell chymase reduces the toxicity of Gila monster venom, scorpion venom, and vasoactive intestinal polypeptide in mice. J. Clin. Investig. 2011, 121, 4180–4191. [Google Scholar] [CrossRef] [Green Version]
- Piliponsky, A.M.; Chen, C.C.; Rios, E.J.; Treuting, P.M.; Lahiri, A.; Abrink, M.; Pejler, G.; Tsai, M.; Galli, S.J. The Chymase Mouse Mast Cell Protease 4 Degrades TNF, Limits Inflammation, and Promotes Survival in a Model of Sepsis. Am. J. Pathol. 2012, 181, 875–886. [Google Scholar] [CrossRef] [Green Version]
- Fu, Z.; Thorpe, M.; Alemayehu, R.; Roy, A.; Kervinen, J.; de Garavilla, L.; Abrink, M.; Hellman, L. Highly Selective Cleavage of Cytokines and Chemokines by the Human Mast Cell Chymase and Neutrophil Cathepsin, G. J. Immunol. 2017, 198, 1474–1483. [Google Scholar] [CrossRef] [Green Version]
- Fu, Z.; Akula, S.; Thorpe, M.; Hellman, L. Highly Selective Cleavage of TH2-Promoting Cytokines by the Human and the Mouse Mast Cell Tryptases, Indicating a Potent Negative Feedback Loop on TH2 Immunity. Int. J. Mol. Sci. 2019, 20, 5147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tchougounova, E.; Forsberg, E.; Angelborg, G.; Kjellen, L.; Pejler, G. Altered processing of fibronectin in mice lacking heparin. a role for heparin-dependent mast cell chymase in fibronectin degradation. J. Biol. Chem. 2001, 276, 3772–3777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tchougounova, E.; Pejler, G. Regulation of extravascular coagulation and fibrinolysis by heparin-dependent mast cell chymase. FASEB J. 2001, 15, 2763–2765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kjellen, L.; Pettersson, I.; Lillhager, P.; Steen, M.L.; Pettersson, U.; Lehtonen, P.; Karlsson, T.; Ruoslahti, E.; Hellman, L. Primary structure of a mouse mastocytoma proteoglycan core protein. Biochem. J. 1989, 263, 105–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angerth, T.; Huang, R.Y.; Aveskogh, M.; Pettersson, I.; Kjellen, L.; Hellman, L. Cloning and structural analysis of a gene encoding a mouse mastocytoma proteoglycan core protein; analysis of its evolutionary relation to three cross hybridizing regions in the mouse genome. Gene 1990, 93, 235–240. [Google Scholar] [CrossRef]
- Ronnberg, E.; Melo, F.R.; Pejler, G. Mast cell proteoglycans. J. Histochem. Cytochem. 2012, 60, 950–962. [Google Scholar] [CrossRef] [Green Version]
- Ohtsu, H.; Tanaka, S.; Terui, T.; Hori, Y.; Makabe-Kobayashi, Y.; Pejler, G.; Tchougounova, E.; Hellman, L.; Gertsenstein, M.; Hirasawa, N.; et al. Mice lacking histidine decarboxylase exhibit abnormal mast cells. FEBS Lett. 2001, 502, 53–56. [Google Scholar] [CrossRef] [Green Version]
- Akula, S.; Mohammadamin, S.; Hellman, L. Fc receptors for immunoglobulins and their appearance during vertebrate evolution. PLoS ONE 2014, 9, e96903. [Google Scholar] [CrossRef] [Green Version]
- Galli, S.J.; Zsebo, K.M.; Geissler, E.N. The kit ligand, stem cell factor. Adv. Immunol. 1994, 55, 1–96. [Google Scholar] [CrossRef]
- Motakis, E.; Guhl, S.; Ishizu, Y.; Itoh, M.; Kawaji, H.; de Hoon, M.; Lassmann, T.; Carninci, P.; Hayashizaki, Y.; Zuberbier, T.; et al. Redefinition of the human mast cell transcriptome by deep-CAGE sequencing. Blood 2014, 123, e58–e67. [Google Scholar] [CrossRef]
- Meurer, S.K.; Ness, M.; Weiskirchen, S.; Kim, P.; Tag, C.G.; Kauffmann, M.; Huber, M.; Weiskirchen, R. Isolation of Mature (Peritoneum-Derived) Mast Cells and Immature (Bone Marrow-Derived) Mast Cell Precursors from Mice. PLoS ONE 2016, 11, e0158104. [Google Scholar] [CrossRef] [PubMed]
- Nabel, G.; Galli, S.J.; Dvorak, A.M.; Dvorak, H.F.; Cantor, H. Inducer T lymphocytes synthesize a factor that stimulates proliferation of cloned mast cells. Nature 1981, 291, 332–334. [Google Scholar] [CrossRef] [PubMed]
- Razin, E.; Cordon-Cardo, C.; Good, R.A. Growth of a pure population of mouse mast cells in vitro with conditioned medium derived from concanavalin A-stimulated splenocytes. Proc. Natl. Acad. Sci. USA 1981, 78, 2559–2561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levi-Schaffer, F.; Austen, K.F.; Gravallese, P.M.; Stevens, R.L. Coculture of interleukin 3-dependent mouse mast cells with fibroblasts results in a phenotypic change of the mast cells. Proc. Natl. Acad. Sci. USA 1986, 83, 6485–6488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosn, E.E.; Cassado, A.A.; Govoni, G.R.; Fukuhara, T.; Yang, Y.; Monack, D.M.; Bortoluci, K.R.; Almeida, S.R.; Herzenberg, L.A.; Herzenberg, L.A. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc. Natl. Acad. Sci. USA 2010, 107, 2568–2573. [Google Scholar] [CrossRef] [Green Version]
- Lunderius, C.; Xiang, Z.; Nilsson, G.; Hellman, L. Murine mast cell lines as indicators of early events in mast cell and basophil development. Eur. J. Immunol. 2000, 30, 3396–3402. [Google Scholar] [CrossRef]
- Forsberg, E.; Pejler, G.; Ringvall, M.; Lunderius, C.; Tomasini-Johansson, B.; Kusche-Gullberg, M.; Eriksson, I.; Ledin, J.; Hellman, L.; Kjellen, L. Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature 1999, 400, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Abrink, M.; Larsson, E.; Hellman, L. Demethylation of ERV3, an endogenous retrovirus regulating the Kruppel-related zinc finger gene H-plk, in several human cell lines arrested during early monocyte development. DNA Cell Biol. 1998, 17, 27–37. [Google Scholar] [CrossRef]
- Williams, R.L.; Hilton, D.J.; Pease, S.; Willson, T.A.; Stewart, C.L.; Gearing, D.P.; Wagner, E.F.; Metcalf, D.; Nicola, N.A.; Gough, N.M. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 1988, 336, 684–687. [Google Scholar] [CrossRef]
- Onishi, K.; Zandstra, P.W. LIF signaling in stem cells and development. Development 2015, 142, 2230–2236. [Google Scholar] [CrossRef] [Green Version]
- Smedsrod, B.; Pertoft, H. Preparation of pure hepatocytes and reticuloendothelial cells in high yield from a single rat liver by means of Percoll centrifugation and selective adherence. J. Leukoc. Biol. 1985, 38, 213–230. [Google Scholar] [CrossRef]
- Hellman, L.; Smedsrod, B.; Sandberg, H.; Pettersson, U. Secretion of coagulant factor VIII activity and antigen by in vitro cultivated rat liver sinusoidal endothelial cells. Br. J. Haematol. 1989, 73, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Abdian, N.; Ghasemi-Dehkordi, P.; Hashemzadeh-Chaleshtori, M.; Ganji-Arjenaki, M.; Doosti, A.; Amiri, B. Comparison of human dermal fibroblasts (HDFs) growth rate in culture media supplemented with or without basic fibroblast growth factor (bFGF). Cell Tissue Bank 2015, 16, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Kamat, R.; Henney, C.S. Studies on T cell clonal expansion. II. The in vitro differentiation of pre-killer and memory T cells. J. Immunol. 1976, 116, 1490–1495. [Google Scholar] [PubMed]
- Miller, H.R.; Wright, S.H.; Knight, P.A.; Thornton, E.M. A novel function for transforming growth factor-beta1: Upregulation of the expression and the IgE-independent extracellular release of a mucosal mast cell granule-specific beta-chymase, mouse mast cell protease-1. Blood 1999, 93, 3473–3486. [Google Scholar] [CrossRef]
- Kakinoki, A.; Kameo, T.; Yamashita, S.; Furuta, K.; Tanaka, S. Establishment and Characterization of a Murine Mucosal Mast Cell Culture Model. Int. J. Mol. Sci. 2019, 21, 236. [Google Scholar] [CrossRef] [Green Version]
- Plaut, M.; Pierce, J.H.; Watson, C.J.; Hanley-Hyde, J.; Nordan, R.P.; Paul, W.E. Mast cell lines produce lymphokines in response to cross-linkage of Fc epsilon RI or to calcium ionophores. Nature 1989, 339, 64–67. [Google Scholar] [CrossRef]
- Mukai, K.; Tsai, M.; Saito, H.; Galli, S.J. Mast cells as sources of cytokines, chemokines, and growth factors. Immunol. Rev. 2018, 282, 121–150. [Google Scholar] [CrossRef]
- Denburg, J.A.; Telizyn, S.; Messner, H.; Lim, B.; Jamal, N.; Ackerman, S.J.; Gleich, G.J.; Bienenstock, J. Heterogeneity of human peripheral blood eosinophil-type colonies: Evidence for a common basophil-eosinophil progenitor. Blood 1985, 66, 312–318. [Google Scholar] [CrossRef] [Green Version]
- Reimer, J.M.; Magnusson, S.; Juremalm, M.; Nilsson, G.; Hellman, L.; Wernersson, S. Isolation of transcriptionally active umbilical cord blood-derived basophils expressing FcepsilonRI, HLA-DR and CD203c. Allergy 2006, 61, 1063–1070. [Google Scholar] [CrossRef]
- Grundstrom, J.; Reimer, J.M.; Magnusson, S.E.; Nilsson, G.; Wernersson, S.; Hellman, L. Human cord blood derived immature basophils show dual characteristics, expressing both basophil and eosinophil associated proteins. PLoS ONE 2012, 7, e48308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abrink, M.; Gobl, A.E.; Huang, R.; Nilsson, K.; Hellman, L. Human cell lines U-937, THP-1 and Mono Mac 6 represent relatively immature cells of the monocyte-macrophage cell lineage. Leukemia 1994, 8, 1579–1584. [Google Scholar] [PubMed]
- Nilsson, G.; Blom, T.; Kusche-Gullberg, M.; Kjellen, L.; Butterfield, J.H.; Sundstrom, C.; Nilsson, K.; Hellman, L. Phenotypic characterization of the human mast-cell line HMC-1. Scand. J. Immunol. 1994, 39, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Blom, T.; Huang, R.; Aveskogh, M.; Nilsson, K.; Hellman, L. Phenotypic characterization of KU812, a cell line identified as an immature human basophilic leukocyte. Eur. J. Immunol. 1992, 22, 2025–2032. [Google Scholar] [CrossRef]
- Blom, T.; Nilsson, G.; Sundstrom, C.; Nilsson, K.; Hellman, L. Characterization of a human basophil-like cell line (LAMA-84). Scand. J. Immunol. 1996, 44, 54–61. [Google Scholar] [CrossRef]
- Blom, T.; Hellman, L. Characterization of a tryptase mRNA expressed in the human basophil cell line KU812. Scand. J. Immunol. 1993, 37, 203–208. [Google Scholar] [CrossRef]
- Harris, P.; Ralph, P. Human leukemic models of myelomonocytic development: A review of the HL-60 and U937 cell lines. J. Leukoc. Biol. 1985, 37, 407–422. [Google Scholar] [CrossRef]
- Mayumi, M. EoL-1, a human eosinophilic cell line. Leuk. Lymphoma 1992, 7, 243–250. [Google Scholar] [CrossRef]
- Kirshenbaum, A.S.; Yin, Y.; Sundstrom, J.B.; Bandara, G.; Metcalfe, D.D. Description and Characterization of a Novel Human Mast Cell Line for Scientific Study. Int. J. Mol. Sci. 2019, 20, 5520. [Google Scholar] [CrossRef] [Green Version]
- Guhl, S.; Babina, M.; Neou, A.; Zuberbier, T.; Artuc, M. Mast cell lines HMC-1 and LAD2 in comparison with mature human skin mast cells--drastically reduced levels of tryptase and chymase in mast cell lines. Exp. Derm. 2010, 19, 845–847. [Google Scholar] [CrossRef]
- Poorafshar, M.; Helmby, H.; Troye-Blomberg, M.; Hellman, L. MMCP-8, the first lineage-specific differentiation marker for mouse basophils. Elevated numbers of potent IL-4-producing and MMCP-8-positive cells in spleens of malaria-infected mice. Eur. J. Immunol. 2000, 30, 2660–2668. [Google Scholar] [CrossRef]
- Sasaki, H.; Kurotaki, D.; Osato, N.; Sato, H.; Sasaki, I.; Koizumi, S.; Wang, H.; Kaneda, C.; Nishiyama, A.; Kaisho, T.; et al. Transcription factor IRF8 plays a critical role in the development of murine basophils and mast cells. Blood 2015, 125, 358–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahlin, J.S.; Hamey, F.K.; Pijuan-Sala, B.; Shepherd, M.; Lau, W.W.Y.; Nestorowa, S.; Weinreb, C.; Wolock, S.; Hannah, R.; Diamanti, E.; et al. A single-cell hematopoietic landscape resolves 8 lineage trajectories and defects in Kit mutant mice. Blood 2018, 131, e1–e11. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Li, Y.; Liu, B. Transcriptional regulation of mast cell and basophil lineage commitment. Semin. Immunopathol. 2016, 38, 539–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, M.Y.; Davidson, D.; Yu, J.; Latour, S.; Veillette, A. Clnk, a novel SLP-76-related adaptor molecule expressed in cytokine-stimulated hemopoietic cells. J. Exp. Med. 1999, 190, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- Utting, O.; Sedgmen, B.J.; Watts, T.H.; Shi, X.; Rottapel, R.; Iulianella, A.; Lohnes, D.; Veillette, A. Immune functions in mice lacking Clnk, an SLP-76-related adaptor expressed in a subset of immune cells. Mol. Cell Biol. 2004, 24, 6067–6075. [Google Scholar] [CrossRef] [Green Version]
- Goitsuka, R.; Kanazashi, H.; Sasanuma, H.; Fujimura, Y.; Hidaka, Y.; Tatsuno, A.; Ra, C.; Hayashi, K.; Kitamura, D. A BASH/SLP-76-related adaptor protein MIST/Clnk involved in IgE receptor-mediated mast cell degranulation. Int. Immunol. 2000, 12, 573–580. [Google Scholar] [CrossRef] [Green Version]
- Fujii, Y.; Wakahara, S.; Nakao, T.; Hara, T.; Ohtake, H.; Komurasaki, T.; Kitamura, K.; Tatsuno, A.; Fujiwara, N.; Hozumi, N.; et al. Targeting of MIST to Src-family kinases via SKAP55-SLAP-130 adaptor complex in mast cells. FEBS Lett. 2003, 540, 111–116. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Errico, D.; Lessmann, E.; Rivera, J. Adapters in the organization of mast cell signaling. Immunol. Rev. 2009, 232, 195–217. [Google Scholar] [CrossRef] [Green Version]
- Tsvilovskyy, V.; Solis-Lopez, A.; Almering, J.; Richter, C.; Birnbaumer, L.; Dietrich, A.; Freichel, M. Analysis of Mrgprb2 Receptor-Evoked Ca (2+) Signaling in Bone Marrow Derived (BMMC) and Peritoneal (PMC) Mast Cells of TRPC-Deficient Mice. Front. Immunol. 2020, 11, 564. [Google Scholar] [CrossRef]
- Ohneda, K.; Ohmori, S.; Yamamoto, M. Mouse Tryptase Gene Expression is Coordinately Regulated by GATA1 and GATA2 in Bone Marrow-Derived Mast Cells. Int. J. Mol. Sci. 2019, 20, 4603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobson, J.T.; Seibert, J.; Teh, E.M.; Da’as, S.; Fraser, R.B.; Paw, B.H.; Lin, T.J.; Berman, J.N. Carboxypeptidase A5 identifies a novel mast cell lineage in the zebrafish providing new insight into mast cell fate determination. Blood 2008, 112, 2969–2972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malbec, O.; Roget, K.; Schiffer, C.; Iannascoli, B.; Dumas, A.R.; Arock, M.; Daeron, M. Peritoneal cell-derived mast cells: An in vitro model of mature serosal-type mouse mast cells. J. Immunol. 2007, 178, 6465–6475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leist, M.; Sunder, C.A.; Drube, S.; Zimmermann, C.; Geldmacher, A.; Metz, M.; Dudeck, A.; Maurer, M. Membrane-bound stem cell factor is the major but not only driver of fibroblast-induced murine skin mast cell differentiation. Exp. Dermatol. 2017, 26, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Gersch, C.; Dewald, O.; Zoerlein, M.; Michael, L.H.; Entman, M.L.; Frangogiannis, N.G. Mast cells and macrophages in normal C57/BL/6 mice. Histochem. Cell Biol. 2002, 118, 41–49. [Google Scholar] [CrossRef]
- Dwyer, D.F.; Barrett, N.A.; Austen, K.F.; Immunological Genome Project, C. Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat. Immunol. 2016, 17, 878–887. [Google Scholar] [CrossRef]
- Babina, M.; Guhl, S.; Artuc, M.; Zuberbier, T. Skin mast cell phenotypes between two highly divergent cohorts-more pronounced variability within than between groups. Exp. Dermatol. 2017, 26, 446–449. [Google Scholar] [CrossRef] [Green Version]
- Babina, M.; Guhl, S.; Artuc, M.; Trivedi, N.N.; Zuberbier, T. Phenotypic variability in human skin mast cells. Exp. Dermatol. 2016, 25, 434–439. [Google Scholar] [CrossRef]
- Frossi, B.; Mion, F.; Sibilano, R.; Danelli, L.; Pucillo, C.E.M. Is it time for a new classification of mast cells? What do we know about mast cell heterogeneity? Immunol. Rev. 2018, 282, 35–46. [Google Scholar] [CrossRef]
| Transcript | BMMCs | P-MCs | MQ | B Cells | Fold Dif |
|---|---|---|---|---|---|
| Mcpt4 (mMCP-4) | 7 | 31,290 | 5 | 10 | (500×) |
| Tpsb2 (Mcpt6) | 3119 | 67,773 | 6 | 17 | (20×) |
| Cma1 (Mcpt5) | 5683 | 45,221 | 4 | 11 | (8×) |
| Mrgprb2 (Substance P rec.) | 8 | 899 | 0.1 | 0.2 | (111×) |
| Gfra2 (Neurotrophin rec.) | 0.1 | 2016 | 7 | 1 | (20,000×) |
| Adamts9 (Protease) | 12 | 370 | 1 | 0 | (31×) |
| C2 (Complement f. 2) | 1.3 | 159 | 5 | 2 | (122×) |
| Milr1 (Allergin) | 48 | 283 | 24 | 23 | (6×) |
| Hdc (Histidine decabo×ylase) | 48 | 936 | 42 | 2 | (19×) |
| CtsE (Cathepsin E) | 336 | 1248 | 75 | 336 | (4×) |
| Serpinb1a (Protease inh.) | 460 | 1037 | 20 | 64 | (2×) |
| Myb (Transcr. f.) | 392 | 2490 | 0 | 9 | (6×) |
| Meis2 (Transcr. f.) | 18 | 300 | 0 | 0 | (17×) |
| Tarm1 (Transcr. f.) | 21 | 156 | 0 | 0 | (7×) |
| Il3ra (IL-3 rec. alpha) | 25 | 159 | 17 | 4 | (6×) |
| Transcript | BMMCs | P-MCs | MQ | B Cells |
|---|---|---|---|---|
| Cpa3 (CPA-3) | 22,478 | 45,604 | 1 | 6 |
| Gata2 (Transcr. factor) | 5205 | 2272 | 0 | 1 |
| Fcer1a (IgE rec. alpha) | 1631 | 345 | 0 | 0 (5× lower in P-MCs) |
| Ms4a2 (IgE rec. beta) | 4288 | 1297 | 0 | 2 (3× lower in P-MCs) |
| Fcgr3 (IgG rec. III) | 538 | 752 | 1968 | 27 |
| Cadm1 (Cell adhesion) | 913 | 1118 | 4 | 9 |
| Rab27b (GTP-ase) | 856 | 858 | 0 | 0.7 |
| Inpp5d (Phospatase) | 826 | 663 | 124 | 287 |
| Cited2 (Trans act.) | 772 | 599 | 17 | 30 |
| Tmem9 (Trans memb. pr.) | 767 | 456 | 52 | 25 |
| Basp1 (Brain acid pr.) | 686 | 1005 | 6 | 2 |
| CD200r3 (Surface rec.) | 640 | 275 | 1 | 0.1 |
| Tuba8 (Tubulin alpha 8) | 622 | 439 | 0.2 | 0.6 |
| Maob (Mono amine ox.) | 577 | 359 | 2 | 0.1 |
| Erv3 (End. retrovirus) | 514 | 141 | 0.2 | 0.7 |
| CD55 (DAF) | 510 | 439 | 12 | 248 |
| Rgs18 (Reg. of G prot.) | 487 | 509 | 79 | 10 |
| Ccl2 (Chemokine) | 427 | 397 | 1.2 | 0.4 |
| Pik3r6 (PI3k Subunit) | 423 | 767 | 179 | 9 |
| Slc45a3 (Solute carrier) | 419 | 673 | 0.8 | 0.2 |
| CstF (Cystatin F) | 388 | 181 | 0 | 0.1 |
| GzmB (Granzyme B) | 386 | 581 | 0 | 0 |
| Dock10 (Dedic. cytok.) | 383 | 389 | 102 | 332 |
| Bmp7 (Bone morf. pr. 7) | 375 | 56 | 0.2 | 0 (7× lower in P-MCs) |
| Slc30a2 (Zinc transp.) | 369 | 220 | 0 | 0 |
| A4galt (enz. galact. cer) | 364 | 71 | 0 | 0 |
| Specc1 (Cytospin-B ) | 300 | 124 | 14 | 18 |
| Cadm3 (Cell adhesion) | 298 | 118 | 0.4 | 0.1 |
| Gata1 (Transcr. f.) | 296 | 74 | 0 | 0 (4× lower in P-MCs) |
| Samsn1 (Neg. reg. B-cl) | 285 | 187 | 22 | 15 |
| Rab44 (Ras fam. memb.) | 276 | 302 | 1.3 | 0.1 |
| Dapp1 (Rec. signaling) | 270 | 389 | 96 | 71 |
| Grap2 (Cell signaling) | 214 | 261 | 5 | 2 |
| Hpgds (Prost.gl. D synt.) | 233 | 450 | 2 | 2 |
| Gm973 (Predicted gene) | 232 | 261 | 0 | 0.4 |
| Slc2a3 (Glucose transp.) | 208 | 102 | 0.5 | 41 |
| Slc6a12 (Solute carrier?) | 205 | 191 | 0 | 0.7 |
| Nrn (Nurin nuclear env.) | 204 | 208 | 22 | 145 |
| Smpx (Small muscle pr.) | 203 | 233 | 0 | 0.1 |
| Nfe2 (Transcr. f.) | 188 | 108 | 126 | 3 |
| Abcb1b (ATP dep. trans.) | 189 | 336 | 37 | 6 |
| Blm (Bloom syndr.) | 175 | 73 | 3 | 10 |
| Mrgpra4 | 163 | 155 | 0 | 9 |
| Tal1 (helix-l-h transc.f.) | 160 | 246 | 3 | 0.1 |
| CD200r1 (Cell surf. rec.) | 150 | 246 | 3 | 0.1 |
| Rnf180 (Ring f. Ubiq. lig.) | 150 | 156 | 5 | 0 |
| Runx3 (Transcr. f.) | 143 | 409 | 7 | 65 (3× lower in BMMCs) |
| Mitf (Transcr. f.) | 144 | 370 | 36 | 3 |
| Tlr4 (TLR-4) | 134 | 61 | 200 | 26 |
| Rgs1 (Reg. G-prot. sign.) | 132 | 51 | 1 | 0 |
| Lat (Signaling) | 130 | 111 | 0.4 | 1.2 |
| Lat2 (Signaling) | 2053 | 1588 | 291 | 546 |
| P2rx1 (ATP rec) | 129 | 327 | 69 | 2 |
| Rgs13 (Reg. G-prot. sign.) | 126 | 289 | 0.2 | 0.5 |
| Dgki (Diacyl glyc. kin.) | 125 | 33 | 0 | 0 |
| Galnt6 (Mucin synth.) | 123 | 122 | 1 | 31 |
| Bcl2 (Anti apoptotic) | 132 | 101 | 10 | 15 |
| CD69 (Leukocyte ag.) | 110 | 45 | 0.5 | 123 |
| Gpr141 (G-prot. c. rec.) | 109 | 47 | 0.3 | 0 |
| Gp1ba (vWF-receptor) | 93 | 61 | 0 | 1 |
| Tespa1 (thymoc. exp.) | 85 | 153 | 0 | 17 |
| Gcsam (Germinal c. ass.) | 82 | 311 | 0 | 0.4 |
| Spn (CD43 Leukosialin) | 72 | 134 | 0 | 5 |
| Cx3cr1 (Fractakine rec.) | 62 | 28 | 0 | 0.2 |
| Ryr3 (Ryanodine rec.) | 59 | 24 | 0 | 0.3 |
| Atp8a2 (Phopholipid tr.) | 59 | 41 | 0.5 | 0.1 |
| Gsg11 (Germ cell ag.) | 58 | 27 | 0 | 0.4 |
| Mcpt8 (Basophil Prot.) | 46 | 38 | 0.1 | 0.1 |
| Hst6st (Heparan s.-O-S) | 44 | 152 | 0 | 0 |
| Kcne3 (Potassium ch.) | 42 | 112 | 2 | 0.4 |
| Prss34 (Mcpt11) | 41 | 257 | 0 | 5 |
| Fcgr2b (Fc-γ rec. 2B) | 40 | 91 | 9 | 72 |
| Mcpt-ps1 | 36 | 210 | 0 | 0 |
| Zfp521 (Zinc finger g.) | 29 | 67 | 0 | 0 |
| Hrh4 (Histamin rec.4) | 26 | 25 | 0 | 0 |
| Il18r1 (IL-18 rec. alpha) | 26 | 24 | 0.1 | 2 |
| Il9r (IL-9 receptor) | 25 | 11 | 0 | 73 |
| Gfi1 (Zinc finger prot.) | 21 | 79 | 0.1 | 0.8 |
| Dfna1 (B lymfo blasts) | 20 | 54 | 1 | 0 |
| Il4 (IL-4) | 14 | 13 | 0 | 0 |
| Il13 (IL-13) | 11 | 4 | 0 | 0 |
| Transcript | BMMCs | P-MCs | MQ | B Cells | Fold Dif. |
|---|---|---|---|---|---|
| Csf2rb (IL-3+GM-CSF rec. beta) | 3485 | 415 | 150 | 84 | (8×) |
| CTLA2a (CTL assoc. prot.) | 1510 | 5 | 2 | 2 | (302×) |
| F2r (Thrombin receptor) | 1463 | 12 | 0 | 0 | (122×) |
| F13a1 (Coagulation factor 13) | 994 | 6 | 122 | 5 | (166×) |
| Clnk (MIST, cell signaling) | 921 | 4 | 0.1 | 0 | (230×) |
| IL10ra (IL-10 rec. alpha) | 919 | 49 | 280 | 268 | (19×) |
| Rnf128 (Ubiquitin ligase) | 889 | 78 | 28 | 0.5 | (11×) |
| Lpar6 (G prot. coupled rec.) | 825 | 70 | 35 | 105 | (12×) |
| Serpina3g (Protease inhibitor) | 710 | 5 | 0.3 | 60 | (142×) |
| Nampt (enzyme) | 706 | 55 | 69 | 48 | (13×) |
| Lif (Leukemia Inh. factor) | 584 | 4 | 0.5 | 0.1 | (146×) |
| Gab2 (Cell signaling) | 583 | 45 | 115 | 17 | (13×) |
| Rnase6 (Anti bacterial) | 557 | 26 | 7 | 153 | (21×) |
| Pik3cd (PI3K delta) | 507 | 64 | 102 | 319 | (8×) |
| CD200r4 (Cell surf. rec.) | 487 | 21 | 37 | 0.6 | (23×) |
| Sema4d (CD100) | 480 | 47 | 3 | 132 | (10×) |
| Anpep (Alanine-amino pep.) | 447 | 118 | 19 | 2 | (4×) |
| Dnm3 (Dynamin) | 435 | 98 | 0 | 0 | (4×) |
| Dtx4 (Ubiquitin Ligase) | 384 | 7 | 18 | 14 | (55×) |
| Rab38 (Ras rel. prot.) | 375 | 6 | 0.3 | 0 | (62×) |
| Akr1c12 (Alpha-keto red.) | 366 | 2 | 0.8 | 0.2 | (183×) |
| Aqp9 (Aquaporin) | 360 | 2 | 46 | 0.5 | (180×) |
| Spns3 (Sphingolipid transp.) | 346 | 12 | 0 | 15 | (29×) |
| Tgfbr1 (TGF beta receptor) | 319 | 96 | 36 | 27 | (3×) |
| Krba1 (KRAB-A cont.) | 304 | 34 | 7 | 57 | (9×) |
| Stap1 (Sign.trans B cells) | 300 | 14 | 1 | 150 | (21×) |
| Stk19 (Ser/Thr kinase) | 299 | 32 | 48 | 52 | (9×) |
| Glipr1 (Cys rich secr. pr.) | 292 | 29 | 5 | 20 | (10×) |
| IL-4ra (IL-4 receptor alpha) | 282 | 12 | 21 | 23 | (24×) |
| Treml2 (Tr. rec. myeloid) | 278 | 4 | 0.3 | 72 | (70×) |
| Avil (Advillin) | 270 | 0.7 | 0 | 0.5 | (386×) |
| Tmem233 (Trans memb. pr.) | 269 | 2 | 0 | 0.1 | (134×) |
| Mthfd2 (Mitoch. enzyme) | 266 | 8 | 6 | 20 | (33×) |
| Tbxas1 (Thrombox. synthase) | 243 | 23 | 103 | 2 | (10×) |
| CD300lf (Membr Glycopr. myeloid) | 239 | 49 | 10 | 63 | (5×) |
| Birc5 (anti-apotosis) | 234 | 41 | 13 | 4 | (6×) |
| Neb (Nebulin actin binding.) | 211 | 0 | 1 | 0.5 | (≈211×) |
| Kcnn4 (Potassium channel) | 202 | 10 | 3 | 63 | (20×) |
| Il2ra (IL-2 rec. alpha) | 196 | 14 | 0.2 | 8 | (14×) |
| Nrop3 (Nucl. rec. interacting) | 165 | 2 | 0 | 0 | (82×) |
| Ptger4 (Prostagl. E2 receptor) | 165 | 11 | 116 | 24 | (15×) |
| Pgr (Progesterone receptor) | 147 | 5 | 0 | 0 | (29×) |
| Gpr174 (G-prot coup rec?) | 145 | 24 | 1 | 137 | (6×) |
| Nlrp3 (NALP3 Inflammasome) | 134 | 8 | 90 | 1.4 | (17×) |
| Ahrr (Aryl hydr. carb. receptor) | 134 | 1.2 | 7 | 0.1 | (112×) |
| Calca (Calcitonin related pept.) | 133 | 1.2 | 0 | 0 | (111×) |
| CD96 (Tactile T-cell act.) | 121 | 3 | 0.5 | 0.5 | (40×) |
| Gfi1b (Zinc finger) | 106 | 25 | 11 | 3 | (4×) |
| Il1b (IL-1 beta) | 109 | 4 | 4 | 0.4 | (27×) |
| Greb1 (Estrogen resp. gene) | 107 | 1.7 | 0 | 0.3 | (63×) |
| Ulbp1 (Stress ind. NK rec.) | 96 | 3 | 2 | 7 | (32×) |
| Rhof (Ras hom. fam.) | 95 | 10 | 0.3 | 251 | (10×) |
| Cited (Trans activator) | 94 | 3 | 0.3 | 1.4 | (31×) |
| Cyp4a12a (Cytochr. P450) | 86 | 0 | 0 | 0 | (≈86×) |
| Aqp8 (Aqua porin) | 64 | 0 | 0 | 0.1 | (≈64×) |
| Il6 (IL-6) | 64 | 12 | 1.2 | 0 | (5×) |
| Transcript | BMMCs | BMMCs + LPS | Fold Dif. |
|---|---|---|---|
| MilR1 (Allergin) | 48 | 332 | (7×) |
| Hdc (Histidine decarb.) | 48 | 279 | (6×) |
| Nfkbiz (NFkB inh.) | 42 | 422 | (10×) |
| Mgat5 (Oligosaccharide enzyme) | 43 | 331 | (8×) |
| Tmem63b (Ion channel) | 59 | 397 | (7×) |
| Il1b (IL-1 beta) | 109 | 1804 | (17×) |
| Il6 (IL-6) | 64 | 246 | (4×) |
| Tnfrsf9 (TNF rec. superf. 9, CD137) | 8 | 459 | (57×) |
| GzmB (Granzyme B) | 386 | 1900 | (5×) |
| Gzmc (Granzyme C) | 0.1 | 110 | (1100×) |
| Calca (Calcitonin related peptide) | 133 | 917 | (7×) |
| Il13 (IL-13) | 11 | 83 | (8×) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akula, S.; Paivandy, A.; Fu, Z.; Thorpe, M.; Pejler, G.; Hellman, L. How Relevant Are Bone Marrow-Derived Mast Cells (BMMCs) as Models for Tissue Mast Cells? A Comparative Transcriptome Analysis of BMMCs and Peritoneal Mast Cells. Cells 2020, 9, 2118. https://doi.org/10.3390/cells9092118
Akula S, Paivandy A, Fu Z, Thorpe M, Pejler G, Hellman L. How Relevant Are Bone Marrow-Derived Mast Cells (BMMCs) as Models for Tissue Mast Cells? A Comparative Transcriptome Analysis of BMMCs and Peritoneal Mast Cells. Cells. 2020; 9(9):2118. https://doi.org/10.3390/cells9092118
Chicago/Turabian StyleAkula, Srinivas, Aida Paivandy, Zhirong Fu, Michael Thorpe, Gunnar Pejler, and Lars Hellman. 2020. "How Relevant Are Bone Marrow-Derived Mast Cells (BMMCs) as Models for Tissue Mast Cells? A Comparative Transcriptome Analysis of BMMCs and Peritoneal Mast Cells" Cells 9, no. 9: 2118. https://doi.org/10.3390/cells9092118
APA StyleAkula, S., Paivandy, A., Fu, Z., Thorpe, M., Pejler, G., & Hellman, L. (2020). How Relevant Are Bone Marrow-Derived Mast Cells (BMMCs) as Models for Tissue Mast Cells? A Comparative Transcriptome Analysis of BMMCs and Peritoneal Mast Cells. Cells, 9(9), 2118. https://doi.org/10.3390/cells9092118






