Abstract
Cells have developed numerous adaptation mechanisms to external cues by controlling signaling-pathway activity, both qualitatively and quantitatively. The Wnt/β-catenin pathway is a highly conserved signaling pathway involved in many biological processes, including cell proliferation, differentiation, somatic cell reprogramming, development, and cancer. The activity of the Wnt/β-catenin pathway and the temporal dynamics of its effector β-catenin are tightly controlled by complex regulations. The latter encompass feedback loops within the pathway (e.g., a negative feedback loop involving Axin2, a β-catenin transcriptional target) and crosstalk interactions with other signaling pathways. Here, we provide a review shedding light on the coupling between Wnt/β-catenin activation levels and fluctuations across processes and cellular systems; in particular, we focus on development, in vitro pluripotency maintenance, and cancer. Possible mechanisms originating Wnt/β-catenin dynamic behaviors and consequently driving different cellular responses are also reviewed, and new avenues for future research are suggested.
1. Introduction
Wnt proteins are key mediators of cell specification and patterning in development, adult tissue homeostasis, and stemness [1]. Mutations of Wnt pathway components can cause a wide range of diseases, including congenital disorders [2,3,4,5,6,7,8,9,10,11,12,13] and cancer [10,14,15,16,17,18,19,20,21,22,23,24]. Wnts, conserved in all metazoan animals, can trigger the activation of two distinct signaling pathways, known as “canonical” and “noncanonical” [25]. The most studied Wnt pathway, and the focus of this review, is the canonical Wnt signaling; β-catenin, the pathway key effector and transcriptional coactivator, mediates canonical Wnt pathway functions. A detailed review about noncanonical Wnt signaling can be found in Reference [26].
β-catenin has a central role in directing diverse intracellular functions. It is involved in cell–cell adhesion through interaction with the E-cadherin cell-adhesion complex and the microtubule network [27,28,29,30,31,32,33,34], and can also trigger gene expression in complex with T-cell transcription factor/lymphocyte enhancer factor (TCF/LEF) family members [35,36,37,38,39,40,41,42,43,44].
The amount of β-catenin protein not complexed with E-cadherin is buffered by the destruction complex. The latter is a multiprotein complex; it consists of scaffolding proteins Axin, tumor suppressor APC, the serine–threonine kinases glycogen synthase kinase 3 (GSK3β), and the casein kinase 1 (CK1), and it is responsible for cytosolic β-catenin phosphorylation, ubiquitination, and degradation [45,46,47,48,49,50,51,52,53,54,55]. Upstream regulations of the destruction complex result in different accumulation of β-catenin in the cytosol.
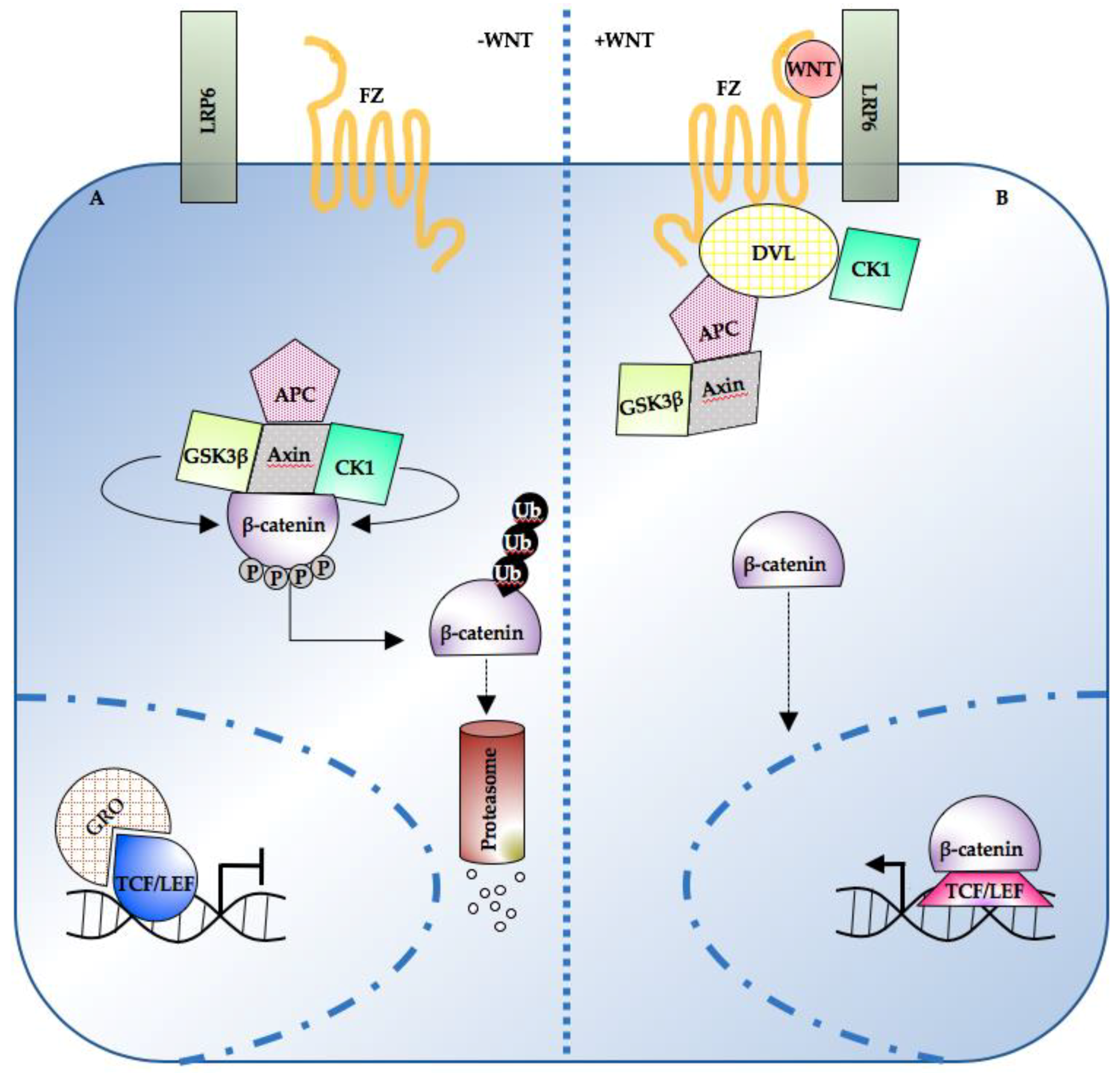
Meanwhile, nuclear TCF proteins associate with transcriptional repressors such as Groucho (Gro) and inhibit the expression of β-catenin target genes [56,57,58] (Figure 1A). When a Wnt ligand (such as Wnt-3a) binds to the seven-pass transmembrane receptor Frizzled (Fz) and its coreceptor, the low-density lipoprotein receptor-related protein 6 (LRP6), the signaling pathway is activated. Axin is recruited to the membrane and the PDZ-containing protein Dishevelled (DVL) is phosphorylated [59,60,61]. β-catenin is then released from the destruction complex; no longer exposed to kinase-mediated phosphorylation, it accumulates and can translocate to the nucleus. Therein, β-catenin has a dual function: it both displaces the transcriptional repressor complex from the DNA and, binding to TCF1 [62], LEF1 [38,43,63,64,65] and TCF4 [57,66,67], it activates the transcription of various target genes (Figure 1B). The latter, in turn, controls genes relevant for cell proliferation [68], stemness [69,70,71] and differentiation [72] (an updated list can be found online on the Wnt homepage, http://web.stanford.edu/group/nusselab/cgi-bin/wnt/).
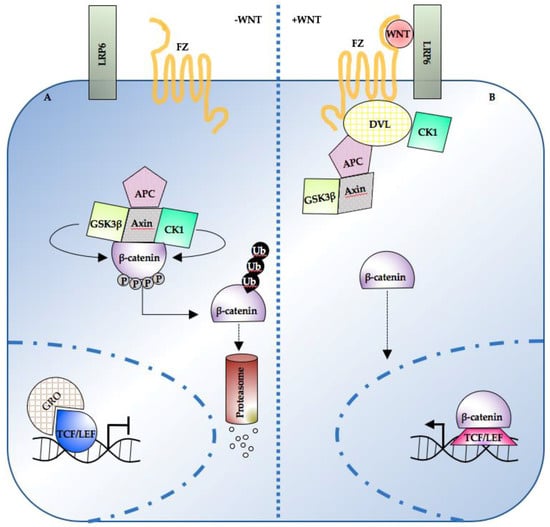
Figure 1.
Overview of the Wnt/β-catenin pathway topology. (A) In the absence of WNT, cytosolic β-catenin is sequestered by the destruction complex and degraded following multiple rounds of phosphorylation and ubiquitination. Low nuclear β-catenin enables the TCF/LEF-mediated repression of target genes. (B) Following WNT ligand stimulation, the destruction complex is inhibited, and β-catenin accumulates. Nuclear β-catenin displaces the repressive complex from the DNA and drives target gene expression in co-operation with TCF/LEF transcription factors. TCF/LEF: T-cell transcription factor/lymphocyte enhancer factor; APC: adenomatous polyposis coli; CK1: casein kinase 1; DVL: dishevelled; FZ: frizzled; GSK3β: glycogen synthase kinase 3; LRP6: low-density lipoprotein receptor-related protein 6; Ub: ubiquitin; GRO: Groucho.
The ability of the Wnt/β-catenin pathway to co-ordinate cell fate and homeostasis during development and in adult tissue has been extensively studied both in vitro and in vivo (reviewed in Reference [73]); furthermore, being the pathway misregulated in various diseases, including cancer and diabetes, various humanized antibodies and small molecules to antagonize its activity have been developed [37].
In this review, we focus on the specific dose- and dynamic-dependent Wnt/β-catenin signaling roles across systems and organisms.
2. Wnt/β-Catenin Pathway Levels, Dynamics, and Spatial Organization in Development
In vivo embryogenesis relies on the interplay of signaling cascades to activate tissue- and organism-specific differentiation programs.
Multicellular-organism development relies on gene-expression patterns and on anterior–posterior (AP) polarity dictated by cellular positioning [74]. The Wnt pathway is one of the signaling pathways involved in the establishment of the AP axis and in supporting proper tissue development [75,76]. Mice lacking functional β-catenin protein are unable to undergo normal gastrulation [77]. Interestingly, excessive β-catenin accumulation is also associated with developmental defects, resulting in misregulated mesodermal differentiation [78]. These results suggest that the levels of β-catenin might need to be controlled and maintained within a certain threshold to properly support development. In this regard, Kemler and colleagues demonstrated that β-catenin levels vary during mouse development, and control all phases of embryogenesis [78,79]. Taking advantage of a conditional β-catenin allele and the ubiquitously active ROSA26 promoter, and generating different Cre lines, the authors showed a clear correlation between protein dosage and developmental progress in postgastrulation embryos, with different types of tissue requiring specific levels of β-catenin [79]. In mice, the Wnt/β-catenin pathway is fundamental for the proliferation of early pancreatic progenitor cells following cellular specification, and supports the undifferentiated state of pancreatic precursors; pancreas-selective β-catenin depletion results in a reduced number of pancreatic islets [80,81,82,83].
In Xenopus, low or high β-catenin accumulation levels in the anterior/posterior endoderm have been shown to maintain a foregut fate or to promote intestinal development, respectively [56,84]. In zebrafish, a key role for timing Wnt/β-catenin pathway activation was reported: upstream-pathway component APC was shown to rely on retinoic acid biosynthesis, and to regulate the transition from early gut endoderm into differentiated epithelial cells specifically postfertilization [85].
The importance of controlled β-catenin protein levels is also evident in murine and human hair-follicle homeostasis, where protein absence or overexpression results in inappropriate differentiation and/or tumor initiation [86,87]. Hair follicles rapidly form within the first two weeks after birth. Epithelial cells first massively proliferate and then differentiate, culminating into a final arrested phase known as telogen. In telogen, the epidermal bulge still retains stem cells that undergo several regenerative cycles during the lifespan [88,89]. In 1994, van Genderen and colleagues reported a detailed phenotypic analysis of the LEF1 transcription factor during development; using a targeted gene-inactivation approach, they demonstrated that different organs, which normally express LEF1 during development, were affected. The most severe phenotype was observed in the mammary gland, the teeth, whiskers, body hair, and the mesencephalic nucleus of the trigeminal nerve (TMN) [90]. Conversely, overexpression of LEF1 was shown to lead to de novo hair-follicle formation, similar to the overexpression of a constitutively stable β-catenin that induced uncontrolled ectopic hair growth and tumorigenesis [91]. Zonal activation of the canonical Wnt pathway was also found in the mesenchyme and epithelium of the developing tooth: using the Wnt/β-catenin pathway reporter Axin2-lacZ, both the epithelium and mesenchyme of the mandible, maxilla and nasal processes, but the frontonasal area showed Axin2 signal [92,93].
More recently, increased β-catenin expression was shown to force the differentiation of the embryonic ectoderm into hair follicles and promote de novo hair-follicle induction in adult skin; on the other hand, β-catenin depletion led to reduced proliferation of epithelial cells and premature catagen (i.e. regression phase prior to telogen) [94]. These observations indicate a temporal “wave” of β-catenin, with high/low levels in the initial/proliferative (and committed) phases, respectively [94]. To tip such balance between proliferation and differentiation [95,96], members of the Wnt family are dynamically expressed in developing hair follicles and skin, and the β-catenin protein itself shows dynamic changes in both accumulation levels and subcellular localization [97,98,99,100,101,102]. β-catenin knockdown experiments showed the canonical Wnt pathway is also important during hair-follicle regeneration; following intradermal injection of β-catenin siRNA into hair-depilated skin, hair growth was delayed of about 40 days [103].
2.1. Somitogenesis
Vertebrae formation starts from cellular precursors in a process known as the segmentation clock [104,105,106,107]; it is an oscillating network controlling the sequential subdivision of the vertebrate embryo elongating the body axis. During this process, somites are progressively formed from the anterior of the presomitic mesoderm (PSM), and elongate to form the body axis [108].
The mutual regulation of various signaling pathways and the resulting gradients and oscillations of molecules guide cell positioning and control somitogenesis [109].
Notch was the first signaling pathway shown to control the process, as the majority of the oscillatory genes are Notch-dependent [110,111,112,113,114,115,116,117,118]. Of note, Notch pathway impairment does not prevent segmentation [119], hinting the involvement of other pathways in somitogenesis. Herrmann’s group was the first reporting about the role of Wnt3a in the murine segmentation clock [119]. They discovered that Axin2, a negative regulator of the Wnt/β-catenin pathway [50,120,121] distributes over the PSM as a gradient and shows oscillatory dynamics in each cycle of somite formation. Axin2 periodic expression in the PSM could be to be due to its rapid and cyclic mRNA degradation, or to periodic production. Considering the topology of the Wnt/β-catenin pathway, the latter hypothesis is more plausible: being a transcriptional target of the canonical Wnt signaling, Axin2 is increased upon pathway activation and, in turns, can reduce pathway activation via its participation to the destruction complex, which reflects on decreased Axin2 transcription via a negative feedback loop [50,120]. Moreover, Axin2, similarly to Axin, might also be destabilized by Wnt signaling [122]. Crosstalk interactions with Notch signaling have been reported: the feedback inhibition of Wnt/β-catenin signaling via Axin2 can trigger Notch target gene activation [123]; thus, Wnt3a stimulation can activate Axin2 expression while inhibiting Notch signaling [119]. Fibroblast growth factor (FGF) signaling has also been observed in the PSM [124,125,126]: Sprouty2 or Dusp6 and Dusp4, all Fgf inhibitors, oscillate in phase with Notch cyclic genes due to further crosstalk interactions between the Notch and FGF pathways [126,127].
Recent in vivo studies from Wilson’s group reported differential levels of Wnt molecules during cell specification. Two subpopulations, both pluripotent, were identified in postimplantation epiblast stem cells (EpiSCs): a partially neuronal-like (Sox1+) fraction, expressing low Wnt/β-catenin levels, and a fraction of progenitor cells, with intermediate activation of the Wnt pathway. Further increase of Wnt/β-catenin signaling activity above a threshold irreversibly promotes mesendodermal and neuromesodermal differentiation [128].
2.2. Colon-Crypt Development and Homeostasis
The intestine has a peculiar functional architecture designed to maximize the available surface for absorbing nutrients and water. Epithelial cells invade the surrounding connective tissue to form tubular glands known as “crypts” [129], which are a reservoir of stem cells (intestinal stem cells, ISCs) supporting intestinal development and epithelium turnover [130,131]. The luminal portion of the mucosa is characterized by villi, fingerlike structures composed of terminally differentiated cells [132].
Intestinal-epithelium regeneration involves a series of events comparable to those taking place during intestinal development at the embryonic stage. ISCs migrate from the bottom of the crypt up to the villi; during this upward migratory process, cells are subjected to different stimuli blocking cell proliferation and promoting differentiation into all cell types required for intestinal functions (enterocytes, secretory goblet cells, Paneth cells, and endoenterocrine cells) [132,133]. In parallel, new crypts are generated through the fission process in order to support consecutive regenerative cycles [134].
The Wnt/β-catenin pathway has been demonstrated to control, in a dose-dependent manner, intestinal epithelium homeostasis in both health and disease. Dickkopf-1 (Dkk-1)-mediated Wnt pathway inhibition and Wnt pathway ablation are detrimental for crypt fission both in vitro and in vivo [135,136], and increased activation of the pathway (i.e., high β-catenin levels) and impaired cell-cell adhesion (i.e., high β-catenin levels and low E-cadherin) can trigger cancer formation [137,138]. In healthy conditions, the Wnt cascade tightly controls ISC overproliferation [139,140,141]: at the crypt base, nuclear β-catenin levels are higher, as well as in Paneth cells (positioned at the bottom of small intestinal crypts), while nuclear Wnt activity progressively decreases up the crypt–villus axis [142]. The resulting expression gradient and the different location of proliferative versus quiescent ISCs cause specific cell responses to Wnt signaling throughout the crypt [143]. Burgess and colleagues, using crypt explants and 3D confocal imaging, reported heterogeneous β-catenin and E-cadherin subcellular localization and patterning within the crypt, and proposed asymmetrical crypt budding in mice [144]; the group observed similar phenotypes in in vitro cultured 3D colonoids [145].
In the crypt, Wnt acts in coordination with other signaling pathways [146], namely: Notch [147] (regulating cell proliferation in the stem-cell niche [148]); Hedgehog (controlling proliferation of the ISC compartment and cell lineage differentiation in both small intestine and colon [149,150,151]); bone morphogenetic proteins (BMPs, belonging to the TGF-β cytokine family and required for intestinal cell-precursor proliferation, maturation, and terminal differentiation [152], and for the prevention of intestinal stem-cell overproliferation through inhibition of the Wnt gradient [146]); and Hippo/YAP (relevant for intestinal tissue regeneration [153]). The complex interaction between these pathways balances the progenitor number in the crypt, as well as their maturation and differentiation while moving up the crypt–villus axis, given cell position and associated signaling concentration gradients [146].
2.3. Central Nervous System
Wnt molecules and gradients regulate different aspects of nervous system development and function in vertebrates.
Exogenous overexpression of a stable (i.e., unresponsive to destruction complex-mediated degradation) form of β-catenin in hippocampal neuronal cultures causes an increased number of dendritic branches in a dose-dependent manner [154]. To test the requirement of endogenous β-catenin for proper dendrite development, Yu and Malenka impaired β-catenin binding to protein partners by overexpressing the intracellular domain of N-cadherin and found substantial reduction in the numbers of dendritic branches; this effect was shown to be independent of β-catenin transcriptional activity, as Lef1 overexpression did not cause a similar phenotype. The study also showed that cultures stimulated with potassium (which mimics the depolarizing effects of neuronal activity) present increased dendritic growth, partially through β-catenin signaling.
The canonical Wnt pathway also guides synaptogenesis [155,156,157]: WNT-7a promotes axonal remodeling by inhibiting the activity of the GSK3 kinase. Indeed, microtubule-associated proteins Tau, MAP-1B, and MAP-2 (microtubules stabilizer) are direct GSK3 substrates. Chemical inhibition of GSK3 recapitulates the phenotype of WNT-7a expression; in contrast, WNT-7a depletion results in reduced synaptic formation [158]. It was also found that, during synaptogenesis, N-cadherin/catenin complexes are initially uniformly distributed across all synaptic sites of neurons but are rapidly redistributed and restricted only to excitatory synaptic sites [159,160]. This process is also controlled by clusters of β-catenin and N-cadherin, which distribute in both pre- and postsynaptic compartments [160]. Additional studies indicated that dendritic morphogenesis depends on Wnt release from neighboring cells, and support the hypothesis of spatial diffusion of Wnt during brain development and activity [154,161,162].
The use of a β-catenin-activated promoter driving the expression of the β-galactosidase reporter helped in defying neurons that respond to activated β-catenin during mouse development. This study confirmed activation of the Wnt pathway in the mid-hindbrain and in the limb apical ectodermal ridge, and identified additional activated regions like the notochord and brain endothelia [75].
3. Wnt/β-Catenin Pathway Levels and Dynamics in Pluripotency, Differentiation, and Somatic-Cell Reprogramming
Embryonic stem cells (ESCs) are characterized by pluripotency (i.e., the potential to differentiate into any somatic cell type) and self-renewal (i.e., the ability of pluripotent cells to divide and maintain such potential). Pluripotency is a transient state in vivo; instead, ESCs can indefinitely be expanded in vitro, maintaining either ground state/naive or primed pluripotency states if isolated from the pre- or postimplantation epiblast, respectively [163]. Culture conditions are also crucial for ESC pluripotency maintenance. Focusing on the Wnt pathway, Sato and colleagues [164] first demonstrated that activation of the canonical pathway by GSK inhibition supports mouse ESC (mESC) in vitro pluripotency maintenance, even in the absence of the leukaemia inhibitory factor (LIF) in the culture medium. It was also demonstrated that Wnt pathway repression leads to mESC differentiation toward epiblast [165]. Such results prompted the establishment of protocols in which the canonical Wnt pathway is constantly activated chemically: the 2i/LIF culture medium combines LIF with two inhibitors (2i), PD0325901 and CHIR99021, repressing MAPK/ERK and GSK3β, respectively [166]. Importantly, 2i/LIF medium is serum free, and enables ground-state pluripotency maintenance, with overall homogeneous transcript levels of key pluripotency genes as compared to their pronounced heterogeneity and temporal fluctuations in FCS/LIF cultures [167,168]. In addition, 2i/LIF confers mESCs high efficiency in chimaera formation [169].
If and how β-catenin transcriptional activity is relevant for pluripotency is debated: in basal conditions (i.e., absence of pathway activators) it is negligible [170], and β-catenin establishes protein complexes with pluripotency master regulators Nanog and Oct4. Such results suggest that Wnt canonical pathway transcriptional signaling might be dispensable for mESC pluripotency [165,171,172,173,174,175]. Nevertheless, ground-state (e.g., 2i/LIF-cultured) mESCs show pronounced nuclear β-catenin accumulation and enhancement, but still heterogeneous transcriptional pathway activity [176]. It is still to be determined if there is a functional relationship between nuclear β-catenin accumulation levels and ground-state pluripotency. Instead, in FCS/LIF cultures, Kielman and colleagues demonstrated that β-catenin doses affect mESC capacity to differentiate into the three germ layers using APC mutants and teratoma-formation assays [177]. Notably, while promoting pluripotency maintenance in mESCs, Wnt/β-catenin signaling also drives the differentiation of primed cells (i.e., EpiSCs) toward the mesendoderm [72].
The canonical Wnt pathway is also relevant for somatic cell reprogramming (i.e., forced conversion of differentiated cells into pluripotent cells [178]). Wnt components are not directly targeted in the original cocktail of overexpressed transcription factors used by Yamanaka and colleagues to reprogram fibroblasts [179]. Nevertheless, is was shown that activation of the pathway can enhance the efficiency of both fusion- and factor-induced mediated reprogramming [180,181,182,183,184]. Interestingly, others and we reported a biphasic role of Wnt/β-catenin pathway in reprogramming, with its activation being beneficial only in the late stages of reprogramming [176,180,181]; this effect seems to not be related to β-catenin-mediated regulation of the cell cycle [68]. Furthermore, we reported detrimental effects of high levels of active β-catenin on cell-fusion-mediated reprogramming (i.e., reprogramming of mouse neural cells upon fusion with mESCs), which, instead, is enhanced by specific β-catenin doses [184].
Regarding human embryonic and induced pluripotent stem cells (hESCs and hiPSCs, respectively), recent studies indicate that their pluripotent state resembles that of mouse EpiSCs [185], as they retain some futures of primed pluripotency [186,187]. As compared to mESCs, hESCs and hiPSCs also respond differently to MEK–ERK pathway inhibition [188]; indeed, when they are cultured in 2i/LIF conditions, naive pluripotency is not supported [189]. Nevertheless, all culture protocols recently proposed to maintain hESCs and hiPSCs in the naïve state of pluripotency rely on Gsk3 inhibition [73]; however, it was also reported that the canonical Wnt pathway promotes hESC differentiation [190]. Ectopic expression of OCT4/KLF4 or KLF2/KLF4 or KLF2/NANOG in human cells can stabilize their pluripotency when cultured in 2i/LIF [191] and 2i/LIF/aPKCi [192]. Recently, a transgene-independent naïve-state medium was proposed: the so-called naive human stem-cell medium (NHSM) contains 2i/LIF further supplemented with p38, Jun N-terminal kinase (JNK), aPKC and RHO-associated protein kinase inhibitors, and a low amount of FGF2 and Activin A or TGFβ1 [188]. However, this culture condition causes loss of DNA imprinting [193]; further studies are needed to define alternative culture protocols that can promote and support naïve pluripotency without erasing germline memory.
WNTs and their effectors are also crucial for potency maintenance in adult stem cells [165,194,195,196,197]. In vivo analysis of various APC mutants suggested that specific signaling levels are associated with cell specification into hematopoietic stem cells, myeloid progenitors, and early thymocytes during haematopoiesis [198]. Bone marrow (BM) from the Axin2LacZ Wnt reporter mouse model showed differential sensitivity in reporter activity when stimulated with the canonical Wnt pathway. Furthermore, when studying hematopoietic stem cell (HSC) differentiation in the presence of varying Wnt/β-catenin activity, mild/intermediate levels were shown to enhance clonogenicity and myeloid differentiation, while high levels strongly reduced the number of colonies; only a mild increase was able to confer increased HSC repopulation potential [198].
4. β-Catenin and Cancer
Homeostasis tightly controls the number of cells within tissue, balancing cell growth and survival. Genetic mutations or sporadic events can result in uncontrolled cell proliferation and/or increased cell survival, which can ultimately lead to cancer initiation [199,200]. Often, dysplastic events involve the stem-cell compartment of the tissue, which is more susceptible to mutagenic events. β-catenin is rarely mutated in cancer, but mutations of its main protein partners and gaining function effects can confer enhanced stability to β-catenin, causing its aberrant accumulation [201,202].
Colorectal cancer (CRC) is the first and most characterized cancer model involving the canonical Wnt pathway. Because of APC mutations, β-catenin levels increase, and pro-proliferative genes are activated following its nuclear translocation [38,202]. A similar cancerogenic phenotype is also induced by mutations in others protein members of the canonical Wnt pathway, such as human naked-cuticle homolog NKD1 [203] and protein phosphate 2A (PP2A) [204,205]. Mutations of NKD1 have been frequently found in CRC: loss-of-function studies, using different mutants of the wild-type gene, showed defective inhibition of Wnt/Dvl signaling [203]. NKD1 is involved in Dvl proteasomal-mediated degradation; therefore, the inability to destabilize Dvl results in aberrant β-catenin accumulation and increased cell proliferation. PP2A functions are related to the control of many signaling cascades by opposing the activity of protein kinases (as reviewed in [206]). PP2A binds to different components of the Wnt pathway, including APC and Axin, and affects its activity both upstream and downstream of β-catenin [207,208,209]. Additionally, PP2A has binding domains for NKD1 [210] and coordinates β-catenin/E-Cadherin binding with a direct effect on the epithelial–mesenchymal transition happening during cancer initiation, ultimately balancing the ratio between complexed and free β-catenin [211]. Indeed, mutants of the PP2A subunit Calpha fail mesoderm differentiation and are lethal [211]. β-catenin accumulation has also been associated with non-CRCs of the gastrointestinal tract, including the liver and the biliary tract [212,213,214], the connective tissue [215], glial cells [216], and the hematopoietic system [217].
A study on a cohort of human gastric adenocarcinomas revealed β-catenin nuclear accumulation in 29% of samples, whereas 71% only showed membrane β-catenin staining; this study did not investigate a possible correlation between β-catenin levels and/or intracellular localization and cancer severity [212]. Similar results were also found in patients with hepatocarcinoma (HCC), where 19% of samples showed β-catenin presence in the nucleus [213]. Desmoid-type fibromatosis showed β-catenin nuclear accumulation in approximately 50% of a tumor, and this observation was consistent across patients with wild-type β-catenin (where accumulation is likely caused by mutations in Wnt partners) or mutated β-catenin [215]. Finally, β-catenin was found to be the driving force of mixed-lineage leukaemia (MLL) stem-cell (LSC) development. So’s research group characterized β-catenin translocation and canonical Wnt pathway activation during the pre-LSCs to LSCs transition (i.e., the process that originates aggressive and drug-resistant leukaemia) and demonstrated that the Wnt pathway is highly activated when tumorigenesis starts, while β-catenin knockdown impairs both murine and human MLL cell proliferation [217].
The role of β-catenin in cancer is not only restricted to the proliferative advantage of cancer cells, but also to their ability to colonize surrounding tissue, potentiating metastasis formation [218,219,220] and immune-system evasion [221,222,223,224]. Transcriptional profiling of metastatic vs. nonmetastatic breast-cancer cells showed overexpression of the canonical Wnt pathway (i.e., β-catenin and LEF1), β-catenin target genes (i.e., c-Myc and cyclin D1), and Wnt ligands (i.e., Wnt3a-7a); secretion of Wnt ligands might be responsible for the activation of the Wnt pathway in cells distant from a primary tumor [218]. An alternative study about the metastatic power of melanoma cells showed that, when overexpressed into mice, β-catenin acts in two phases: it initially reduces cell migration and only in the second stage it promotes metastatic spread [219]. Reduced cell migration can be explained by an autonomous property of cancer cells by which they need to be less motile in order to colonize the primary tissue before spreading to others [219]. These observations can be reconsidered in view of recent findings of β-catenin-mediated immune-system elusion of cancer cells: melanoma specifically expressing constitutive active β-catenin does not present any T-cell infiltration [223]. Manicassamy and colleagues recently proposed a molecular mechanism for tumor-induced immunosuppression, in which increased β-catenin activity in tumor-resident dendritic cells (DCs) can lead to enhanced activity of vitamin A-metabolizing enzymes. This results in faster retinoic acid (RA) metabolism, with RA driving regulatory T-cell responses and immune tolerance [225]. Interestingly, in the same work, β-catenin inhibition was shown to reduce tumor growth, opening new therapeutic avenues for combined targeting of Wnt/β-catenin and RA metabolism pathways.
Although β-catenin is involved in many phases of cancer progression and higher levels worsen prognosis [220,226], little is known on whether different amounts of Wnt protein accumulation relate to specific phases of carcinogenesis.
5. Conclusions and Future Directions
The pleiotropic roles of the Wnt/β-catenin pathway in regulating multiple processes, including embryogenesis, pluripotency, differentiation, and cancer, have been reviewed. It appears clear that the functions of the pathway are highly cell- and context-dependent [227]; across different systems, dose- and dynamic-dependent functions have also been shown (Figure 2).
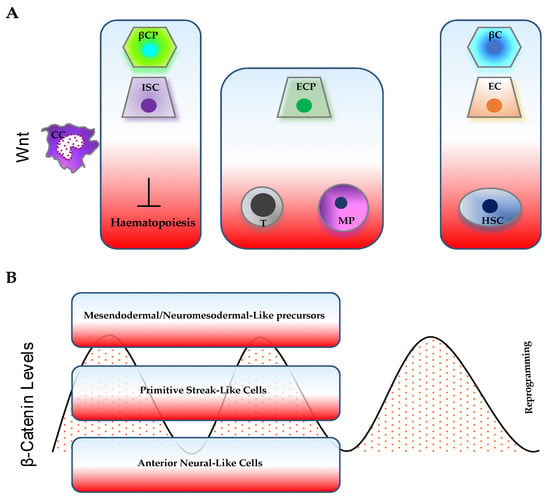
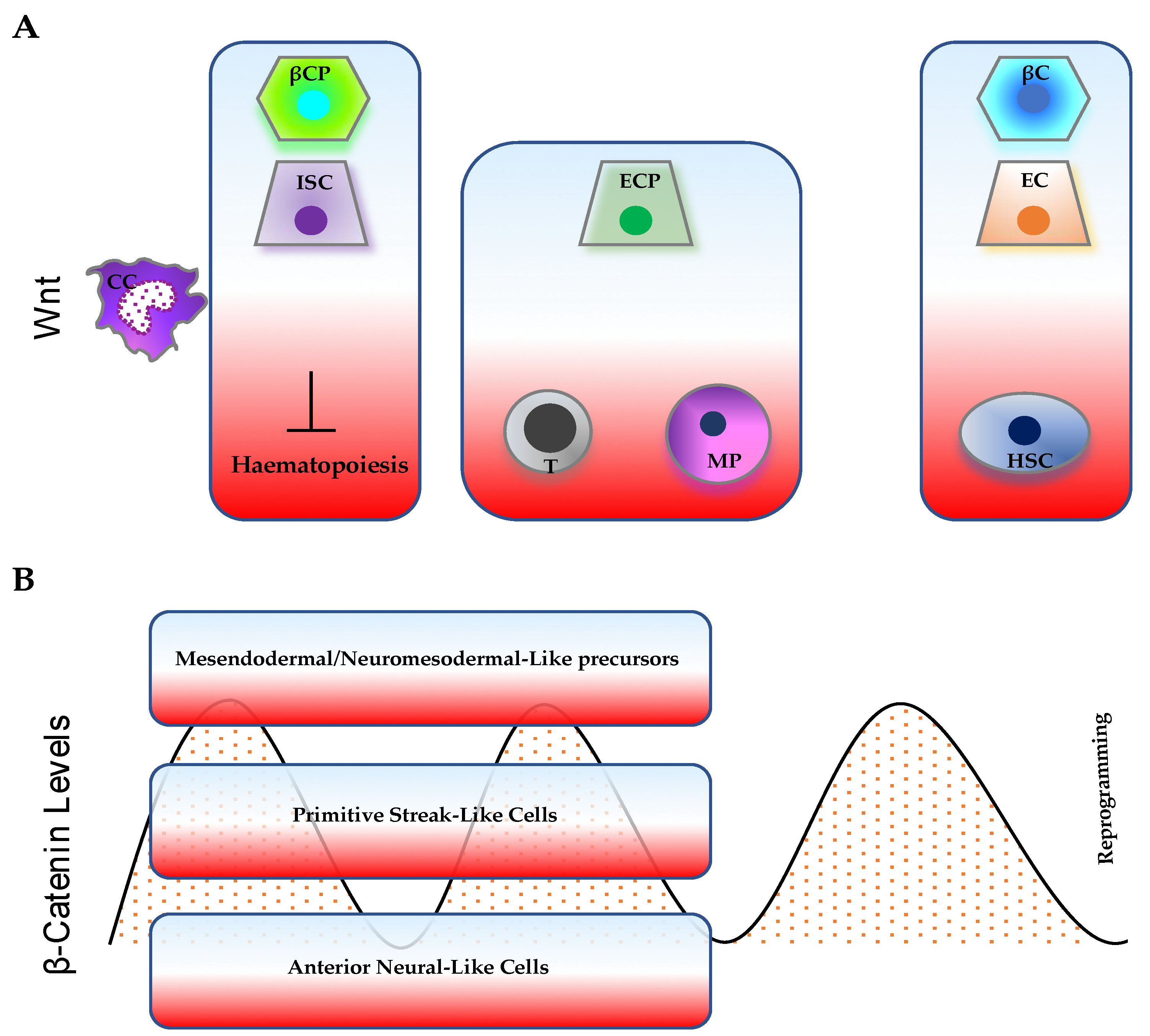
Figure 2.
Representative cellular processes influenced by (A) Wnt gradient and (B) time-varying β-catenin levels. The cellular response can depend on both the levels of Wnt/β-catenin pathway activity and on cellular/tissue context. (A) High/intermediate Wnt levels support both intestinal stem-cell (ISC) and β-cell progenitor (βCP) expansion; low Wnt levels stimulate terminal differentiation of enterocytes (EC) and β cells (βC), but sustain hematopoietic stem-cell (HSC) maintenance. Intermediate Wnt levels are mostly associated with blood-cell commitment (T cells, T; myeloid progenitors, MP) and enterocyte-progenitor (ECP) differentiation. (B) β-catenin oscillations control embryo patterning. High or low levels of β-catenin can either promote or impair somatic cell reprogramming, respectively.
Feedback loops in pathway topology [120,228] and crosstalk interactions with other pathways across species and systems [125,229,230,231,232,233] might be the cause of switch and oscillatory-like pathway behaviors. Such nonlinear dynamics could explain β-catenin’s dual role in systems for which its heterogeneous levels have been reported but not functionally characterized, such as mESCs cultured in both naïve (i.e., serum-based) and ground-state (i.e., serum free) pluripotency media [176,234].
Mathematical models can be instrumental in providing quantitative insights into the characteristics and dynamics of signaling pathways and coupled dynamic processes, allowing to test hypotheses and generate in silico predictions [235]. While the validity of modeling results inevitably depends on model parameters, assumptions, and structure [236], computational representations of cells can be instrumental when combined with ad hoc experimental validations. An elegant work confirmed experimentally in mammalian cells predictions of a computational model (developed using Xenopus embryos data [237]) that the canonical Wnt transcriptional system can respond to β-catenin fold changes instead of its absolute levels, possibly due to incoherent feed-forward loops in pathway topology [238]. The canonical Wnt pathway is coupled to the cell cycle (see References [239,240] for a review). Despite its established promitotic role, recent studies [102] have shown it can also promote expression of cell-cycle negative regulators in mESCs. Given this, and the aforementioned role of pathway gradients in tissue organogenesis and homeostasis, as well as in cell-cell adhesion, multiscale computational models and agent-based cell-simulation frameworks might be needed to better formalize signaling-pathway dynamics, thanks to their ability to describe cell mechanics, 2D/3D tissue geometries, single-cell gene expression and its coupling to cell proliferation [241,242,243].
Novel experimental techniques can also be instrumental to developing a better and quantitative understanding of the role of spatial and temporal Wnt/β-catenin (and other signaling) pathway dynamics in controlling tissue homeostasis. Three-dimensional in vitro cell clusters and organoids enable studying processes like organogenesis and cancer development in a fully controllable setting [244], especially when combined with genome-modification strategies such as viral transgene delivery and CRISPR/Cas9 technology. Successful examples include the development of bladder cancer-cell-derived organoid cultures to study the link between Wnt/β-catenin pathway activation and cellular organization and proliferation [245]; analysis of precardiac spheroids to study Wnt and BMP role in the specification of two-cardiac origin [246]; and the use of intestinal organoids to study epithelial self-organization in the gut [244,247,248].
Live-cell imaging, combined with precise perturbations of signaling-pathway dynamics, might contribute significantly to dissect how the latter control cell fate. In a recent work, a microfluidics/imaging-based approach was developed to entrain oscillations of Notch and Wnt signaling to predetermined external periodic forces in the anterior monolayer PSM [123]; this experimental setup allowed to quantitatively address important questions about the crosstalk between the two pathways, and the role of relative timing between individual pathway oscillations in the control of mesoderm segmentation.
The use of feedback control, recently combined with microfluidics/imaging platforms to precisely regulate gene expression in living cells [249], and with synthetic biology tools to recreate, in a fully controllable and reproducible way, signaling-pathway activity [250,251], might open important avenues to quantitatively study dose- and dynamic-dependent signaling functions across biological systems, and develop ad hoc interventions in case of malfunction.
Author Contributions
E.P. and L.M. drafted, edited, and revised the manuscript.
Funding
This work was supported by the Medical Research Council, grant MR/N021444/1 to L.M., by the Engineering and Physical Sciences Research Council grant EP/R041695/1 to L.M., and by BrisSynBio, a BBSRC/EPSRC Synthetic Biology Research Centre (BB/L01386X/1), to L.M.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Clevers, H.; Loh, K.M.; Nusse, R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 2014, 346, 1248012. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R.; Clevers, H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Slee, R.B.; Fukai, N.; Rawadi, G.; Roman-Roman, S.; Reginato, A.M.; Wang, H.; Cundy, T.; Glorieux, F.H.; Lev, D.; et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 2001, 107, 513–523. [Google Scholar] [CrossRef]
- Toomes, C.; Bottomley, H.M.; Jackson, R.M.; Towns, K.V.; Scott, S.; Mackey, D.A.; Craig, J.E.; Jiang, L.; Yang, Z.; Trembath, R.; et al. Mutations in LRP5 or FZD4 underlie the common familial exudative vitreoretinopathy locus on chromosome 11q. Am. J. Hum. Genet. 2004, 74, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Pyott, S.M.; Tran, T.T.; Leistritz, D.F.; Pepin, M.G.; Mendelsohn, N.J.; Temme, R.T.; Fernandez, B.A.; Elsayed, S.M.; Elsobky, E.; Verma, I.; et al. WNT1 mutations in families affected by moderately severe and progressive recessive osteogenesis imperfecta. Am. J. Hum. Genet. 2013, 92, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Person, A.D.; Beiraghi, S.; Sieben, C.M.; Hermanson, S.; Neumann, A.N.; Robu, M.E.; Schleiffarth, J.R.; Billington, C.J., Jr.; van Bokhoven, H.; Hoogeboom, J.M.; et al. WNT5A mutations in patients with autosomal dominant Robinow syndrome. Dev. Dyn. 2010, 239, 327–337. [Google Scholar] [CrossRef] [PubMed]
- White, J.; Mazzeu, J.F.; Hoischen, A.; Jhangiani, S.N.; Gambin, T.; Alcino, M.C.; Penney, S.; Saraiva, J.M.; Hove, H.; Skovby, F.; et al. DVL1 frameshift mutations clustering in the penultimate exon cause autosomal-dominant Robinow syndrome. Am. J. Hum. Genet. 2015, 96, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Massink, M.P.; Creton, M.A.; Spanevello, F.; Fennis, W.M.; Cune, M.S.; Savelberg, S.M.; Nijman, I.J.; Maurice, M.M.; van den Boogaard, M.J.; van Haaften, G. Loss-of-Function Mutations in the WNT Co-receptor LRP6 Cause Autosomal-Dominant Oligodontia. Am. J. Hum. Genet. 2015, 97, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Adaimy, L.; Chouery, E.; Megarbane, H.; Mroueh, S.; Delague, V.; Nicolas, E.; Belguith, H.; de Mazancourt, P.; Megarbane, A. Mutation in WNT10A is associated with an autosomal recessive ectodermal dysplasia: The odonto-onycho-dermal dysplasia. Am. J. Hum. Genet. 2007, 81, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Lammi, L.; Arte, S.; Somer, M.; Jarvinen, H.; Lahermo, P.; Thesleff, I.; Pirinen, S.; Nieminen, P. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am. J. Hum. Genet. 2004, 74, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Mani, A.; Radhakrishnan, J.; Wang, H.; Mani, A.; Mani, M.A.; Nelson-Williams, C.; Carew, K.S.; Mane, S.; Najmabadi, H.; Wu, D.; et al. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science 2007, 315, 1278–1282. [Google Scholar] [CrossRef] [PubMed]
- Robitaille, J.; MacDonald, M.L.; Kaykas, A.; Sheldahl, L.C.; Zeisler, J.; Dube, M.P.; Zhang, L.H.; Singaraja, R.R.; Guernsey, D.L.; Zheng, B.; et al. Mutant frizzled-4 disrupts retinal angiogenesis in familial exudative vitreoretinopathy. Nat. Genet. 2002, 32, 326–330. [Google Scholar] [CrossRef] [PubMed]
- De Ferrari, G.V.; Papassotiropoulos, A.; Biechele, T.; Wavrant De-Vrieze, F.; Avila, M.E.; Major, M.B.; Myers, A.; Saez, K.; Henriquez, J.P.; Zhao, A.; et al. Common genetic variation within the low-density lipoprotein receptor-related protein 6 and late-onset Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2007, 104, 9434–9439. [Google Scholar] [CrossRef] [PubMed]
- Anastas, J.N.; Moon, R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 2013, 13, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, A.S.; Grosschedl, R.; Guzman, R.C.; Parslow, T.; Varmus, H.E. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell 1988, 55, 619–625. [Google Scholar] [CrossRef]
- Morin, P.J.; Sparks, A.B.; Korinek, V.; Barker, N.; Clevers, H.; Vogelstein, B.; Kinzler, K.W. Activation of β-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 1997, 275, 1787–1790. [Google Scholar] [CrossRef] [PubMed]
- Bass, A.J.; Lawrence, M.S.; Brace, L.E.; Ramos, A.H.; Drier, Y.; Cibulskis, K.; Sougnez, C.; Voet, D.; Saksena, G.; Sivachenko, A.; et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat. Genet. 2011, 43, 964–968. [Google Scholar] [CrossRef] [PubMed]
- Zurawel, R.H.; Chiappa, S.A.; Allen, C.; Raffel, C. Sporadic medulloblastomas contain oncogenic β-catenin mutations. Cancer Res. 1998, 58, 896–899. [Google Scholar] [PubMed]
- Wu, J.; Jiao, Y.; Dal Molin, M.; Maitra, A.; de Wilde, R.F.; Wood, L.D.; Eshleman, J.R.; Goggins, M.G.; Wolfgang, C.L.; Canto, M.I.; et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 21188–21193. [Google Scholar] [CrossRef] [PubMed]
- Palacios, J.; Gamallo, C. Mutations in the β-catenin gene (CTNNB1) in endometrioid ovarian carcinomas. Cancer Res. 1998, 58, 1344–1347. [Google Scholar] [PubMed]
- Satoh, S.; Daigo, Y.; Furukawa, Y.; Kato, T.; Miwa, N.; Nishiwaki, T.; Kawasoe, T.; Ishiguro, H.; Fujita, M.; Tokino, T.; et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat. Genet. 2000, 24, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Rubinfeld, B.; Albert, I.; Porfiri, E.; Fiol, C.; Munemitsu, S.; Polakis, P. Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly. Science 1996, 272, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Dong, X.; Mai, M.; Seelan, R.S.; Taniguchi, K.; Krishnadath, K.K.; Halling, K.C.; Cunningham, J.M.; Boardman, L.A.; Qian, C.; et al. Mutations in AXIN2 cause colorectal cancer with defective mismatch repair by activating beta-catenin/TCF signalling. Nat. Genet. 2000, 26, 146–147. [Google Scholar] [CrossRef] [PubMed]
- Seshagiri, S.; Stawiski, E.W.; Durinck, S.; Modrusan, Z.; Storm, E.E.; Conboy, C.B.; Chaudhuri, S.; Guan, Y.; Janakiraman, V.; Jaiswal, B.S.; et al. Recurrent R-spondin fusions in colon cancer. Nature 2012, 488, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Komiya, Y.; Habas, R. Wnt signal transduction pathways. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Kohn, A.D.; Moon, R.T. Wnt and calcium signaling: β-catenin-independent pathways. Cell Calcium 2005, 38, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Mbom, B.C.; Nelson, W.J.; Barth, A. beta-catenin at the centrosome: Discrete pools of beta-catenin communicate during mitosis and may co-ordinate centrosome functions and cell cycle progression. Bioessays 2013, 35, 804–809. [Google Scholar] [CrossRef] [PubMed]
- Yap, A.S.; Brieher, W.M.; Gumbiner, B.M. Molecular and functional analysis of cadherin-based adherens junctions. Annu. Rev. Cell Dev. Biol. 1997, 13, 119–146. [Google Scholar] [CrossRef] [PubMed]
- Aberle, H.; Butz, S.; Stappert, J.; Weissig, H.; Kemler, R.; Hoschuetzky, H. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J. Cell Sci. 1994, 107 (Pt 12), 3655–3663. [Google Scholar]
- Davis, M.A.; Ireton, R.C.; Reynolds, A.B. A core function for p120-catenin in cadherin turnover. J. Cell Biol. 2003, 163, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Bek, S.; Kemler, R. Protein kinase CKII regulates the interaction of β-catenin with alpha-catenin and its protein stability. J. Cell Sci. 2002, 115, 4743–4753. [Google Scholar] [CrossRef] [PubMed]
- Lickert, H.; Bauer, A.; Kemler, R.; Stappert, J. Casein kinase II phosphorylation of E-cadherin increases E-cadherin/beta-catenin interaction and strengthens cell-cell adhesion. J. Biol. Chem. 2000, 275, 5090–5095. [Google Scholar] [CrossRef] [PubMed]
- Bahmanyar, S.; Kaplan, D.D.; Deluca, J.G.; Giddings, T.H., Jr.; O’Toole, E.T.; Winey, M.; Salmon, E.D.; Casey, P.J.; Nelson, W.J.; Barth, A.I. beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 2008, 22, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, D.D.; Meigs, T.E.; Kelly, P.; Casey, P.J. Identification of a role for β-catenin in the establishment of a bipolar mitotic spindle. J. Biol. Chem. 2004, 279, 10829–10832. [Google Scholar] [CrossRef] [PubMed]
- Hoppler, S.; Kavanagh, C.L. Wnt signalling: Variety at the core. J. Cell Sci. 2007, 120, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Arce, L.; Yokoyama, N.N.; Waterman, M.L. Diversity of LEF/TCF action in development and disease. Oncogene 2006, 25, 7492–7504. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/β-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Behrens, J.; von Kries, J.P.; Kuhl, M.; Bruhn, L.; Wedlich, D.; Grosschedl, R.; Birchmeier, W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature 1996, 382, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, M.; van de Wetering, M.; Oosterwegel, M.; Peterson-Maduro, J.; Godsave, S.; Korinek, V.; Roose, J.; Destree, O.; Clevers, H. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell 1996, 86, 391–399. [Google Scholar] [CrossRef]
- van de Wetering, M.; Cavallo, R.; Dooijes, D.; van Beest, M.; van Es, J.; Loureiro, J.; Ypma, A.; Hursh, D.; Jones, T.; Bejsovec, A.; et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 1997, 88, 789–799. [Google Scholar] [CrossRef]
- Stadeli, R.; Basler, K. Dissecting nuclear Wingless signalling: Recruitment of the transcriptional co-activator Pygopus by a chain of adaptor proteins. Mech. Dev. 2005, 122, 1171–1182. [Google Scholar] [CrossRef] [PubMed]
- Van Beest, M.; Dooijes, D.; van De Wetering, M.; Kjaerulff, S.; Bonvin, A.; Nielsen, O.; Clevers, H. Sequence-specific high mobility group box factors recognize 10-12-base pair minor groove motifs. J. Biol. Chem. 2000, 275, 27266–27273. [Google Scholar] [CrossRef] [PubMed]
- Graham, T.A.; Weaver, C.; Mao, F.; Kimelman, D.; Xu, W. Crystal structure of a beta-catenin/Tcf complex. Cell 2000, 103, 885–896. [Google Scholar] [CrossRef]
- Graham, T.A.; Ferkey, D.M.; Mao, F.; Kimelman, D.; Xu, W. Tcf4 can specifically recognize β-catenin using alternative conformations. Nat. Struct. Biol. 2001, 8, 1048–1052. [Google Scholar] [CrossRef] [PubMed]
- Stamos, J.L.; Weis, W.I. The β-catenin destruction complex. Cold Spring Harb. Perspect. Biol. 2013, 5, a007898. [Google Scholar] [CrossRef] [PubMed]
- Heisenberg, C.P.; Houart, C.; Take-Uchi, M.; Rauch, G.J.; Young, N.; Coutinho, P.; Masai, I.; Caneparo, L.; Concha, M.L.; Geisler, R.; et al. A mutation in the Gsk3-binding domain of zebrafish Masterblind/Axin1 leads to a fate transformation of telencephalon and eyes to diencephalon. Genes Dev. 2001, 15, 1427–1434. [Google Scholar] [CrossRef] [PubMed]
- Orford, K.; Crockett, C.; Jensen, J.P.; Weissman, A.M.; Byers, S.W. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J. Biol. Chem. 1997, 272, 24735–24738. [Google Scholar] [CrossRef] [PubMed]
- Amit, S.; Hatzubai, A.; Birman, Y.; Andersen, J.S.; Ben-Shushan, E.; Mann, M.; Ben-Neriah, Y.; Alkalay, I. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: A molecular switch for the Wnt pathway. Genes Dev. 2002, 16, 1066–1076. [Google Scholar] [CrossRef] [PubMed]
- Frame, S.; Cohen, P.; Biondi, R.M. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol. Cell 2001, 7, 1321–1327. [Google Scholar] [CrossRef]
- Jho, E.H.; Zhang, T.; Domon, C.; Joo, C.K.; Freund, J.N.; Costantini, F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 2002, 22, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Chia, I.V.; Costantini, F. Mouse axin and axin2/conductin proteins are functionally equivalent in vivo. Mol. Cell. Biol. 2005, 25, 4371–4376. [Google Scholar] [CrossRef] [PubMed]
- Aberle, H.; Bauer, A.; Stappert, J.; Kispert, A.; Kemler, R. β-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997, 16, 3797–3804. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.Y.; Kolligs, F.T.; Wu, R.; Zhai, Y.; Kuick, R.; Hanash, S.; Cho, K.R.; Fearon, E.R. Activation of AXIN2 expression by β-catenin-T cell factor. A feedback repressor pathway regulating Wnt signaling. J. Biol. Chem. 2002, 277, 21657–21665. [Google Scholar] [CrossRef] [PubMed]
- Eklof Spink, K.; Fridman, S.G.; Weis, W.I. Molecular mechanisms of β-catenin recognition by adenomatous polyposis coli revealed by the structure of an APC-β-catenin complex. EMBO J. 2001, 20, 6203–6212. [Google Scholar] [CrossRef] [PubMed]
- Day, C.L.; Alber, T. Crystal structure of the amino-terminal coiled-coil domain of the APC tumor suppressor. J. Mol. Biol. 2000, 301, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Cadigan, K.M.; Liu, Y.I. Wnt signaling: Complexity at the surface. J. Cell Sci. 2006, 119, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Daniels, D.L.; Weis, W.I. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 2005, 12, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Range, R.C.; Venuti, J.M.; McClay, D.R. LvGroucho and nuclear β-catenin functionally compete for Tcf binding to influence activation of the endomesoderm gene regulatory network in the sea urchin embryo. Dev. Biol. 2005, 279, 252–267. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, S.; van Leeuwen, F.; Wodarz, A.; Klingensmith, J.; Nusse, R. The dishevelled protein is modified by wingless signaling in Drosophila. Genes Dev. 1995, 9, 1087–1097. [Google Scholar] [CrossRef] [PubMed]
- Bryja, V.; Schulte, G.; Rawal, N.; Grahn, A.; Arenas, E. Wnt-5a induces Dishevelled phosphorylation and dopaminergic differentiation via a CK1-dependent mechanism. J. Cell Sci. 2007, 120, 586–595. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Sancho, J.M.; Brennan, K.R.; Castelo-Soccio, L.A.; Brown, A.M. Wnt proteins induce dishevelled phosphorylation via an LRP5/6- independent mechanism, irrespective of their ability to stabilize β-catenin. Mol. Cell. Biol. 2004, 24, 4757–4768. [Google Scholar] [CrossRef] [PubMed]
- Waterman, M.L.; Fischer, W.H.; Jones, K.A. A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes Dev. 1991, 5, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Travis, A.; Amsterdam, A.; Belanger, C.; Grosschedl, R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor α enhancer function. Genes Dev. 1991, 5, 880–894. [Google Scholar] [CrossRef] [PubMed]
- van de Wetering, M.; Oosterwegel, M.; Dooijes, D.; Clevers, H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991, 10, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Korinek, V.; Barker, N.; Willert, K.; Molenaar, M.; Roose, J.; Wagenaar, G.; Markman, M.; Lamers, W.; Destree, O.; Clevers, H. Two members of the Tcf family implicated in Wnt/β-catenin signaling during embryogenesis in the mouse. Mol. Cell. Biol. 1998, 18, 1248–1256. [Google Scholar] [CrossRef] [PubMed]
- Cadigan, K.M.; Waterman, M.L. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef] [PubMed]
- Poy, F.; Lepourcelet, M.; Shivdasani, R.A.; Eck, M.J. Structure of a human Tcf4-β-catenin complex. Nat. Struct. Biol. 2001, 8, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- De Jaime-Soguero, A.; Aulicino, F.; Ertaylan, G.; Griego, A.; Cerrato, A.; Tallam, A.; Del Sol, A.; Cosma, M.P.; Lluis, F. Wnt/Tcf1 pathway restricts embryonic stem cell cycle through activation of the Ink4/Arf locus. PLoS Genet. 2017, 13, e1006682. [Google Scholar] [CrossRef] [PubMed]
- Wray, J.; Kalkan, T.; Gomez-Lopez, S.; Eckardt, D.; Cook, A.; Kemler, R.; Smith, A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 2011, 13, 838–845. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K.; Nishinakamura, R.; Iwamatsu, Y.; Shimosato, D.; Niwa, H. Synergistic action of Wnt and LIF in maintaining pluripotency of mouse ES cells. Biochem. Biophys. Res. Commun. 2006, 343, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Takao, Y.; Yokota, T.; Koide, H. Beta-catenin up-regulates Nanog expression through interaction with Oct-3/4 in embryonic stem cells. Biochem. Biophys. Res. Commun. 2007, 353, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Kurek, D.; Neagu, A.; Tastemel, M.; Tuysuz, N.; Lehmann, J.; van de Werken, H.J.; Philipsen, S.; van der Linden, R.; Maas, A.; van, I.W.F.; et al. Endogenous WNT signals mediate BMP-induced and spontaneous differentiation of epiblast stem cells and human embryonic stem cells. Stem Cell Rep. 2015, 4, 114–128. [Google Scholar] [CrossRef] [PubMed]
- de Jaime-Soguero, A.; Abreu de Oliveira, W.A.; Lluis, F. The Pleiotropic Effects of the Canonical Wnt Pathway in Early Development and Pluripotency. Genes (Basel) 2018, 9, 93. [Google Scholar] [CrossRef] [PubMed]
- Kimelman, D.; Martin, B.L. Anterior-posterior patterning in early development: Three strategies. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Maretto, S.; Cordenonsi, M.; Dupont, S.; Braghetta, P.; Broccoli, V.; Hassan, A.B.; Volpin, D.; Bressan, G.M.; Piccolo, S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc. Natl. Acad. Sci. USA 2003, 100, 3299–3304. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Descalzo, S.; Hadjantonakis, A.K.; Arias, A.M. Wnt/ss-catenin signalling and the dynamics of fate decisions in early mouse embryos and embryonic stem (ES) cells. Semin. Cell Dev. Biol. 2015, 47–48, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Huelsken, J.; Vogel, R.; Brinkmann, V.; Erdmann, B.; Birchmeier, C.; Birchmeier, W. Requirement for β-catenin in anterior-posterior axis formation in mice. J. Cell Biol. 2000, 148, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Kemler, R.; Hierholzer, A.; Kanzler, B.; Kuppig, S.; Hansen, K.; Taketo, M.M.; de Vries, W.N.; Knowles, B.B.; Solter, D. Stabilization of β-catenin in the mouse zygote leads to premature epithelial-mesenchymal transition in the epiblast. Development 2004, 131, 5817–5824. [Google Scholar] [CrossRef] [PubMed]
- Rudloff, S.; Kemler, R. Differential requirements for β-catenin during mouse development. Development 2012, 139, 3711–3721. [Google Scholar] [CrossRef] [PubMed]
- Murtaugh, L.C.; Law, A.C.; Dor, Y.; Melton, D.A. β-catenin is essential for pancreatic acinar but not islet development. Development 2005, 132, 4663–4674. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, S.; Edlund, H. Attenuated Wnt signaling perturbs pancreatic growth but not pancreatic function. Diabetes 2005, 54, 2844–2851. [Google Scholar] [CrossRef] [PubMed]
- Dessimoz, J.; Bonnard, C.; Huelsken, J.; Grapin-Botton, A. Pancreas-specific deletion of β-catenin reveals Wnt-dependent and Wnt-independent functions during development. Curr. Biol. 2005, 15, 1677–1683. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.M.; Esni, F.; Boivin, G.P.; Aronow, B.J.; Stuart, W.; Combs, C.; Sklenka, A.; Leach, S.D.; Lowy, A.M. Wnt/beta-catenin signaling is required for development of the exocrine pancreas. BMC Dev. Biol. 2007, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- McLin, V.A.; Rankin, S.A.; Zorn, A.M. Repression of Wnt/β-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development 2007, 134, 2207–2217. [Google Scholar] [CrossRef] [PubMed]
- Nadauld, L.D.; Sandoval, I.T.; Chidester, S.; Yost, H.J.; Jones, D.A. Adenomatous polyposis coli control of retinoic acid biosynthesis is critical for zebrafish intestinal development and differentiation. J. Biol. Chem. 2004, 279, 51581–51589. [Google Scholar] [CrossRef] [PubMed]
- Kajino, Y.; Yamaguchi, A.; Hashimoto, N.; Matsuura, A.; Sato, N.; Kikuchi, K. beta-Catenin gene mutation in human hair follicle-related tumors. Pathol. Int. 2001, 51, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Huelsken, J.; Vogel, R.; Erdmann, B.; Cotsarelis, G.; Birchmeier, W. β-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell 2001, 105, 533–545. [Google Scholar] [CrossRef]
- Ridanpaa, M.; Fodde, R.; Kielman, M. Dynamic expression and nuclear accumulation of β-catenin during the development of hair follicle-derived structures. Mech. Dev. 2001, 109, 173–181. [Google Scholar] [CrossRef]
- Blanpain, C.; Fuchs, E. Epidermal stem cells of the skin. Annu. Rev. Cell Dev. Biol. 2006, 22, 339–373. [Google Scholar] [CrossRef] [PubMed]
- van Genderen, C.; Okamura, R.M.; Farinas, I.; Quo, R.G.; Parslow, T.G.; Bruhn, L.; Grosschedl, R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994, 8, 2691–2703. [Google Scholar] [CrossRef] [PubMed]
- Alonso, L.; Fuchs, E. Stem cells in the skin: Waste not, Wnt not. Genes Dev. 2003, 17, 1189–1200. [Google Scholar] [CrossRef] [PubMed]
- Lohi, M.; Tucker, A.S.; Sharpe, P.T. Expression of Axin2 indicates a role for canonical Wnt signaling in development of the crown and root during pre- and postnatal tooth development. Dev. Dyn. 2010, 239, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Brugmann, S.A.; Goodnough, L.H.; Gregorieff, A.; Leucht, P.; ten Berge, D.; Fuerer, C.; Clevers, H.; Nusse, R.; Helms, J.A. Wnt signaling mediates regional specification in the vertebrate face. Development 2007, 134, 3283–3295. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.S.; Zhang, Y.; Xu, M.; Yang, Y.; Ito, M.; Peng, T.; Cui, Z.; Nagy, A.; Hadjantonakis, A.K.; Lang, R.A.; et al. Distinct functions for Wnt/beta-catenin in hair follicle stem cell proliferation and survival and interfollicular epidermal homeostasis. Cell Stem Cell 2013, 13, 720–733. [Google Scholar] [CrossRef] [PubMed]
- Millar, S.E.; Willert, K.; Salinas, P.C.; Roelink, H.; Nusse, R.; Sussman, D.J.; Barsh, G.S. WNT signaling in the control of hair growth and structure. Dev. Biol. 1999, 207, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shackleford, G.M. Murine Wnt10a and Wnt10b: Cloning and expression in developing limbs, face and skin of embryos and in adults. Oncogene 1996, 13, 1537–1544. [Google Scholar] [PubMed]
- Nusse, R.; Varmus, H.E. Wnt genes. Cell 1992, 69, 1073–1087. [Google Scholar] [CrossRef]
- Christiansen, J.H.; Dennis, C.L.; Wicking, C.A.; Monkley, S.J.; Wilkinson, D.G.; Wainwright, B.J. Murine Wnt-11 and Wnt-12 have temporally and spatially restricted expression patterns during embryonic development. Mech. Dev. 1995, 51, 341–350. [Google Scholar] [CrossRef]
- Tanda, N.; Ohuchi, H.; Yoshioka, H.; Noji, S.; Nohno, T. A chicken Wnt gene, Wnt-11, is involved in dermal development. Biochem. Biophys. Res. Commun. 1995, 211, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Chuong, C.M.; Widelitz, R.B.; Ting-Berreth, S.; Jiang, T.X. Early events during avian skin appendage regeneration: Dependence on epithelial-mesenchymal interaction and order of molecular reappearance. J. Investig. Dermatol. 1996, 107, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, A.; Hansen, L.A.; Vogel, J.C.; Udey, M.C. Characterization of Wnt gene expression in murine skin: Possible involvement of epidermis-derived Wnt-4 in cutaneous epithelial-mesenchymal interactions. Exp. Cell Res. 1998, 243, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, J.; Burgeson, R.E.; Morgan, B.A. Wnt signaling maintains the hair-inducing activity of the dermal papilla. Genes Dev. 2000, 14, 1181–1185. [Google Scholar] [PubMed]
- Xing, Y.; Ma, X.; Guo, H.; Deng, F.; Yang, J.; Li, Y. Wnt5a Suppresses β-catenin Signaling during Hair Follicle Regeneration. Int. J. Med. Sci. 2016, 13, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Cooke, J.; Zeeman, E.C. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J. Theor. Biol. 1976, 58, 455–476. [Google Scholar] [CrossRef]
- Kerszberg, M.; Wolpert, L. A clock and trail model for somite formation, specialization and polarization. J. Theor. Biol. 2000, 205, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Meinhardt, H. Hierarchical inductions of cell states: A model for segmentation in Drosophila. J. Cell Sci. Suppl 1986, 4, 357–381. [Google Scholar] [CrossRef] [PubMed]
- Stern, C.D.; Fraser, S.E.; Keynes, R.J.; Primmett, D.R. A cell lineage analysis of segmentation in the chick embryo. Development 1988, 104, 231–244. [Google Scholar] [PubMed]
- Richardson, M.K.; Allen, S.P.; Wright, G.M.; Raynaud, A.; Hanken, J. Somite number and vertebrate evolution. Development 1998, 125, 151–160. [Google Scholar] [PubMed]
- Aulehla, A.; Pourquie, O. Oscillating signaling pathways during embryonic development. Curr. Opin. Cell Biol. 2008, 20, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Bessho, Y.; Sakata, R.; Komatsu, S.; Shiota, K.; Yamada, S.; Kageyama, R. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 2001, 15, 2642–2647. [Google Scholar] [CrossRef] [PubMed]
- Aulehla, A.; Johnson, R.L. Dynamic expression of lunatic fringe suggests a link between notch signaling and an autonomous cellular oscillator driving somite segmentation. Dev. Biol. 1999, 207, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Holley, S.A.; Geisler, R.; Nusslein-Volhard, C. Control of her1 expression during zebrafish somitogenesis by a delta-dependent oscillator and an independent wave-front activity. Genes Dev. 2000, 14, 1678–1690. [Google Scholar] [PubMed]
- Jiang, Y.J.; Aerne, B.L.; Smithers, L.; Haddon, C.; Ish-Horowicz, D.; Lewis, J. Notch signalling and the synchronization of the somite segmentation clock. Nature 2000, 408, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Jouve, C.; Palmeirim, I.; Henrique, D.; Beckers, J.; Gossler, A.; Ish-Horowicz, D.; Pourquie, O. Notch signalling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development 2000, 127, 1421–1429. [Google Scholar] [PubMed]
- Leimeister, C.; Dale, K.; Fischer, A.; Klamt, B.; Hrabe de Angelis, M.; Radtke, F.; McGrew, M.J.; Pourquie, O.; Gessler, M. Oscillating expression of c-Hey2 in the presomitic mesoderm suggests that the segmentation clock may use combinatorial signaling through multiple interacting bHLH factors. Dev. Biol. 2000, 227, 91–103. [Google Scholar] [CrossRef] [PubMed]
- McGrew, M.J.; Dale, J.K.; Fraboulet, S.; Pourquie, O. The lunatic fringe gene is a target of the molecular clock linked to somite segmentation in avian embryos. Curr. Biol. 1998, 8, 979–982. [Google Scholar] [CrossRef]
- Palmeirim, I.; Henrique, D.; Ish-Horowicz, D.; Pourquie, O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 1997, 91, 639–648. [Google Scholar] [CrossRef]
- Sawada, A.; Fritz, A.; Jiang, Y.J.; Yamamoto, A.; Yamasu, K.; Kuroiwa, A.; Saga, Y.; Takeda, H. Zebrafish Mesp family genes, mesp-a and mesp-b are segmentally expressed in the presomitic mesoderm, and Mesp-b confers the anterior identity to the developing somites. Development 2000, 127, 1691–1702. [Google Scholar] [PubMed]
- Aulehla, A.; Wehrle, C.; Brand-Saberi, B.; Kemler, R.; Gossler, A.; Kanzler, B.; Herrmann, B.G. Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell 2003, 4, 395–406. [Google Scholar] [CrossRef]
- Lustig, B.; Jerchow, B.; Sachs, M.; Weiler, S.; Pietsch, T.; Karsten, U.; van de Wetering, M.; Clevers, H.; Schlag, P.M.; Birchmeier, W.; et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol. 2002, 22, 1184–1193. [Google Scholar] [CrossRef] [PubMed]
- Bernkopf, D.B.; Hadjihannas, M.V.; Behrens, J. Negative-feedback regulation of the Wnt pathway by conductin/axin2 involves insensitivity to upstream signalling. J. Cell Sci. 2015, 128, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Seidensticker, M.J.; Behrens, J. Biochemical interactions in the wnt pathway. Biochim. Biophys. Acta 2000, 1495, 168–182. [Google Scholar] [CrossRef]
- Sonnen, K.F.; Lauschke, V.M.; Uraji, J.; Falk, H.J.; Petersen, Y.; Funk, M.C.; Beaupeux, M.; Francois, P.; Merten, C.A.; Aulehla, A. Modulation of Phase Shift between Wnt and Notch Signaling Oscillations Controls Mesoderm Segmentation. Cell 2018, 172, 1079–1090. [Google Scholar] [CrossRef] [PubMed]
- Dale, J.K.; Malapert, P.; Chal, J.; Vilhais-Neto, G.; Maroto, M.; Johnson, T.; Jayasinghe, S.; Trainor, P.; Herrmann, B.; Pourquie, O. Oscillations of the snail genes in the presomitic mesoderm coordinate segmental patterning and morphogenesis in vertebrate somitogenesis. Dev. Cell 2006, 10, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Dequeant, M.L.; Glynn, E.; Gaudenz, K.; Wahl, M.; Chen, J.; Mushegian, A.; Pourquie, O. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science 2006, 314, 1595–1598. [Google Scholar] [CrossRef] [PubMed]
- Niwa, Y.; Masamizu, Y.; Liu, T.; Nakayama, R.; Deng, C.X.; Kageyama, R. The initiation and propagation of Hes7 oscillation are cooperatively regulated by Fgf and notch signaling in the somite segmentation clock. Dev. Cell 2007, 13, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Goldbeter, A.; Pourquie, O. Modeling the segmentation clock as a network of coupled oscillations in the Notch, Wnt and FGF signaling pathways. J. Theor. Biol. 2008, 252, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Tsakiridis, A.; Huang, Y.; Blin, G.; Skylaki, S.; Wymeersch, F.; Osorno, R.; Economou, C.; Karagianni, E.; Zhao, S.; Lowell, S.; et al. Distinct Wnt-driven primitive streak-like populations reflect in vivo lineage precursors. Development 2014, 141, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H.; Batlle, E. SnapShot: The intestinal crypt. Cell 2013, 152, 1198. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. The intestinal crypt, a prototype stem cell compartment. Cell 2013, 154, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Clevers, H. Coexistence of quiescent and active adult stem cells in mammals. Science 2010, 327, 542–545. [Google Scholar] [CrossRef] [PubMed]
- Marshman, E.; Booth, C.; Potten, C.S. The intestinal epithelial stem cell. Bioessays 2002, 24, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Bjerknes, M.; Cheng, H. Clonal analysis of mouse intestinal epithelial progenitors. Gastroenterology 1999, 116, 7–14. [Google Scholar] [CrossRef]
- Langlands, A.J.; Almet, A.A.; Appleton, P.L.; Newton, I.P.; Osborne, J.M.; Nathke, I.S. Paneth Cell-Rich Regions Separated by a Cluster of Lgr5+ Cells Initiate Crypt Fission in the Intestinal Stem Cell Niche. PLoS Biol. 2016, 14, e1002491. [Google Scholar] [CrossRef] [PubMed]
- Fauser, J.K.; Donato, R.P.; Woenig, J.A.; Proctor, S.J.; Trotta, A.P.; Grover, P.K.; Howarth, G.S.; Penttila, I.A.; Cummins, A.G. Wnt blockade with dickkopf reduces intestinal crypt fission and intestinal growth in infant rats. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Ootani, A.; Li, X.; Sangiorgi, E.; Ho, Q.T.; Ueno, H.; Toda, S.; Sugihara, H.; Fujimoto, K.; Weissman, I.L.; Capecchi, M.R.; et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 2009, 15, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.A.; Inamura, K.; Yamauchi, M.; Nishihara, R.; Mima, K.; Sukawa, Y.; Li, T.; Yasunari, M.; Morikawa, T.; Fitzgerald, K.C.; et al. Loss of CDH1 (E-cadherin) expression is associated with infiltrative tumour growth and lymph node metastasis. Br. J. Cancer 2016, 114, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.M.; Fang, C.M.; Chuah, L.H.; Leong, C.O.; Ngai, S.C. E-cadherin: Its dysregulation in carcinogenesis and clinical implications. Crit. Rev. Oncol. Hematol. 2018, 121, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Krausova, M.; Korinek, V. Signal transduction pathways participating in homeostasis and malignant transformation of the intestinal tissue. Neoplasma 2012, 59, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; Ridgway, R.A.; van Es, J.H.; van de Wetering, M.; Begthel, H.; van den Born, M.; Danenberg, E.; Clarke, A.R.; Sansom, O.J.; Clevers, H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 2009, 457, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Powell, A.E.; Vlacich, G.; Zhao, Z.Y.; McKinley, E.T.; Washington, M.K.; Manning, H.C.; Coffey, R.J. Inducible loss of one Apc allele in Lrig1-expressing progenitor cells results in multiple distal colonic tumors with features of familial adenomatous polyposis. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G16–G23. [Google Scholar] [CrossRef] [PubMed]
- Scoville, D.H.; Sato, T.; He, X.C.; Li, L. Current view: Intestinal stem cells and signaling. Gastroenterology 2008, 134, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Mah, A.T.; Yan, K.S.; Kuo, C.J. Wnt pathway regulation of intestinal stem cells. J. Physiol. 2016, 594, 4837–4847. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.W.; Hirokawa, Y.; Gardiner, B.S.; Smith, D.W.; Burgess, A.W. Colon cryptogenesis: Asymmetric budding. PLoS ONE 2013, 8, e78519. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.W.; Hirokawa, Y.; Burgess, A.W. Analysis of Wnt signalling dynamics during colon crypt development in 3D culture. Sci. Rep. 2015, 5, 11036. [Google Scholar] [CrossRef] [PubMed]
- Vanuytsel, T.; Senger, S.; Fasano, A.; Shea-Donohue, T. Major signaling pathways in intestinal stem cells. Biochim. Biophys. Acta 2013, 1830, 2410–2426. [Google Scholar] [CrossRef] [PubMed]
- Fre, S.; Huyghe, M.; Mourikis, P.; Robine, S.; Louvard, D.; Artavanis-Tsakonas, S. Notch signals control the fate of immature progenitor cells in the intestine. Nature 2005, 435, 964–968. [Google Scholar] [CrossRef] [PubMed]
- Crosnier, C.; Stamataki, D.; Lewis, J. Organizing cell renewal in the intestine: Stem cells, signals and combinatorial control. Nat. Rev. Genet. 2006, 7, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Kosinski, C.; Stange, D.E.; Xu, C.; Chan, A.S.; Ho, C.; Yuen, S.T.; Mifflin, R.C.; Powell, D.W.; Clevers, H.; Leung, S.Y.; et al. Indian hedgehog regulates intestinal stem cell fate through epithelial-mesenchymal interactions during development. Gastroenterology 2010, 139, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Madison, B.B.; Braunstein, K.; Kuizon, E.; Portman, K.; Qiao, X.T.; Gumucio, D.L. Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development 2005, 132, 279–289. [Google Scholar] [CrossRef] [PubMed]
- van den Brink, G.R.; Bleuming, S.A.; Hardwick, J.C.; Schepman, B.L.; Offerhaus, G.J.; Keller, J.J.; Nielsen, C.; Gaffield, W.; van Deventer, S.J.; Roberts, D.J.; et al. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat. Genet. 2004, 36, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Auclair, B.A.; Benoit, Y.D.; Rivard, N.; Mishina, Y.; Perreault, N. Bone morphogenetic protein signaling is essential for terminal differentiation of the intestinal secretory cell lineage. Gastroenterology 2007, 133, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Zhang, N.; Zheng, Y.; de Wilde, R.F.; Maitra, A.; Pan, D. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010, 24, 2383–2388. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Malenka, R.C. β-catenin is critical for dendritic morphogenesis. Nat. Neurosci. 2003, 6, 1169–1177. [Google Scholar] [CrossRef] [PubMed]
- Lucas, F.R.; Salinas, P.C. WNT-7a induces axonal remodeling and increases synapsin I levels in cerebellar neurons. Dev. Biol. 1997, 192, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Salinas, P.C.; Fletcher, C.; Copeland, N.G.; Jenkins, N.A.; Nusse, R. Maintenance of Wnt-3 expression in Purkinje cells of the mouse cerebellum depends on interactions with granule cells. Development 1994, 120, 1277–1286. [Google Scholar] [PubMed]
- Togashi, H.; Abe, K.; Mizoguchi, A.; Takaoka, K.; Chisaka, O.; Takeichi, M. Cadherin regulates dendritic spine morphogenesis. Neuron 2002, 35, 77–89. [Google Scholar] [CrossRef]
- Hall, A.C.; Lucas, F.R.; Salinas, P.C. Axonal remodeling and synaptic differentiation in the cerebellum is regulated by WNT-7a signaling. Cell 2000, 100, 525–535. [Google Scholar] [CrossRef]
- Uchida, N.; Honjo, Y.; Johnson, K.R.; Wheelock, M.J.; Takeichi, M. The catenin/cadherin adhesion system is localized in synaptic junctions bordering transmitter release zones. J. Cell Biol. 1996, 135, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.L.; Tanaka, H. N-cadherin redistribution during synaptogenesis in hippocampal neurons. J. Neurosci. 1998, 18, 6892–6904. [Google Scholar] [CrossRef] [PubMed]
- Cadigan, K.M.; Nusse, R. Wnt signaling: A common theme in animal development. Genes Dev. 1997, 11, 3286–3305. [Google Scholar] [CrossRef] [PubMed]
- Packard, M.; Koo, E.S.; Gorczyca, M.; Sharpe, J.; Cumberledge, S.; Budnik, V. The Drosophila Wnt, wingless, provides an essential signal for pre- and postsynaptic differentiation. Cell 2002, 111, 319–330. [Google Scholar] [CrossRef]
- Nichols, J.; Smith, A. Naive and primed pluripotent states. Cell Stem Cell 2009, 4, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Meijer, L.; Skaltsounis, L.; Greengard, P.; Brivanlou, A.H. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 2004, 10, 55–63. [Google Scholar] [CrossRef] [PubMed]
- ten Berge, D.; Kurek, D.; Blauwkamp, T.; Koole, W.; Maas, A.; Eroglu, E.; Siu, R.K.; Nusse, R. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat. Cell Biol. 2011, 13, 1070–1075. [Google Scholar] [CrossRef] [PubMed]
- Ying, Q.L.; Wray, J.; Nichols, J.; Batlle-Morera, L.; Doble, B.; Woodgett, J.; Cohen, P.; Smith, A. The ground state of embryonic stem cell self-renewal. Nature 2008, 453, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Godwin, S.; Ward, D.; Pedone, E.; Homer, M.; Fletcher, A.G.; Marucci, L. An extended model for culture-dependent heterogenous gene expression and proliferation dynamics in mouse embryonic stem cells. NPJ Syst. Biol. Appl. 2017, 3, 19. [Google Scholar] [CrossRef] [PubMed]
- Marks, H.; Kalkan, T.; Menafra, R.; Denissov, S.; Jones, K.; Hofemeister, H.; Nichols, J.; Kranz, A.; Stewart, A.F.; Smith, A.; et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 2012, 149, 590–604. [Google Scholar] [CrossRef] [PubMed]
- Alexandrova, S.; Kalkan, T.; Humphreys, P.; Riddell, A.; Scognamiglio, R.; Trumpp, A.; Nichols, J. Selection and dynamics of embryonic stem cell integration into early mouse embryos. Development 2016, 143, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Faunes, F.; Hayward, P.; Descalzo, S.M.; Chatterjee, S.S.; Balayo, T.; Trott, J.; Christoforou, A.; Ferrer-Vaquer, A.; Hadjantonakis, A.K.; Dasgupta, R.; et al. A membrane-associated beta-catenin/Oct4 complex correlates with ground-state pluripotency in mouse embryonic stem cells. Development 2013, 140, 1171–1183. [Google Scholar] [CrossRef] [PubMed]
- Miyabayashi, T.; Teo, J.L.; Yamamoto, M.; McMillan, M.; Nguyen, C.; Kahn, M. Wnt/beta-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. USA 2007, 104, 5668–5673. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.F.; Johnstone, S.E.; Newman, J.J.; Kagey, M.H.; Young, R.A. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008, 22, 746–755. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R.; Fuerer, C.; Ching, W.; Harnish, K.; Logan, C.; Zeng, A.; ten Berge, D.; Kalani, Y. Wnt signaling and stem cell control. Cold Spring Harb. Symp. Quant. Biol. 2008, 73, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.S.; Saj, A.; Gocha, T.; Murphy, M.; Gonsalves, F.C.; Zhang, X.; Hayward, P.; Akgol Oksuz, B.; Shen, S.S.; Madar, A.; et al. Inhibition of β-catenin-TCF1 interaction delays differentiation of mouse embryonic stem cells. J. Cell Biol. 2015, 211, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Munoz Descalzo, S.; Rue, P.; Faunes, F.; Hayward, P.; Jakt, L.M.; Balayo, T.; Garcia-Ojalvo, J.; Martinez Arias, A. A competitive protein interaction network buffers Oct4-mediated differentiation to promote pluripotency in embryonic stem cells. Mol. Syst. Biol. 2013, 9, 694. [Google Scholar] [CrossRef] [PubMed]
- Marucci, L.; Pedone, E.; Di Vicino, U.; Sanuy-Escribano, B.; Isalan, M.; Cosma, M.P. beta-catenin fluctuates in mouse ESCs and is essential for Nanog-mediated reprogramming of somatic cells to pluripotency. Cell Rep. 2014, 8, 1686–1696. [Google Scholar] [CrossRef] [PubMed]
- Kielman, M.F.; Rindapaa, M.; Gaspar, C.; van Poppel, N.; Breukel, C.; van Leeuwen, S.; Taketo, M.M.; Roberts, S.; Smits, R.; Fodde, R. Apc modulates embryonic stem-cell differentiation by controlling the dosage of beta-catenin signaling. Nat. Genet. 2002, 32, 594–605. [Google Scholar] [CrossRef] [PubMed]
- Buganim, Y.; Faddah, D.A.; Jaenisch, R. Mechanisms and models of somatic cell reprogramming. Nat. Rev. Genet. 2013, 14, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Aulicino, F.; Theka, I.; Ombrato, L.; Lluis, F.; Cosma, M.P. Temporal perturbation of the Wnt signaling pathway in the control of cell reprogramming is modulated by TCF1. Stem Cell Rep. 2014, 2, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Ho, R.; Papp, B.; Hoffman, J.A.; Merrill, B.J.; Plath, K. Stage-specific regulation of reprogramming to induced pluripotent stem cells by Wnt signaling and T cell factor proteins. Cell Rep. 2013, 3, 2113–2126. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Nakajima-Koyama, M.; Lee, J.; Nishida, E. Transient Expression of WNT2 Promotes Somatic Cell Reprogramming by Inducing beta-Catenin Nuclear Accumulation. Stem Cell Rep. 2016, 6, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Marson, A.; Levine, S.S.; Cole, M.F.; Frampton, G.M.; Brambrink, T.; Johnstone, S.; Guenther, M.G.; Johnston, W.K.; Wernig, M.; Newman, J.; et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 2008, 134, 521–533. [Google Scholar] [CrossRef] [PubMed]
- Lluis, F.; Pedone, E.; Pepe, S.; Cosma, M.P. Periodic activation of Wnt/β-catenin signaling enhances somatic cell reprogramming mediated by cell fusion. Cell Stem Cell 2008, 3, 493–507. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, L.; Ayyash, M.; Novershtern, N.; Hanna, J.H. Dynamic stem cell states: Naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016, 17, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Mekhoubad, S.; Bock, C.; de Boer, A.S.; Kiskinis, E.; Meissner, A.; Eggan, K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell 2012, 10, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Chan, M.M.; Humm, K.C.; Karnik, R.; Mekhoubad, S.; Regev, A.; Eggan, K.; Meissner, A. DNA methylation dynamics of the human preimplantation embryo. Nature 2014, 511, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Gafni, O.; Weinberger, L.; Mansour, A.A.; Manor, Y.S.; Chomsky, E.; Ben-Yosef, D.; Kalma, Y.; Viukov, S.; Maza, I.; Zviran, A.; et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013, 504, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Cheng, A.W.; Saha, K.; Kim, J.; Lengner, C.J.; Soldner, F.; Cassady, J.P.; Muffat, J.; Carey, B.W.; Jaenisch, R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. USA 2010, 107, 9222–9227. [Google Scholar] [CrossRef] [PubMed]
- Davidson, K.C.; Adams, A.M.; Goodson, J.M.; McDonald, C.E.; Potter, J.C.; Berndt, J.D.; Biechele, T.L.; Taylor, R.J.; Moon, R.T. Wnt/β-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proc. Natl. Acad. Sci. USA 2012, 109, 4485–4490. [Google Scholar] [CrossRef] [PubMed]
- Theunissen, T.W.; Powell, B.E.; Wang, H.; Mitalipova, M.; Faddah, D.A.; Reddy, J.; Fan, Z.P.; Maetzel, D.; Ganz, K.; Shi, L.; et al. Systematic Identification of Culture Conditions for Induction and Maintenance of Naive Human Pluripotency. Cell Stem Cell 2014, 15, 524–526. [Google Scholar] [CrossRef] [PubMed]
- Takashima, Y.; Guo, G.; Loos, R.; Nichols, J.; Ficz, G.; Krueger, F.; Oxley, D.; Santos, F.; Clarke, J.; Mansfield, W.; et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 2014, 158, 1254–1269. [Google Scholar] [CrossRef] [PubMed]
- Pastor, W.A.; Chen, D.; Liu, W.; Kim, R.; Sahakyan, A.; Lukianchikov, A.; Plath, K.; Jacobsen, S.E.; Clark, A.T. Naive Human Pluripotent Cells Feature a Methylation Landscape Devoid of Blastocyst or Germline Memory. Cell Stem Cell 2016, 18, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Korinek, V.; Barker, N.; Moerer, P.; van Donselaar, E.; Huls, G.; Peters, P.J.; Clevers, H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 1998, 19, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Willert, K.; Brown, J.D.; Danenberg, E.; Duncan, A.W.; Weissman, I.L.; Reya, T.; Yates, J.R., 3rd; Nusse, R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 2003, 423, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Reya, T.; Duncan, A.W.; Ailles, L.; Domen, J.; Scherer, D.C.; Willert, K.; Hintz, L.; Nusse, R.; Weissman, I.L. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature 2003, 423, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R. Wnt signaling and stem cell control. Cell Res. 2008, 18, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Luis, T.C.; Naber, B.A.; Roozen, P.P.; Brugman, M.H.; de Haas, E.F.; Ghazvini, M.; Fibbe, W.E.; van Dongen, J.J.; Fodde, R.; Staal, F.J. Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell Stem Cell 2011, 9, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Dai, W. Genomic Instability and Cancer. J. Carcinog. Mutagen. 2014, 5. [Google Scholar] [CrossRef]
- Nowell, P.C. The clonal evolution of tumor cell populations. Science 1976, 194, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, A.M.; Fuentes, D.; Morshid, A.I.; Burke, M.R.; Kaseb, A.O.; Hassan, M.; Hazle, J.D.; Elsayes, K.M. Role of Wnt/beta-catenin signaling in hepatocellular carcinoma, pathogenesis, and clinical significance. J. Hepatocell. Carcinoma 2018, 5, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Morin, P.J. beta-catenin signaling and cancer. Bioessays 1999, 21, 1021–1030. [Google Scholar] [CrossRef]
- Guo, J.; Cagatay, T.; Zhou, G.; Chan, C.C.; Blythe, S.; Suyama, K.; Zheng, L.; Pan, K.; Qian, C.; Hamelin, R.; et al. Mutations in the human naked cuticle homolog NKD1 found in colorectal cancer alter Wnt/Dvl/beta-catenin signaling. PLoS ONE 2009, 4, e7982. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Hahn, W.C. Involvement of PP2A in viral and cellular transformation. Oncogene 2005, 24, 7746–7755. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Possemato, R.; Campbell, K.T.; Plattner, C.A.; Pallas, D.C.; Hahn, W.C. Identification of specific PP2A complexes involved in human cell transformation. Cancer Cell 2004, 5, 127–136. [Google Scholar] [CrossRef]
- Millward, T.A.; Zolnierowicz, S.; Hemmings, B.A. Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem. Sci. 1999, 24, 186–191. [Google Scholar] [CrossRef]
- Yamamoto, H.; Hinoi, T.; Michiue, T.; Fukui, A.; Usui, H.; Janssens, V.; Van Hoof, C.; Goris, J.; Asashima, M.; Kikuchi, A. Inhibition of the Wnt signaling pathway by the PR61 subunit of protein phosphatase 2A. J. Biol. Chem. 2001, 276, 26875–26882. [Google Scholar] [CrossRef] [PubMed]
- Seeling, J.M.; Miller, J.R.; Gil, R.; Moon, R.T.; White, R.; Virshup, D.M. Regulation of β-catenin signaling by the B56 subunit of protein phosphatase 2A. Science 1999, 283, 2089–2091. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.; Zeng, L.; Costantini, F. Identification of a domain of Axin that binds to the serine/threonine protein phosphatase 2A and a self-binding domain. J. Biol. Chem. 1999, 274, 3439–3445. [Google Scholar] [CrossRef] [PubMed]
- Creyghton, M.P.; Roel, G.; Eichhorn, P.J.; Hijmans, E.M.; Maurer, I.; Destree, O.; Bernards, R. PR72, a novel regulator of Wnt signaling required for Naked cuticle function. Genes Dev. 2005, 19, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Gotz, J.; Probst, A.; Mistl, C.; Nitsch, R.M.; Ehler, E. Distinct role of protein phosphatase 2A subunit Calpha in the regulation of E-cadherin and β-catenin during development. Mech. Dev. 2000, 93, 83–93. [Google Scholar] [CrossRef]
- Clements, W.M.; Wang, J.; Sarnaik, A.; Kim, O.J.; MacDonald, J.; Fenoglio-Preiser, C.; Groden, J.; Lowy, A.M. beta-Catenin mutation is a frequent cause of Wnt pathway activation in gastric cancer. Cancer Res. 2002, 62, 3503–3506. [Google Scholar] [PubMed]
- Laurent-Puig, P.; Legoix, P.; Bluteau, O.; Belghiti, J.; Franco, D.; Binot, F.; Monges, G.; Thomas, G.; Bioulac-Sage, P.; Zucman-Rossi, J. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology 2001, 120, 1763–1773. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhong, W.; Yuan, J.; Yan, C.; Hu, S.; Tong, Y.; Mao, Y.; Hu, T.; Zhang, B.; Song, G. Involvement of Wnt/β-catenin signaling in the mesenchymal stem cells promote metastatic growth and chemoresistance of cholangiocarcinoma. Oncotarget 2015, 6, 42276–42289. [Google Scholar] [CrossRef] [PubMed]
- Crago, A.M.; Chmielecki, J.; Rosenberg, M.; O’Connor, R.; Byrne, C.; Wilder, F.G.; Thorn, K.; Agius, P.; Kuk, D.; Socci, N.D.; et al. Near universal detection of alterations in CTNNB1 and Wnt pathway regulators in desmoid-type fibromatosis by whole-exome sequencing and genomic analysis. Genes Chromosomes Cancer 2015, 54, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Lee, J.K.; Ahn, S.H.; Lee, J.; Nam, D.H. WNT signaling in glioblastoma and therapeutic opportunities. Lab. Investig. 2016, 96, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Yeung, J.; Esposito, M.T.; Gandillet, A.; Zeisig, B.B.; Griessinger, E.; Bonnet, D.; So, C.W. β-Catenin mediates the establishment and drug resistance of MLL leukemic stem cells. Cancer Cell 2010, 18, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.B.; Kim, J.Y.; Cho, S.D.; Park, K.S.; Jung, J.Y.; Lee, H.Y.; Hong, I.S.; Nam, J.S. Blockade of Wnt/β-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci. Rep. 2015, 5, 12465. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, S.J.; Rambow, F.; Kumasaka, M.; Champeval, D.; Bellacosa, A.; Delmas, V.; Larue, L. β-catenin inhibits melanocyte migration but induces melanoma metastasis. Oncogene 2013, 32, 2230–2238. [Google Scholar] [CrossRef] [PubMed]
- Kruck, S.; Eyrich, C.; Scharpf, M.; Sievert, K.D.; Fend, F.; Stenzl, A.; Bedke, J. Impact of an altered Wnt1/β-catenin expression on clinicopathology and prognosis in clear cell renal cell carcinoma. Int. J. Mol. Sci. 2013, 14, 10944–10957. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Liang, X.; Cui, W.; Ober-Blobaum, J.L.; Vazzana, J.; Shrikant, P.A.; Lee, K.P.; Clausen, B.E.; Mellman, I.; Jiang, A. β-Catenin in dendritic cells exerts opposite functions in cross-priming and maintenance of CD8+ T cells through regulation of IL-10. Proc. Natl. Acad. Sci. USA 2015, 112, 2823–2828. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S.; Gajewski, T.F. A new paradigm for tumor immune escape: β-catenin-driven immune exclusion. J. Immunother. Cancer 2015, 3, 43. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S.; Bao, R.; Gajewski, T.F. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 2015, 523, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Swafford, D.; Manicassamy, S. Wnt signaling in dendritic cells: Its role in regulation of immunity and tolerance. Discov. Med. 2015, 19, 303–310. [Google Scholar] [PubMed]
- Hong, Y.; Manoharan, I.; Suryawanshi, A.; Majumdar, T.; Angus-Hill, M.L.; Koni, P.A.; Manicassamy, B.; Mellor, A.L.; Munn, D.H.; Manicassamy, S. beta-catenin promotes regulatory T-cell responses in tumors by inducing vitamin A metabolism in dendritic cells. Cancer Res. 2015, 75, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Gaujoux, S.; Grabar, S.; Fassnacht, M.; Ragazzon, B.; Launay, P.; Libe, R.; Chokri, I.; Audebourg, A.; Royer, B.; Sbiera, S.; et al. β-catenin activation is associated with specific clinical and pathologic characteristics and a poor outcome in adrenocortical carcinoma. Clin. Cancer Res. 2011, 17, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Logan, C.Y.; Nusse, R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004, 20, 781–810. [Google Scholar] [CrossRef] [PubMed]
- Larraguibel, J.; Weiss, A.R.; Pasula, D.J.; Dhaliwal, R.S.; Kondra, R.; Van Raay, T.J. Wnt ligand-dependent activation of the negative feedback regulator Nkd1. Mol. Biol. Cell 2015, 26, 2375–2384. [Google Scholar] [CrossRef] [PubMed]
- Fliniaux, I.; Mikkola, M.L.; Lefebvre, S.; Thesleff, I. Identification of dkk4 as a target of Eda-A1/Edar pathway reveals an unexpected role of ectodysplasin as inhibitor of Wnt signalling in ectodermal placodes. Dev. Biol. 2008, 320, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Aurrekoetxea, M.; Irastorza, I.; Garcia-Gallastegui, P.; Jimenez-Rojo, L.; Nakamura, T.; Yamada, Y.; Ibarretxe, G.; Unda, F.J. Wnt/β-Catenin Regulates the Activity of Epiprofin/Sp6, SHH, FGF, and BMP to Coordinate the Stages of Odontogenesis. Front. Cell Dev. Biol. 2016, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.Y.; Rath, O.; Zebisch, A.; Choo, S.M.; Kolch, W.; Cho, K.H. Functional roles of multiple feedback loops in extracellular signal-regulated kinase and Wnt signaling pathways that regulate epithelial-mesenchymal transition. Cancer Res. 2010, 70, 6715–6724. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, P.K.; Bouchie, M.P.; Nita-Lazar, M.; Yang, H.Y.; Kukuruzinska, M.A. Coordinate regulation of N-glycosylation gene DPAGT1, canonical Wnt signaling and E-cadherin adhesion. J. Cell Sci. 2013, 126, 484–496. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Hottiger, M.O. Crosstalk between Wnt/beta-Catenin and NF-kappaB Signaling Pathway during Inflammation. Front. Immunol. 2016, 7, 378. [Google Scholar] [CrossRef] [PubMed]
- Fagnocchi, L.; Mazzoleni, S.; Zippo, A. Integration of Signaling Pathways with the Epigenetic Machinery in the Maintenance of Stem Cells. Stem Cells Int. 2016, 2016, 8652748. [Google Scholar] [CrossRef] [PubMed]
- Kühl, M.K.B.; Kestler, H.A. Mathematical Models of Wnt Signaling Pathways. In Wnt Signaling in Development and Disease: Molecular Mechanisms and Biological Functions; Hoppler, S.M.R.T., Ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2014. [Google Scholar]
- Marucci, L. Nanog Dynamics in Mouse Embryonic Stem Cells: Results from Systems Biology Approaches. Stem Cells Int. 2017, 2017, 7160419. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Salic, A.; Kruger, R.; Heinrich, R.; Kirschner, M.W. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol. 2003, 1, E10. [Google Scholar] [CrossRef] [PubMed]
- Goentoro, L.; Shoval, O.; Kirschner, M.W.; Alon, U. The incoherent feedforward loop can provide fold-change detection in gene regulation. Mol. Cell 2009, 36, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Bryja, V.; Cervenka, I.; Cajanek, L. The connections of Wnt pathway components with cell cycle and centrosome: Side effects or a hidden logic? Crit. Rev. Biochem. Mol. Biol. 2017, 52, 614–637. [Google Scholar] [CrossRef] [PubMed]
- Davidson, G. The cell cycle and Wnt. Cell Cycle 2010, 9, 1667–1668. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Lewis, B.; Fletcher, A.G.; Dale, T.C.; Byrne, H.M. Toward a quantitative understanding of the Wnt/beta-catenin pathway through simulation and experiment. Wiley Interdiscip. Rev. Syst. Biol. Med. 2013, 5, 391–407. [Google Scholar] [CrossRef] [PubMed]
- Dunn, S.J.; Osborne, J.M.; Appleton, P.L.; Nathke, I. Combined changes in Wnt signaling response and contact inhibition induce altered proliferation in radiation-treated intestinal crypts. Mol. Biol. Cell 2016, 27, 1863–1874. [Google Scholar] [CrossRef] [PubMed]
- Matyjaszkiewicz, A.; Fiore, G.; Annunziata, F.; Grierson, C.S.; Savery, N.J.; Marucci, L.; di Bernardo, M. BSim 2.0: An Advanced Agent-Based Cell Simulator. ACS Synth. Biol. 2017, 6, 1969–1972. [Google Scholar] [CrossRef] [PubMed]
- Kretzschmar, K.; Clevers, H. Organoids: Modeling Development and the Stem Cell Niche in a Dish. Dev. Cell 2016, 38, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Sopko, N.A.; Kates, M.; Liu, X.; Joice, G.; McConkey, D.J.; Bivalacqua, T.J. Three-dimensional organoid culture reveals involvement of Wnt/beta-catenin pathway in proliferation of bladder cancer cells. Oncotarget 2018, 9, 11060–11070. [Google Scholar] [CrossRef] [PubMed]
- Andersen, P.; Tampakakis, E.; Jimenez, D.V.; Kannan, S.; Miyamoto, M.; Shin, H.K.; Saberi, A.; Murphy, S.; Sulistio, E.; Chelko, S.P.; et al. Precardiac organoids form two heart fields via Bmp/Wnt signaling. Nat. Commun. 2018, 9, 3140. [Google Scholar] [CrossRef] [PubMed]
- Koch, S. Extrinsic control of Wnt signaling in the intestine. Differentiation 2017, 97, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Clevers, H. Growing self-organizing mini-guts from a single intestinal stem cell: Mechanism and applications. Science 2013, 340, 1190–1194. [Google Scholar] [CrossRef] [PubMed]
- Postiglione, L.; Napolitano, S.; Pedone, E.; Rocca, D.L.; Aulicino, F.; Santorelli, M.; Tumaini, B.; Marucci, L.; di Bernardo, D. Regulation of Gene Expression and Signaling Pathway Activity in Mammalian Cells by Automated Microfluidics Feedback Control. ACS Synth. Biol. 2018, 7, 2558–2565. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.S.; Collins, J.J. Synthetic biology: Applications come of age. Nat. Rev. Genet. 2010, 11, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Fussenegger, M. Synthetic therapeutic gene circuits in mammalian cells. FEBS Lett. 2014, 588, 2537–2544. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).