Germline-Specific Repetitive Elements in Programmatically Eliminated Chromosomes of the Sea Lamprey (Petromyzon marinus)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Research Animals
2.2. Production of Lamprey Embryos
2.3. PACT Clearing
2.4. Preparation of Metaphase Spreads
2.5. C0t DNA Isolation
2.6. Computational Prediction of Germline-Specific Repeats
2.7. Fluorescence in Situ Hybridization
3. Results and Discussion
3.1. Comparative Hybridization Indicates the Presence of Germline-Specific Repeats
3.2. Computational Identification of Germline-Specific and Centromeric Repeats
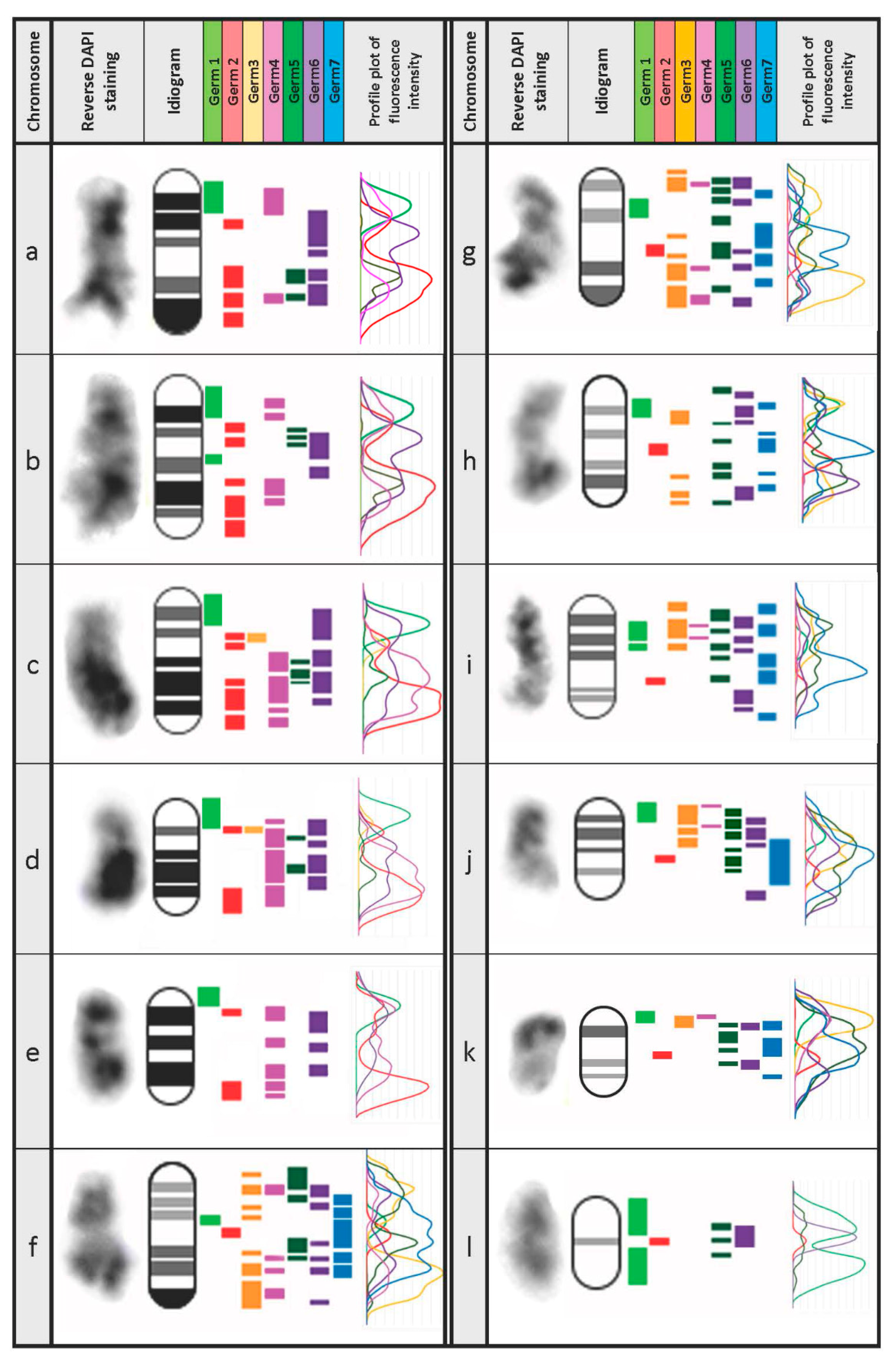
3.3. Defining the Chromosomal Localization of Germline-Restricted Repeats
3.4. Chromosomal Localization of Germline-Restricted Repeats in Elimination Anaphases
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smith, J.J.; Baker, C.; Eichler, E.E.; Amemiya, C.T. Genetic consequences of programmed genome rearrangement. Curr. Biol. 2012, 22, 1524–1529. [Google Scholar] [CrossRef]
- Timoshevskiy, V.A.; Herdy, J.R.; Keinath, M.C.; Smith, J.J. Cellular and molecular features of developmentally programmed genome rearrangement in a vertebrate (sea lamprey: Petromyzon marinus). PLoS Genet. 2016, 12, e1006103. [Google Scholar] [CrossRef]
- Wang, J.; Davis, R.E. Programmed DNA elimination in multicellular organisms. Curr. Opin. Genet. Dev. 2014, 27, 26–34. [Google Scholar] [CrossRef] [Green Version]
- Bracht, J.R.; Fang, W.; Goldman, A.D.; Dolzhenko, E.; Stein, E.M.; Landweber, L.F. Genomes on the edge: Programmed genome instability in ciliates. Cell 2013, 152, 406–416. [Google Scholar] [CrossRef]
- Mochizuki, K.; Gorovsky, M.A. Small rnas in genome rearrangement in tetrahymena. Curr. Opin. Genet. Dev. 2004, 14, 181–187. [Google Scholar] [CrossRef]
- Mochizuki, K. Developmentally programmed, rna-directed genome rearrangement in tetrahymena. Dev. Growth Differ. 2012, 54, 108–119. [Google Scholar] [CrossRef]
- Boveri, T. Uber differenzierung der zellkerne wahrend der furchung des eies von ascaris megalocephala. Anat. Anz. 1887, 2, 688–693. [Google Scholar]
- Streit, A.; Wang, J.; Kang, Y.; Davis, R.E. Gene silencing and sex determination by programmed DNA elimination in parasitic nematodes. Curr. Opin. Microbiol. 2016, 32, 120–127. [Google Scholar] [CrossRef] [Green Version]
- Goday, C.; Esteban, M.R. Chromosome elimination in sciarid flies. Bioessays 2001, 23, 242–250. [Google Scholar] [CrossRef]
- Singh, P.B.; Belyakin, S.N. L chromosome behaviour and chromosomal imprinting in sciara coprophila. Genes 2018, 9, 440. [Google Scholar] [CrossRef]
- Grishanin, A. Chromatin diminution in copepoda (crustacea): Pattern, biological role and evolutionary aspects. Comp. Cytogenet. 2014, 8, 1–10. [Google Scholar] [CrossRef]
- Staiber, W. Chromosome elimination in germ line-soma differentiation of acricotopus lucidus (diptera, chironomidae). Genome 2006, 49, 269–274. [Google Scholar] [CrossRef]
- Kohno, S.; Nakai, Y.; Satoh, S.; Yoshida, M.; Kobayashi, H. Chromosome elimination in the japanese hagfish, eptatretus burgeri (agnatha, cyclostomata). Cytogenet. Genome Res. 1986, 41, 209–214. [Google Scholar] [CrossRef]
- Kubota, S.; Nakai, Y.; Sato, N.; Kuroo, M.; Kohno, S. Chromosome elimination in northeast pacific hagfish, eptatretus-stoutii (cyclostomata, agnatha). J. Hered. 1994, 85, 413–415. [Google Scholar] [CrossRef]
- Nakai, Y.; Kubota, S.; Goto, Y.; Ishibashi, T.; Davison, W.; Kohno, S. Chromosome elimination in three baltic, south pacific and north-east pacific hagfish species. Chromosome Res. 1995, 3, 321–330. [Google Scholar] [CrossRef]
- Nakai, Y.; Kubota, S.; Kohno, S. Chromatin diminution and chromosome elimination in four japanese hagfish species. Cytogenet. Genome Res. 1991, 56, 196–198. [Google Scholar] [CrossRef]
- Pigozzi, M.I.; Solari, A.J. Germ cell restriction and regular transmission of an accessory chromosome that mimics a sex body in the zebra finch, taeniopygia guttata. Chromosome Res. 1998, 6, 105–113. [Google Scholar] [CrossRef]
- Torgasheva, A.A.; Malinovskaya, L.P.; Zadesenets, K.S.; Karamysheva, T.V.; Kizilova, E.A.; Akberdina, E.A.; Pristyazhnyuk, I.E.; Shnaider, E.P.; Volodkina, V.A.; Saifitdinova, A.F.; et al. Germline-restricted chromosome (grc) is widespread among songbirds. Proc. Natl Acad. Sci. USA 2019, 116, 11845–11850. [Google Scholar] [CrossRef]
- Smith, J.J.; Antonacci, F.; Eichler, E.E.; Amemiya, C.T. Programmed loss of millions of base pairs from a vertebrate genome. Proc. Natl. Acad. Sci. USA 2009, 106, 11212–11217. [Google Scholar] [CrossRef] [Green Version]
- Timoshevskiy, V.A.; Lampman, R.T.; Hess, J.E.; Porter, L.L.; Smith, J.J. Deep ancestry of programmed genome rearrangement in lampreys. Dev. Biol. 2017, 429, 31–34. [Google Scholar] [CrossRef]
- Yao, M.C.; Chao, J.L. Rna-guided DNA deletion in tetrahymena: An rnai-based mechanism for programmed genome rearrangements. Annu. Rev. Genet. 2005, 39, 537–559. [Google Scholar] [CrossRef] [PubMed]
- Zufall, R.A.; Robinson, T.; Katz, L.A. Evolution of developmentally regulated genome rearrangements in eukaryotes. J. Exp. Zool. Part B 2005, 304, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Kohno, S.I.; Kubota, S.; Nakai, Y. Chromatin diminution and chromosome elimination in hagfishes. In The Biology of Hagfishes; Jørgensen, J.M., Lomholt, J.P., Eds.; Chapman and Hall: London, UK, 1998; pp. 81–100. [Google Scholar]
- Del Priore, L.; Pigozzi, M.I. Histone modifications related to chromosome silencing and elimination during male meiosis in bengalese finch. Chromosoma 2014, 123, 293–302. [Google Scholar] [CrossRef]
- Muller, F.; Bernard, V.; Tobler, H. Chromatin diminution in nematodes. Bioessays 1996, 18, 133–138. [Google Scholar] [CrossRef]
- Wang, J.; Gao, S.; Mostovoy, Y.; Kang, Y.; Zagoskin, M.; Sun, Y.; Zhang, B.; White, L.K.; Easton, A.; Nutman, T.B.; et al. Comparative genome analysis of programmed DNA elimination in nematodes. Genome Res. 2017, 27, 2001–2014. [Google Scholar] [CrossRef]
- Standiford, D.M. The effects of chromatin diminution on the pattern of c-banding in the chromosomes of acanthocyclops vernalis fischer (copepoda: Crustacea). Genetica 1989, 79, 207–214. [Google Scholar] [CrossRef]
- Wyngaard, G.A.; Gregory, T.R. Temporal control of DNA replication and the adaptive value of chromatin diminution in copepods. J. Exp. Zool. 2001, 291, 310–316. [Google Scholar] [CrossRef]
- Staiber, W.; Schiffkowski, C. Structural evolution of the germ line-limited chromosomes in acricotopus. Chromosoma 2000, 109, 343–349. [Google Scholar] [CrossRef]
- Kojima, N.F.; Kojima, K.K.; Kobayakawa, S.; Higashide, N.; Hamanaka, C.; Nitta, A.; Koeda, I.; Yamaguchi, T.; Shichiri, M.; Kohno, S.; et al. Whole chromosome elimination and chromosome terminus elimination both contribute to somatic differentiation in taiwanese hagfish paramyxine sheni. Chromosome Res. 2010, 18, 383–400. [Google Scholar] [CrossRef]
- Chmielewska, M.; Dedukh, D.; Haczkiewicz, K.; Rozenblut-Koscisty, B.; Kazmierczak, M.; Kolenda, K.; Serwa, E.; Pietras-Lebioda, A.; Krasikova, A.; Ogielska, M. The programmed DNA elimination and formation of micronuclei in germ line cells of the natural hybridogenetic water frog pelophylax esculentus. Sci. Rep. 2018, 8, 7870. [Google Scholar] [CrossRef]
- Tobler, H. The differentiation of germ and somatic cell lines in nematodes. In Germ Line-Soma Differentiation: Results and Problems in Cell Differentiation; Hennig, W., Ed.; Springer: Berlin, Germany, 1986; Volume 13, pp. 1–69. [Google Scholar]
- de Saint Phalle, B.; Sullivan, W. Incomplete sister chromatid separation is the mechanism of programmed chromosome elimination during early sciara coprophila embryogenesis. Development 1996, 122, 3775–3784. [Google Scholar] [PubMed]
- Beermann, S. The diminution of heterochromatic chromosomal segments in cyclops (crustacea, copepoda). Chromosoma 1977, 60, 297–344. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Timoshevskaya, N.; Ye, C.; Holt, C.; Keinath, M.C.; Parker, H.J.; Cook, M.E.; Hess, J.E.; Narum, S.R.; Lamanna, F.; et al. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat. Genet. 2018, 50, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Biederman, M.K.; Nelson, M.M.; Asalone, K.C.; Pedersen, A.L.; Saldanha, C.J.; Bracht, J.R. Discovery of the first germline-restricted gene by subtractive transcriptomic analysis in the zebra finch, taeniopygia guttata. Curr. Biol. 2018, 28, 1620–1627. [Google Scholar] [CrossRef] [PubMed]
- Potter, I.C.; Rothwell, B. The mitotic chromosomes of the lamprey, petromyzon marinus L. Experientia 1970, 26, 429–430. [Google Scholar] [CrossRef] [PubMed]
- Covelo-Soto, L.; Moran, P.; Pasantes, J.J.; Perez-Garcia, C. Cytogenetic evidences of genome rearrangement and differential epigenetic chromatin modification in the sea lamprey (petromyzon marinus). Genetica 2014, 142, 545–554. [Google Scholar] [CrossRef]
- Smith, J.J.; Stuart, A.B.; Sauka-Spengler, T.; Clifton, S.W.; Amemiya, C.T. Development and analysis of a germline bac resource for the sea lamprey, a vertebrate that undergoes substantial chromatin diminution. Chromosoma 2010, 119, 381–389. [Google Scholar] [CrossRef]
- Nikitina, N.; Bronner-Fraser, M.; Sauka-Spengler, T. The sea lamprey petromyzon marinus: A model for evolutionary and developmental biology. In Emerging Model Organisms: A Laboratory Manual; CSHL Press: Cold Spring Harbor, NY, USA, 2009; Volume 1, pp. 405–429. [Google Scholar]
- Piavis, G.W. Embryological Stages in the Sea Lamprey and Effects of Temperature on Development; U. S. Fish and Wildlife Service: Washington, DC, USA, 1961; pp. 111–143.
- Harland, R.M. In situ hybridization: An improved whole-mount method for xenopus embryos. Methods Cell Biol. 1991, 36, 685–695. [Google Scholar]
- Yang, B.; Treweek, J.B.; Kulkarni, R.P.; Deverman, B.E.; Chen, C.K.; Lubeck, E.; Shah, S.; Cai, L.; Gradinaru, V. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 2014, 158, 945–958. [Google Scholar] [CrossRef]
- Claussen, U.; Michel, S.; Muhlig, P.; Westermann, M.; Grummt, U.W.; Kromeyer-Hauschild, K.; Liehr, T. Demystifying chromosome preparation and the implications for the concept of chromosome condensation during mitosis. Cytogenet. Genome Res. 2002, 98, 136–146. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; Volume 3, p. 2344. [Google Scholar]
- Timoshevskiy, V.A.; Severson, D.W.; Debruyn, B.S.; Black, W.C.; Sharakhov, I.V.; Sharakhova, M.V. An integrated linkage, chromosome, and genome map for the yellow fever mosquito aedes aegypti. PLoS Negl. Trop. Dis. 2013, 7, e2052. [Google Scholar] [CrossRef] [PubMed]
- Marcais, G.; Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.H.R. Repeatmodeler Open-1.0. Available online: http://www.repeatmasker.org (accessed on 12 September 2019).
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Timoshevskaya, N. Difcover. Available online: https://github.com/timnat/DifCover (accessed on 12 September 2019).
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. Blast+: Architecture and applications. BMC Bioinf. 2009, 10, 421. [Google Scholar] [CrossRef]
- Timoshevskiy, V.A.; Sharma, A.; Sharakhov, I.V.; Sharakhova, M.V. Fluorescent in situ hybridization on mitotic chromosomes of mosquitoes. J. Vis. Exp. 2012, 67, e4215. [Google Scholar] [CrossRef]
- Solovei, I. Fluorescence in situ hybridization (fish) on tissue cryosections. Methods Mol. Biol. 2010, 659, 71–82. [Google Scholar]
- Rooney, D.E.; Czepulkowski, B.H. Human Cytogenetics: A Practical Approach, 2nd ed.; IRL Press: Oxford, UK, 1992. [Google Scholar]
- Keinath, M.C.; Timoshevskiy, V.A.; Timoshevskaya, N.Y.; Tsonis, P.A.; Voss, S.R.; Smith, J.J. Initial characterization of the large genome of the salamander ambystoma mexicanum using shotgun and laser capture chromosome sequencing. Sci. Rep. 2015, 5, 16413. [Google Scholar] [CrossRef]
- Cleland, R. The cytogenetics of oenothera. Adv. Genet. 1963, 11, 147–237. [Google Scholar]
- Syren, R.M.; Luykx, P. Permanent segmental interchange complex in the termite incisitermes schwarzi. Nature 1977, 266, 167–168. [Google Scholar] [CrossRef]
- Gazoni, T.; Gruber, S.L.; Silva, A.P.; Araujo, O.G.; Narimatsu, H.; Strussmann, C.; Haddad, C.F.; Kasahara, S. Cytogenetic analyses of eight species in the genus leptodactylus fitzinger, 1843 (amphibia, anura, leptodactylidae), including a new diploid number and a karyotype with multiple translocations. BMC Genet. 2012, 13, 109. [Google Scholar] [CrossRef] [PubMed]
- Grutzner, F.; Rens, W.; Tsend-Ayush, E.; El-Mogharbel, N.; O’Brien, P.C.; Jones, R.C.; Ferguson-Smith, M.A.; Marshall Graves, J.A. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird z and mammal x chromosomes. Nature 2004, 432, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Rens, W.; O’Brien, P.C.; Grutzner, F.; Clarke, O.; Graphodatskaya, D.; Tsend-Ayush, E.; Trifonov, V.A.; Skelton, H.; Wallis, M.C.; Johnston, S.; et al. The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian z. Genome Biol. 2007, 8, R243. [Google Scholar] [CrossRef] [PubMed]
- Ashley, T. Chromosome chains and platypus sex: Kinky connections. Bioessays 2005, 27, 681–684. [Google Scholar] [CrossRef]
- Eaker, S.; Pyle, A.; Cobb, J.; Handel, M.A. Evidence for meiotic spindle checkpoint from analysis of spermatocytes from robertsonian-chromosome heterozygous mice. J. Cell Sci. 2001, 114, 2953–2965. [Google Scholar]
- Johannisson, R.; Winking, H. Synaptonemal complexes of chains and rings in mice heterozygous for multiple robertsonian translocations. Chromosome Res. 1994, 2, 137–145. [Google Scholar] [CrossRef]
- Cenci, G.; Ciapponi, L.; Gatti, M. The mechanism of telomere protection: A comparison between drosophila and humans. Chromosoma 2005, 114, 135–145. [Google Scholar] [CrossRef]
- Cenci, G.; Siriaco, G.; Raffa, G.D.; Kellum, R.; Gatti, M. The drosophila hoap protein is required for telomere capping. Nat. Cell Biol. 2003, 5, 82–84. [Google Scholar] [CrossRef]
- Iglesias, N.; Moazed, D. Silencing repetitive DNA. eLife 2017, 6, e29503. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timoshevskiy, V.A.; Timoshevskaya, N.Y.; Smith, J.J. Germline-Specific Repetitive Elements in Programmatically Eliminated Chromosomes of the Sea Lamprey (Petromyzon marinus). Genes 2019, 10, 832. https://doi.org/10.3390/genes10100832
Timoshevskiy VA, Timoshevskaya NY, Smith JJ. Germline-Specific Repetitive Elements in Programmatically Eliminated Chromosomes of the Sea Lamprey (Petromyzon marinus). Genes. 2019; 10(10):832. https://doi.org/10.3390/genes10100832
Chicago/Turabian StyleTimoshevskiy, Vladimir A., Nataliya Y. Timoshevskaya, and Jeramiah J. Smith. 2019. "Germline-Specific Repetitive Elements in Programmatically Eliminated Chromosomes of the Sea Lamprey (Petromyzon marinus)" Genes 10, no. 10: 832. https://doi.org/10.3390/genes10100832
APA StyleTimoshevskiy, V. A., Timoshevskaya, N. Y., & Smith, J. J. (2019). Germline-Specific Repetitive Elements in Programmatically Eliminated Chromosomes of the Sea Lamprey (Petromyzon marinus). Genes, 10(10), 832. https://doi.org/10.3390/genes10100832




