The Role of the Late Embryogenesis-Abundant (LEA) Protein Family in Development and the Abiotic Stress Response: A Comprehensive Expression Analysis of Potato (Solanum Tuberosum)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Late Embryogenesis-Abundant (LEA) Genes in Potato Genomes
2.2. Distribution of LEA Genes on Potato Chromosomes
2.3. Structural Characterisation of Potato LEA
2.4. Phylogenetic Analysis of Solanum Tuberosum LEA (StLEA)
2.5. Expression Profile Analysis of Potato LEA Genes
2.6. Quantitative Real-Time Polymerase Chain Reaction Analysis of StLEA Proteins
3. Results
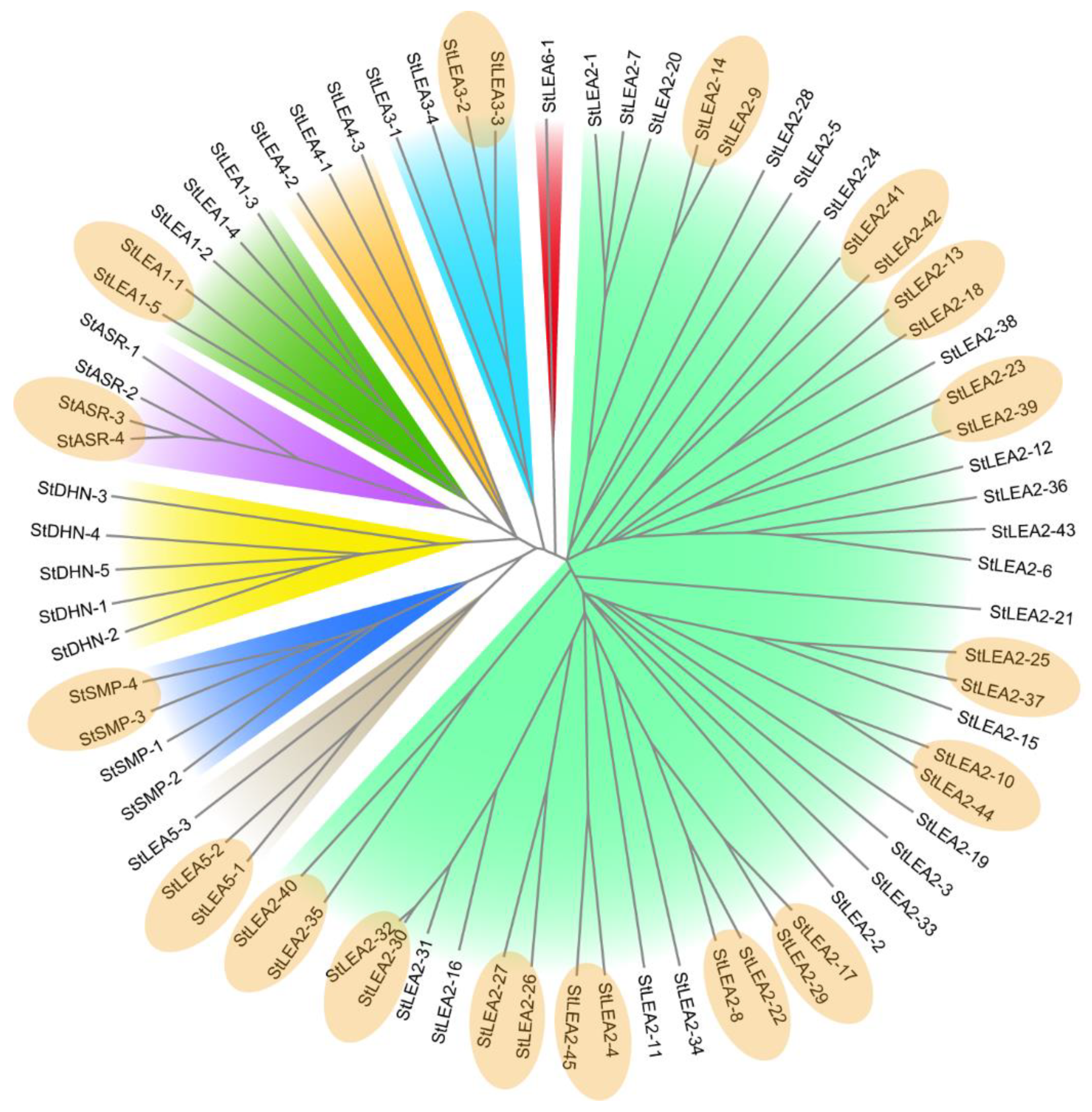
3.1. Genome-Wide Identification and Phylogenetic Analysis of Potato LEA Genes
3.2. Structural Characterisation of Potato LEA
3.3. Chromosomal Location and Duplication of Potato LEA Genes
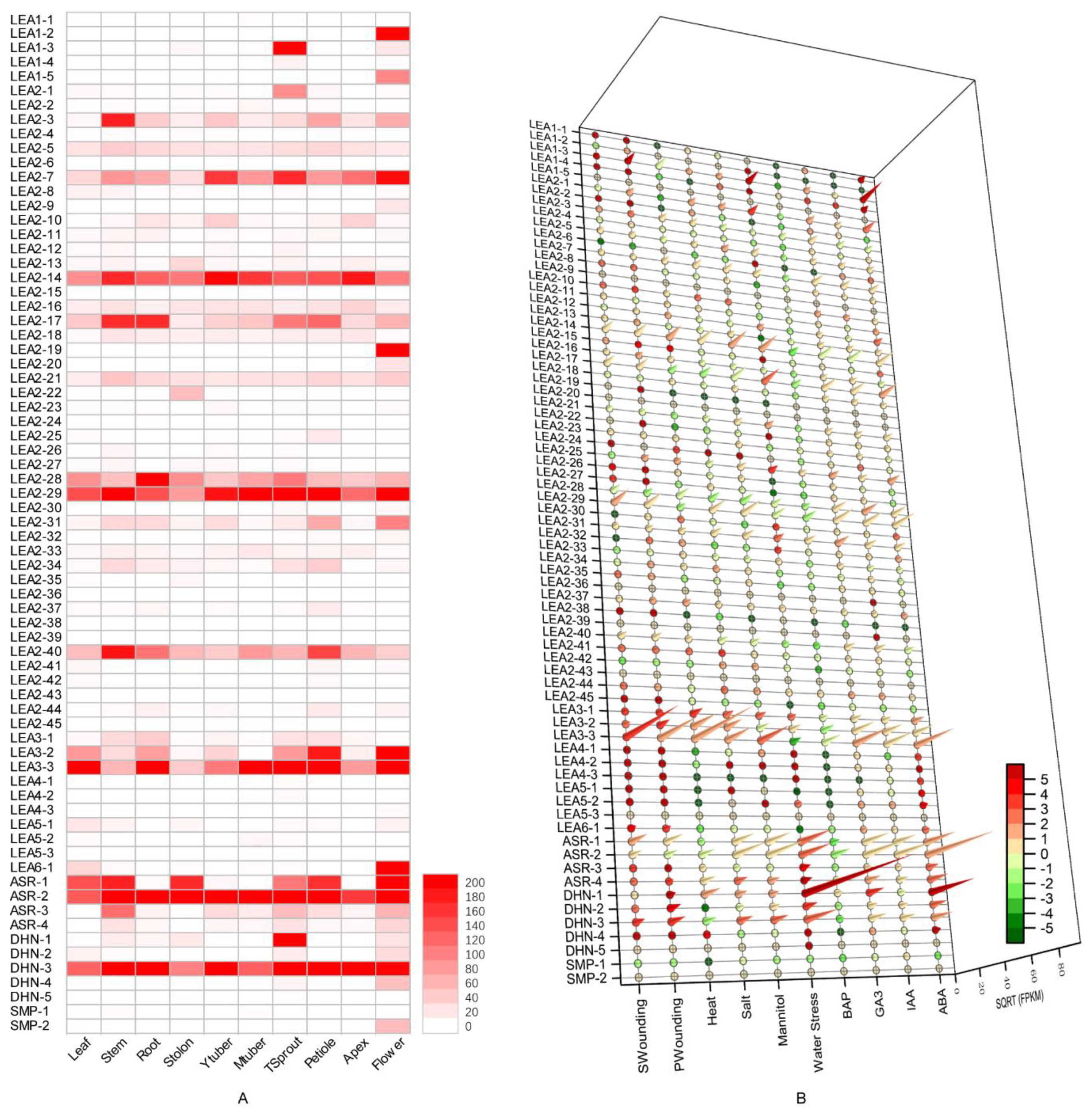
3.4. Expression Profile Analysis of Potato LEA Genes in Different Tissues
3.5. Effect of Exogenous Hormone Treatment on Potato LEA Expression
3.6. Expression Profiles of Potato LEA Genes under Abiotic Stresses
3.7. qRT-PCR Analysis of Potato LEA Genes under Abiotic Stresses
4. Discussion
4.1. Molecular Characteristics and Evolution of the Potato LEA Gene Family
4.2. Expression and Function of Potato LEA in Response to Abiotic Stress
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lan, T.; Gao, J.; Zeng, Q.Y. Genome-wide analysis of the LEA (late embryogenesis abundant) protein gene family in Populus trichocarpa. Tree Genet. Genomes 2013, 9, 253–264. [Google Scholar] [CrossRef]
- Huang, A.; Zhong, X.J.; He, J.; Jin, S.H.; Guo, H.D.; Yu, X.F.; Zhou, Y.J.; Li, X.; Ma, M.D.; Chen, Q.B.; et al. Genome-wide identification, characterization, and stress-responsive expression profiling of genes encoding LEA (late embryogenesis abundant) proteins in Moso bamboo (Phyllostachys edulis). PLoS ONE 2016, 1, e0165953. [Google Scholar] [CrossRef] [PubMed]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Lu, H.; Wang, X.; Cai, X.; Zhou, Z.; Zhang, Z.; Salih, H.; Wang, K.; et al. Characterization of the late embryogenesis abundant (LEA) proteins family and their role in drought stress tolerance in upland cotton. BMC Genet. 2018, 19, 6. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.; Covarrubias, A.A. Late embryogenesis abundant (LEA) proteins in legumes. Front. Plant Sci. 2013, 4, 190. [Google Scholar] [CrossRef]
- Gao, J.; Lan, T. Functional characterization of the late embryogenesis abundant (LEA) protein gene family from Pinus tabuliformis (Pinaceae) in Escherichia coli. Sci. Rep. 2016, 6, 19467. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Zeng, X.; Guo, S. Functional insights into the late embryogenesis abundant (LEA) protein family from Dendrobium officinale (Orchidaceae) using an Escherichia coli system. Sci. Rep. 2016, 6, 39693. [Google Scholar] [CrossRef] [PubMed]
- Hunault, G.; Jaspard, E. LEAPdb: A database for the late embryogenesis abundant proteins. BMC Genom. 2010, 11, 221. [Google Scholar] [CrossRef] [PubMed]
- González, R.M.; Iusem, N.D. Twenty years of research on Asr, (ABA-stress-ripening) genes and proteins. Planta 2014, 239, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Wetzler, D.E.; Wightman, F.F.; Bucci, H.A.; Rinaldi, J.; Caramelo, J.J.; Iusem, N.D.; Ricardi, M.M. Conformational plasticity of the intrinsically disordered protein ASR1 modulates its function as a drought stress-responsive gene. PLoS ONE 2018, 13, e0202808. [Google Scholar] [CrossRef]
- Wang, X.S.; Zhu, H.B.; Jin, G.L.; Liu, H.L.; Wu, W.R.; Zhu, J. Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.). Plant Sci. 2007, 172, 414–420. [Google Scholar] [CrossRef]
- Choi, D.W.; Zhu, B.; Close, T.J. The barley (Hordeum vulgare L.) dehydrin multigene family: Sequences, allele types, chromosome assignments, and expression characteristics of 11 Dhn genes of cv Dicktoo. Theor. Appl. Genet. 2007, 98, 1234–1247. [Google Scholar] [CrossRef]
- Hundertmark, M.; Hincha, D.K. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Zhang, Q.; Cheng, T.; Pan, H.; Yang, W.; Sun, L. Genome-wide identification and analysis of late embryogenesis abundant (LEA) genes in Prunus mume. Mol. Biol. Rep. 2013, 40, 1937–1946. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Xia, H.; Wu, S.; Ma, F.W. Genome-wide identification and expression profiling of dehydrin gene family in Malus domestica. Mol. Biol. Rep. 2012, 39, 10759–10768. [Google Scholar] [CrossRef]
- Cao, J.; Li, X. Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum). Planta 2015, 241, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, R.; Gusev, O.; Cornette, R.; Shimura, S.; Kikuta, S.; Okada, J.; Okuda, T. Diversity of the expression profiles of late embryogenesis abundant (LEA) protein encoding genes in the anhydrobiotic midge Polypedilum vanderplanki. Planta 2015, 242, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Olvera-Carrillo, Y.; Campos, F.; Reyes, L.; Garciarrubio, A.; Covarrubias, A.A. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol. 2010, 154, 373–390. [Google Scholar] [CrossRef]
- Tolleter, D.; Hincha, D.K.; Macherel, D. A mitochondrial late embryogenesis abundant protein stabilizes model membranes in the dry state. BBA-Mol. Basis Dis. 2010, 1798, 1926–1933. [Google Scholar] [CrossRef] [Green Version]
- Charfeddine, S.; Saïdi, M.N.; Charfeddine, M.; Gargouri-Bouzid, R. Genome-wide identification and expression profiling of the late embryogenesis abundant genes in potato with emphasis on dehydrins. Mol. Biol. Rep. 2015, 42, 1163–1174. [Google Scholar] [CrossRef]
- Zamora-Briseño, J.A.; de Jiménez, E.S. A LEA 4 protein up-regulated by ABA is involved in drought response in maize roots. Mol. Biol. Rep. 2016, 43, 221–228. [Google Scholar] [CrossRef]
- Jeon, O.S.; Kim, C.S.; Lee, S.P.; Kang, S.K.; Kim, C.M.; Kang, B.G.; Hur, Y.; Kim, I.J. Fruit ripening-related expression of a gene encoding group 5 late embryogenesis abundant protein in Citrus. J. Plant Biol. 2006, 49, 403–408. [Google Scholar] [CrossRef]
- Kim, I.J.; Lee, J.; Han, J.A.; Kim, C.S.; Hur, Y. Citrus Lea promoter confers fruit-preferential and stress-inducible gene expression in Arabidopsis. Can. J. Plant Sci. 2011, 91, 459–466. [Google Scholar] [CrossRef]
- Zhao, Q.; Liu, J.; Xu, X.; Xu, Q.; Gao, W.; Xu, J. PicW orthologs from spruce with differential freezing tolerance expressed in Escherichia coli. Int. J. Biol. Macromol. 2017, 101, 595–602. [Google Scholar] [CrossRef]
- Zhang, H.; Shi, Y.; Liu, X.; Wang, R.; Li, J.; Xu, J. Transgenic creeping bentgrass plants expressing a Picea wilsonii dehydrin gene (PicW) demonstrate improved freezing tolerance. Mol. Biol. Rep. 2018, 45, 1627–1635. [Google Scholar] [CrossRef] [PubMed]
- De Jong, H. Impact of the potato on society. Am. J. Potato Res. 2016, 93, 415–429. [Google Scholar] [CrossRef]
- Iwama, K.; Yamaguchi, J. Abiotic stresses. In Handbook of Potato Production, Improvement and Post-Harvest Management; Gopal, J., Khurana, S.M.P., Eds.; Food Product Press: New York, NY, USA, 2006; pp. 231–278. [Google Scholar]
- Cabello, R.; Monneveux, P.; de Mendiburu, F.; Bonierbale, M. Comparison of yield based drought tolerance indices in improved varieties, genetic stocks and landraces of potato (Solanum tuberosum L.). Euphytica 2013, 193, 147–156. [Google Scholar] [CrossRef]
- Rykaczewska, K. Impact of heat and drought stresses on size and quality of the potato yield. Plant Soil Environ. 2017, 63, 40–46. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef]
- Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.H. Importance of lineage specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008, 148, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Cavalcanti, A.; Chen, F.C.; Bouman, P.; Li, W.H. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef] [PubMed]
- George, T.S.; Taylor, M.A.; Dodd, I.C.; White, P.J. Climate change and consequences for potato production: A review of tolerance to emerging abiotic stress. Potato Res. 2017, 60, 239–268. [Google Scholar] [CrossRef]
- Bies-Ethève, N.; Gaubier-Comella, P.; Debures, A.; Lasserre, E.; Jobet, E.; Raynal, M.; Cooke, R.; Delseny, M. Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol. 2008, 67, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Xiong, Z.; Zheng, J.; Xu, D.; Zhu, Z.; Xiang, J.; Gan, J.; Raboanatahiry, N.; Yin, Y.; Li, M. Genome-wide identification, structural analysis and new insights into late embryogenesis abundant (LEA) gene family formation pattern in Brassica napus. Sci. Rep. 2016, 6, 24265. [Google Scholar] [CrossRef]
- Xie, D.W.; Wang, X.N.; Fu, L.S.; Sun, J.; Zheng, W.; Li, Z.F. Identification of the trehalose-6-phosphate synthase gene family in winter wheat and expression analysis under conditions of freezing stress. J. Genet. 2015, 94, 55–65. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef]
- Jue, D.; Sang, X.; Lu, S.; Dong, C.; Zhao, Q.; Chen, H.; Jia, L. Genome-wide identification, phylogenetic and expression analyses of the ubiquitin-conjugating enzyme gene family in maize. PLoS ONE 2015, 10, e0143488. [Google Scholar] [CrossRef]
- Shimizu, T.; Kanamori, Y.; Furuki, T.; Kikawada, T.; Okuda, T.; Takahashi, T.; Mihara, H.; Sakurai, M. Desiccation-induced structuralization and glass formation of group 3 late embryogenesis abundant protein model peptides. Biochemistry 2010, 49, 1093–1104. [Google Scholar] [CrossRef] [PubMed]
- Caramelo, J.J.; Iusem, N.D. When cells lose water: Lessons from biophysics and molecular biology. Prog. Biophys. Mol. Biol. 2009, 99, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Majumder, A.L. Insight into the salt tolerance factors of a wild halophytic rice, Porteresia coarctata: A physiological and proteomic approach. Planta 2009, 229, 911–929. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Lai, Y.; Wu, X.; Wu, G.; Guo, C. Overexpression of OsEm1 encoding a group I LEA protein confers enhanced drought tolerance in rice. Biochem. Biophys. Res. Commun. 2016, 478, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, Y.X.; Wei, W.; Han, L.; Guan, Z.Q.; Wang, Z.; Chai, T.Y. BjDHNs confer heavy-metal tolerance in plants. Mol. Biotechnol. 2008, 38, 91–98. [Google Scholar] [CrossRef] [PubMed]



| Spud ID | Name | Amino Acid Number | MW | pI | Instability Index | Aliphatic Index | GRAVY |
|---|---|---|---|---|---|---|---|
| PGSC0003DMG400002093 | LEA1-1 | 123 | 13,268.01 | 9.4 | 48 | 71.54 | −0.715 |
| PGSC0003DMG400011437 | LEA1-2 | 96 | 10,361.49 | 9.22 | 28.3 | 38.85 | −1.161 |
| PGSC0003DMG400011438 | LEA1-3 | 88 | 9261.33 | 9.22 | 17.2 | 30.34 | −1.116 |
| PGSC0003DMG400011439 | LEA1-4 | 271 | 26,628 | 8.19 | 3.02 | 44.06 | −0.589 |
| PGSC0003DMG400001819 | LEA1-5 | 104 | 11,515.89 | 5.94 | 29.76 | 65.96 | −0.828 |
| PGSC0003DMG400000066 | LEA2-1 | 163 | 17,950.67 | 4.74 | 20.54 | 96.2 | −0.153 |
| PGSC0003DMG400002090 | LEA2-2 | 191 | 22,310.06 | 9.47 | 31.39 | 102.88 | 0.103 |
| PGSC0003DMG400002586 | LEA2-3 | 267 | 29,063.08 | 10.27 | 36.24 | 83 | −0.136 |
| PGSC0003DMG400005073 | LEA2-4 | 227 | 25,846.5 | 9.93 | 52.17 | 82.03 | −0.292 |
| PGSC0003DMG400005685 | LEA2-5 | 221 | 25,174.21 | 9.06 | 35.71 | 103.62 | −0.015 |
| PGSC0003DMG400005783 | LEA2-6 | 235 | 26,022.56 | 9.87 | 43.17 | 100.72 | −0.039 |
| PGSC0003DMG400000067 | LEA2-7 | 175 | 19,281.12 | 4.57 | 33.54 | 101.83 | −0.189 |
| PGSC0003DMG400005898 | LEA2-8 | 208 | 23,404.16 | 9.08 | 48.26 | 103.51 | 0.089 |
| PGSC0003DMG400006460 | LEA2-9 | 320 | 35,591.61 | 5.34 | 26.71 | 89.47 | −0.463 |
| PGSC0003DMG400008711 | LEA2-10 | 270 | 29,709.53 | 9.77 | 45.43 | 87.63 | −0.15 |
| PGSC0003DMG400011296 | LEA2-11 | 228 | 25,291.92 | 9.69 | 50.24 | 105.88 | 0.133 |
| PGSC0003DMG400000069 | LEA2-12 | 223 | 24,906.2 | 9.68 | 19.77 | 94.71 | −0.111 |
| PGSC0003DMG400012142 | LEA2-13 | 197 | 21,631.87 | 9.1 | 35.79 | 85.53 | 0.197 |
| PGSC0003DMG400013715 | LEA2-14 | 320 | 35,609.51 | 4.83 | 24.23 | 96.19 | −0.41 |
| PGSC0003DMG400015214 | LEA2-15 | 258 | 29,131.51 | 10.1 | 36.09 | 102.33 | −0.092 |
| PGSC0003DMG400016390 | LEA2-16 | 229 | 26,053.18 | 8.97 | 43.15 | 77.42 | −0.154 |
| PGSC0003DMG400016420 | LEA2-17 | 211 | 23,915.69 | 9.44 | 40.96 | 88.67 | −0.132 |
| PGSC0003DMG400000115 | LEA2-18 | 198 | 21,541.04 | 9.69 | 20.03 | 85 | 0.171 |
| PGSC0003DMG400018355 | LEA2-19 | 224 | 25,280.38 | 8.72 | 49.75 | 87.95 | −0.169 |
| PGSC0003DMG400019407 | LEA2-20 | 175 | 19,541.28 | 4.79 | 25.24 | 87.94 | −0.362 |
| PGSC0003DMG400019632 | LEA2-21 | 257 | 28,453.37 | 10.19 | 46.62 | 72.18 | −0.325 |
| PGSC0003DMG400019639 | LEA2-22 | 209 | 23,687.5 | 9.36 | 38.63 | 108.56 | 0.115 |
| PGSC0003DMG400020863 | LEA2-23 | 204 | 22,542.59 | 9.53 | 34.85 | 118.43 | 0.245 |
| PGSC0003DMG400020886 | LEA2-24 | 204 | 23,102.85 | 9.7 | 34.46 | 87.94 | −0.19 |
| PGSC0003DMG400021454 | LEA2-25 | 259 | 29,397.61 | 10.06 | 40.41 | 92.12 | −0.344 |
| PGSC0003DMG400000476 | LEA2-26 | 209 | 24,030.01 | 10.05 | 40.72 | 98.76 | −0.191 |
| PGSC0003DMG400024324 | LEA2-27 | 193 | 22,002.62 | 9.83 | 36.55 | 103.52 | −0.051 |
| PGSC0003DMG400026335 | LEA2-28 | 107 | 12,026.84 | 6.58 | 43.24 | 93.74 | −0.077 |
| PGSC0003DMG400028151 | LEA2-29 | 211 | 24,106.86 | 9.59 | 52.84 | 91.8 | −0.153 |
| PGSC0003DMG400028152 | LEA2-30 | 225 | 26,077.18 | 9.29 | 42.62 | 86.18 | −0.279 |
| PGSC0003DMG400028153 | LEA2-31 | 230 | 26,121.29 | 9.11 | 46.83 | 89 | −0.219 |
| PGSC0003DMG400028235 | LEA2-32 | 225 | 26,126.21 | 9.3 | 43.04 | 86.62 | −0.318 |
| PGSC0003DMG400029777 | LEA2-33 | 227 | 24,670.78 | 9.41 | 28.14 | 105.15 | 0.257 |
| PGSC0003DMG400000583 | LEA2-34 | 213 | 23,996.84 | 9.41 | 31.45 | 90.56 | −0.098 |
| PGSC0003DMG400032803 | LEA2-35 | 302 | 34,188.98 | 9.11 | 42.42 | 86.46 | −0.198 |
| PGSC0003DMG400037897 | LEA2-36 | 235 | 26,782.17 | 9.59 | 51.48 | 95.7 | −0.196 |
| PGSC0003DMG401019715 | LEA2-37 | 260 | 29,590.68 | 9.57 | 46.77 | 91.08 | −0.217 |
| PGSC0003DMG401021419 | LEA2-38 | 192 | 21,050.61 | 9.53 | 25.77 | 100.05 | 0.177 |
| PGSC0003DMG401027691 | LEA2-39 | 214 | 24,317.33 | 9.55 | 40.92 | 92.94 | −0.204 |
| PGSC0003DMG402002623 | LEA2-40 | 310 | 34,258.25 | 9.75 | 55.92 | 76.03 | −0.324 |
| PGSC0003DMG400012315 | LEA2-41 | 197 | 22,684.33 | 10.42 | 52.16 | 90.56 | −0.23 |
| PGSC0003DMG400012355 | LEA2-42 | 223 | 25,274 | 9.68 | 33.41 | 87.4 | −0.217 |
| PGSC0003DMG400042041 | LEA2-43 | 101 | 11,316.65 | 10.02 | 18.01 | 107.03 | 0.242 |
| PGSC0003DMG400002014 | LEA2-44 | 263 | 29,204.88 | 9.97 | 43.83 | 87 | −0.222 |
| PGSC0003DMG400002082 | LEA2-45 | 222 | 25,335.22 | 9.51 | 41.89 | 91.76 | −0.106 |
| PGSC0003DMG400004703 | LEA3-1 | 97 | 10,887.16 | 9.2 | 41.7 | 57.22 | −0.903 |
| PGSC0003DMG400017936 | LEA3-2 | 89 | 9786.03 | 9.66 | 45.73 | 66.85 | −0.473 |
| PGSC0003DMG400031788 | LEA3-3 | 98 | 10,402.73 | 9.89 | 51.67 | 73.67 | −0.351 |
| Sotub08g019440.1.1 | LEA3-4 | 84 | 9575.08 | 9.66 | 49.98 | 65 | −0.554 |
| PGSC0003DMG400002731 | LEA4-1 | 400 | 43,122.45 | 5.46 | 26.56 | 51 | −1.093 |
| PGSC0003DMG400019976 | LEA4-2 | 501 | 53,302.67 | 5.32 | 29.5 | 57.56 | −0.816 |
| PGSC0003DMG400029706 | LEA4-3 | 255 | 28,437.98 | 8.84 | 39.03 | 47.61 | −1.112 |
| PGSC0003DMG400006648 | LEA5-1 | 93 | 10,011.84 | 5.91 | 55.08 | 40.97 | −1.446 |
| PGSC0003DMG400008497 | LEA5-2 | 100 | 10,855.84 | 9.1 | 60.31 | 38.1 | −1.469 |
| PGSC0003DMG400041241 | LEA5-3 | 105 | 11,740.47 | 10.07 | 68.21 | 68.67 | −1.017 |
| PGSC0003DMG400024942 | LEA6-1 | 88 | 9221.92 | 5.11 | 42.02 | 46.7 | −1.125 |
| PGSC0003DMG400003530 | DHN-1 | 140 | 14,534.01 | 7.07 | 34.82 | 15.36 | −1.268 |
| PGSC0003DMG400003531 | DHN-2 | 157 | 16,659.06 | 7.23 | 21.86 | 36.69 | −1.214 |
| PGSC0003DMG400009968 | DHN-3 | 209 | 23,673.14 | 5.24 | 63.91 | 47.56 | −1.499 |
| PGSC0003DMG400015495 | DHN-4 | 80 | 8544.27 | 5.9 | 29.91 | 37.88 | −1.31 |
| PGSC0003DMG400030949 | DHN-5 | 243 | 25,121.94 | 7.38 | 23.94 | 59.92 | −0.715 |
| PGSC0003DMG400019328 | SMP-1 | 257 | 26,335.96 | 4.47 | 40.16 | 80.54 | −0.251 |
| PGSC0003DMG400022470 | SMP-2 | 206 | 21,737.03 | 6.78 | 48.15 | 74.03 | −0.729 |
| Sotub09g023980.1.1 | SMP-3 | 259 | 26,563.44 | 4.7 | 41.15 | 74.09 | −0.353 |
| Sotub09g023990.1.1 | SMP-4 | 263 | 25,839.64 | 4.62 | 35.47 | 73.57 | −0.184 |
| PGSC0003DMG400006661 | ASR-1 | 306 | 33,956.66 | 4.92 | 28.91 | 19.87 | −1.629 |
| PGSC0003DMG400006662 | ASR-2 | 109 | 12,370.74 | 6.57 | 40.22 | 54.86 | −1.294 |
| PGSC0003DMG400006663 | ASR-3 | 111 | 12,481.95 | 9.25 | 41.47 | 52.97 | −1.31 |
| PGSC0003DMG400024093 | ASR-4 | 108 | 12,158.49 | 6.65 | 46.37 | 52.59 | −1.235 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Li, C.; Zhang, B.; Yi, J.; Yang, Y.; Kong, C.; Lei, C.; Gong, M. The Role of the Late Embryogenesis-Abundant (LEA) Protein Family in Development and the Abiotic Stress Response: A Comprehensive Expression Analysis of Potato (Solanum Tuberosum). Genes 2019, 10, 148. https://doi.org/10.3390/genes10020148
Chen Y, Li C, Zhang B, Yi J, Yang Y, Kong C, Lei C, Gong M. The Role of the Late Embryogenesis-Abundant (LEA) Protein Family in Development and the Abiotic Stress Response: A Comprehensive Expression Analysis of Potato (Solanum Tuberosum). Genes. 2019; 10(2):148. https://doi.org/10.3390/genes10020148
Chicago/Turabian StyleChen, Yongkun, Canhui Li, Bo Zhang, Jing Yi, Yu Yang, Chunyan Kong, Chunxia Lei, and Ming Gong. 2019. "The Role of the Late Embryogenesis-Abundant (LEA) Protein Family in Development and the Abiotic Stress Response: A Comprehensive Expression Analysis of Potato (Solanum Tuberosum)" Genes 10, no. 2: 148. https://doi.org/10.3390/genes10020148
APA StyleChen, Y., Li, C., Zhang, B., Yi, J., Yang, Y., Kong, C., Lei, C., & Gong, M. (2019). The Role of the Late Embryogenesis-Abundant (LEA) Protein Family in Development and the Abiotic Stress Response: A Comprehensive Expression Analysis of Potato (Solanum Tuberosum). Genes, 10(2), 148. https://doi.org/10.3390/genes10020148





