Centromere Repeats: Hidden Gems of the Genome
Abstract
:1. Introduction
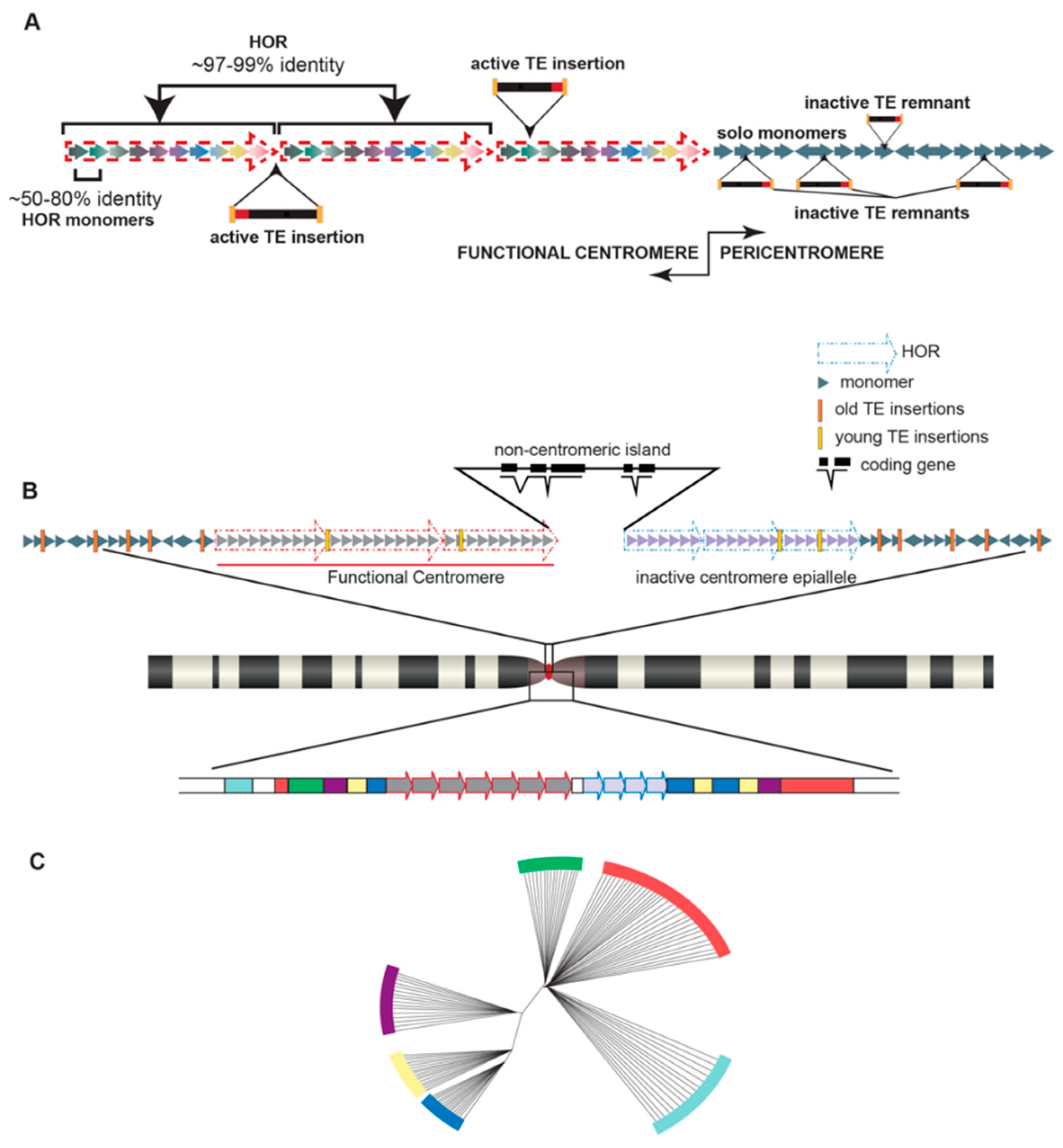
2. A Brief Primer on Satellite DNA in a Chromosomal Context
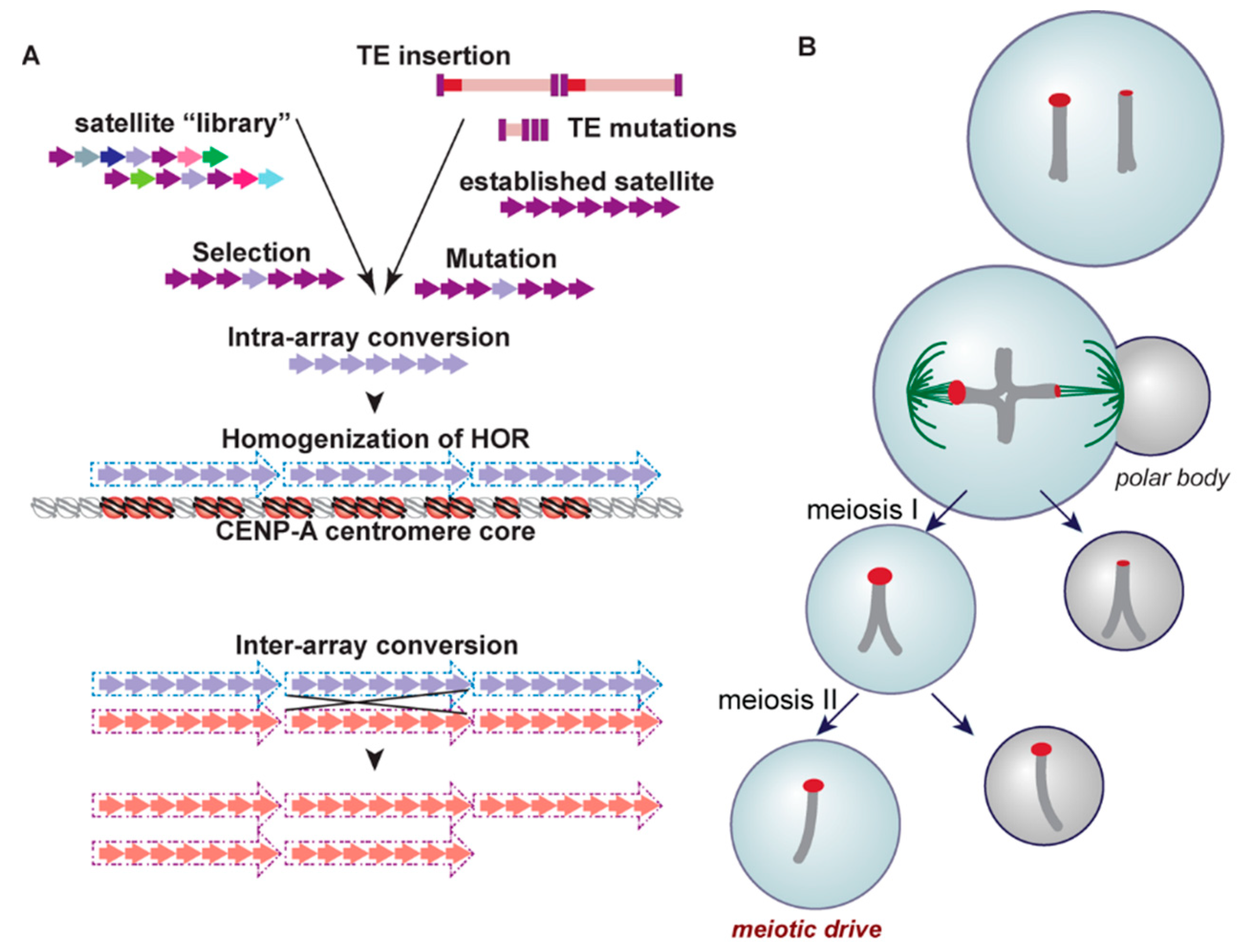
3. Centromere Repeats Endure Unique Evolutionary Processes
4. Satellites and Their Party Friends—Transposable Elements
5. Transcription in the Centromere—Let’s Get the Party Started!
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015, 23, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B.; Sniegowski, P.; Stephan, W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 1994, 371, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Flores, I.; Garrido-Ramos, M.A. The repetitive DNA content of eukaryotic genomes. Genome Dyn. 2012, 7, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvak, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Ramos, M.A. Satellite DNA: An Evolving Topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L.L.; Chew, K.; Sullivan, B.A. α satellite DNA variation and function of the human centromere. Nucleus 2017, 8, 331–339. [Google Scholar] [CrossRef] [Green Version]
- Jagannathan, M.; Cummings, R.; Yamashita, Y.M. A conserved function for pericentromeric satellite DNA. eLife 2018, 7, e34122. [Google Scholar] [CrossRef] [Green Version]
- Kit, S. Equilibrium sedimentation in density gradients of DNA preparations from animal tissues. J. Mol. Biol. 1961, 3, 711–716. [Google Scholar] [CrossRef]
- Sueoka, N. Variation and heterogeneity of base composition of deoxyribonucleic acids: A compilation of old and new data. J. Mol. Biol. 1961, 3, 31–40. [Google Scholar] [CrossRef]
- Navashin, S. On the nuclear dimorphism in somatic cells of Galtonia candicans. Bull. Acad. Imp. Sci 1912, 6, 375–385. [Google Scholar]
- Singer, M.F. Highly repeated sequences in mammalian genomes. Int. Rev. Cytol. 1982, 76, 67–112. [Google Scholar] [PubMed]
- Waring, M.; Britten, R.J. Nucleotide sequence repetition: A rapidly reassociating fraction of mouse DNA. Science 1966, 154, 791–794. [Google Scholar] [CrossRef]
- Horz, W.; Zachau, H.G. Characterization of distinct segments in mouse satellite DNA by restriction nucleases. Eur. J. Biochem. 1977, 73, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz Cde, F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Mishra, R.K.; Singh, L. Genome-wide analysis of microsatellite repeats in humans: Their abundance and density in specific genomic regions. Genome Biol. 2003, 4, R13. [Google Scholar] [CrossRef] [PubMed]
- Ramel, C. Mini- and microsatellites. Environ Health Perspect 1997, 105, 781–789. [Google Scholar] [PubMed]
- Naslund, K.; Saetre, P.; von Salome, J.; Bergstrom, T.F.; Jareborg, N.; Jazin, E. Genome-wide prediction of human VNTRs. Genomics 2005, 85, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Vergnaud, G.; Gauguier, D.; Schott, J.J.; Lepetit, D.; Lauthier, V.; Mariat, D.; Buard, J. Detection, cloning, and distribution of minisatellites in some mammalian genomes. EXS 1993, 67, 47–57. [Google Scholar]
- O’Sullivan, R.J.; Karlseder, J. Telomeres: Protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010, 11, 171. [Google Scholar] [CrossRef]
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626. [Google Scholar] [CrossRef]
- Bandaria, J.N.; Qin, P.; Berk, V.; Chu, S.; Yildiz, A. Shelterin protects chromosome ends by compacting telomeric chromatin. Cell 2016, 164, 735–746. [Google Scholar] [CrossRef]
- Wyatt, H.D.; West, S.C.; Beattie, T.L. InTERTpreting telomerase structure and function. Nucleic Acids Res. 2010, 38, 5609–5622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cusanelli, E.; Chartrand, P. Telomeric repeat-containing RNA TERRA: A noncoding RNA connecting telomere biology to genome integrity. Front. Genet. 2015, 6, 143. [Google Scholar] [CrossRef]
- Maddar, H.; Ratzkovsky, N.; Krauskopf, A. Role for telomere cap structure in meiosis. Mol. Biol. Cell. 2001, 12, 3191–3203. [Google Scholar] [CrossRef] [PubMed]
- Willard, H.F. Chromosome-specific organization of human α satellite DNA. Am. J. Hum. Genet. 1985, 37, 524–532. [Google Scholar] [PubMed]
- Van Hooser, A.A.; Ouspenski, I.I.; Gregson, H.C.; Starr, D.A.; Yen, T.J.; Goldberg, M.L.; Yokomori, K.; Earnshaw, W.C.; Sullivan, K.F.; Brinkley, B.R. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 2001, 114, 3529–3542. [Google Scholar] [PubMed]
- Willard, H.F.; Waye, J.S.; Skolnick, M.H.; Schwartz, C.E.; Powers, V.E.; England, S.B. Detection of restriction fragment length polymorphisms at the centromeres of human chromosomes by using chromosome-specific α satellite DNA probes: Implications for development of centromere-based genetic linkage maps. Proc. Natl. Acad. Sci. USA 1986, 83, 5611–5615. [Google Scholar] [CrossRef]
- Alexandrov, I.; Kazakov, A.; Tumeneva, I.; Shepelev, V.; Yurov, Y. α-satellite DNA of primates: Old and new families. Chromosoma 2001, 110, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, I.A.; Medvedev, L.I.; Mashkova, T.D.; Kisselev, L.L.; Romanova, L.Y.; Yurov, Y.B. Definition of a new α satellite suprachromosomal family characterized by monomeric organization. Nucleic Acids Res. 1993, 21, 2209–2215. [Google Scholar] [CrossRef] [PubMed]
- Shepelev, V.A.; Uralsky, L.I.; Alexandrov, A.A.; Yurov, Y.B.; Rogaev, E.I.; Alexandrov, I.A. Annotation of suprachromosomal families reveals uncommon types of α satellite organization in pericentromeric regions of hg38 human genome assembly. Genom. Data 2015, 5, 139–146. [Google Scholar] [CrossRef]
- McNulty, S.M.; Sullivan, B.A. α satellite DNA biology: Finding function in the recesses of the genome. Chromosome Res 2018, 26, 115–138. [Google Scholar] [CrossRef] [PubMed]
- Carine, K.; Jacquemin-Sablon, A.; Waltzer, E.; Mascarello, J.; Scheffler, I.E. Molecular characterization of human minichromosomes with centromere from chromosome 1 in human-hamster hybrid cells. Somat. Cell Mol. Genet. 1989, 15, 445–460. [Google Scholar] [CrossRef] [PubMed]
- Waye, J.S.; England, S.B.; Willard, H.F. Genomic organization of α satellite DNA on human chromosome 7: Evidence for two distinct alphoid domains on a single chromosome. Mol. Cell. Biol. 1987, 7, 349–356. [Google Scholar] [CrossRef]
- Tyler-Smith, C.; Brown, W.R. Structure of the major block of alphoid satellite DNA on the human Y chromosome. J. Mol. Biol. 1987, 195, 457–470. [Google Scholar] [CrossRef]
- Roizès, G. Human centromeric alphoid domains are periodically homogenized so that they vary substantially between homologues. Mechanism and implications for centromere functioning. Nucleic Acids Res. 2006, 34, 1912–1924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aldrup-Macdonald, M.E.; Sullivan, B.A. The past, present, and future of human centromere genomics. Genes 2014, 5, 33–50. [Google Scholar] [CrossRef]
- Alexandrov, I.A.; Mashkova, T.D.; Akopian, T.A.; Medvedev, L.I.; Kisselev, L.L.; Mitkevich, S.P.; Yurov, Y.B. Chromosome-specific α satellites: Two distinct families on human chromosome 18. Genomics 1991, 11, 15–23. [Google Scholar] [CrossRef]
- Alexandrov, I.A.; Mitkevich, S.P.; Yurov, Y.B. The phylogeny of human chromosome specific α satellites. Chromosoma 1988, 96, 443–453. [Google Scholar] [CrossRef]
- Rosandic, M.; Paar, V.; Basar, I.; Gluncic, M.; Pavin, N.; Pilas, I. CENP-B box and pJalpha sequence distribution in human α satellite higher-order repeats (HOR). Chromosome Res 2006, 14, 735–753. [Google Scholar] [CrossRef]
- Shepelev, V.A.; Alexandrov, A.A.; Yurov, Y.B.; Alexandrov, I.A. The evolutionary origin of man can be traced in the layers of defunct ancestral α satellites flanking the active centromeres of human chromosomes. PLoS Genet. 2009, 5, e1000641. [Google Scholar] [CrossRef]
- Aldrup-MacDonald, M.E.; Kuo, M.E.; Sullivan, L.L.; Chew, K.; Sullivan, B.A. Genomic variation within α satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genome Res. 2016, 26, 1301–1311. [Google Scholar] [CrossRef] [PubMed]
- Miga, K.H.; Newton, Y.; Jain, M.; Altemose, N.; Willard, H.F.; Kent, W.J. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 2014, 24, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Waye, J.S.; Willard, H.F. Structure, organization, and sequence of α satellite DNA from human chromosome 17: Evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol. Cell. Biol. 1986, 6, 3156–3165. [Google Scholar] [CrossRef]
- Rudd, M.K.; Willard, H.F. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004, 20, 529–533. [Google Scholar] [CrossRef]
- Warburton, P.E.; Willard, H.F. Interhomologue sequence variation of α satellite DNA from human chromosome 17: Evidence for concerted evolution along haplotypic lineages. J. Mol. Evol. 1995, 41, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Maloney, K.A.; Sullivan, L.L.; Matheny, J.E.; Strome, E.D.; Merrett, S.L.; Ferris, A.; Sullivan, B.A. Functional epialleles at an endogenous human centromere. Proc. Natl. Acad. Sci. USA 2012, 109, 13704–13709. [Google Scholar] [CrossRef] [Green Version]
- Jain, M.; Olsen, H.E.; Turner, D.J.; Stoddart, D.; Bulazel, K.V.; Paten, B.; Haussler, D.; Willard, H.F.; Akeson, M.; Miga, K.H. Linear assembly of a human centromere on the Y chromosome. Nat. Biotechnol. 2018, 36, 321–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durfy, S.J.; Willard, H.F. Molecular analysis of a polymorphic domain of α satellite from the human X chromosome. Am. J. Hum. Genet. 1987, 41, 391–401. [Google Scholar] [PubMed]
- Schindelhauer, D.; Schwarz, T. Evidence for a fast, intrachromosomal conversion mechanism from mapping of nucleotide variants within a homogeneous α-satellite DNA array. Genome Res. 2002, 12, 1815–1826. [Google Scholar] [CrossRef]
- Alkan, C.; Cardone, M.F.; Catacchio, C.R.; Antonacci, F.; O’Brien, S.J.; Ryder, O.A.; Purgato, S.; Zoli, M.; Della Valle, G.; Eichler, E.E.; et al. Genome-wide characterization of centromeric satellites from multiple mammalian genomes. Genome Res. 2011, 21, 137–145. [Google Scholar] [CrossRef]
- Henikoff, S.; Ahmad, K.; Malik, H. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 2001, 293, 1098–1102. [Google Scholar] [CrossRef] [PubMed]
- Malik, H.S.; Henikoff, S. Conflict begets complexity: The evolution of centromeres. Curr. Opin. Genet. Dev. 2002, 12, 711–718. [Google Scholar] [CrossRef]
- Melters, D.P.; Bradnam, K.R.; Young, H.A.; Telis, N.; May, M.R.; Ruby, J.G.; Sebra, R.; Peluso, P.; Eid, J.; Rank, D.; et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 2013, 14, R10. [Google Scholar] [CrossRef]
- Plohl, M.; Mestrovic, N.; Mravinac, B. Centromere identity from the DNA point of view. Chromosoma 2014, 123, 313–325. [Google Scholar] [CrossRef] [Green Version]
- Plohl, M.; Mestrovic, N.; Mravinac, B. Satellite DNA evolution. Genome Dyn. 2012, 7, 126–152. [Google Scholar] [CrossRef] [PubMed]
- Dover, G. Molecular drive: A cohesive mode of species evolution. Nature 1982, 299, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Evolution of repeated DNA sequences by unequal crossover. Science 1976, 191, 528–535. [Google Scholar] [CrossRef]
- Walsh, J.B. Persistence of tandem arrays: Implications for satellite and simple-sequence DNAs. Genetics 1987, 115, 553–567. [Google Scholar]
- Shi, J.; Wolf, S.E.; Burke, J.M.; Presting, G.G.; Ross-Ibarra, J.; Dawe, R.K. Widespread gene conversion in centromere cores. PLoS Biol. 2010, 8, e1000327. [Google Scholar] [CrossRef]
- Bertelsen, A.H.; Humayun, M.Z.; Karfopoulos, S.G.; Rush, M.G. Molecular characterization of small polydisperse circular deoxyribonucleic acid from an African green monkey cell line. Biochemistry 1982, 21, 2076–2085. [Google Scholar] [CrossRef]
- Gaubatz, J.W. Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells. Mutat. Res. 1990, 237, 271–292. [Google Scholar] [CrossRef]
- Baldini, A.; Smith, D.I.; Rocchi, M.; Miller, O.J.; Miller, D.A. A human alphoid DNA clone from the EcoRI dimeric family: Genomic and internal organization and chromosomal assignment. Genomics 1989, 5, 822–828. [Google Scholar] [CrossRef]
- Pironon, N.; Puechberty, J.; Roizès, G. Molecular and evolutionary characteristics of the fraction of human α satellite DNA associated with CENP-A at the centromeres of chromosomes 1, 5, 19, and 21. BMC Genom. 2010, 11, 195. [Google Scholar] [CrossRef]
- Greig, G.M.; Warburton, P.E.; Willard, H.F. Organization and evolution of an α satellite DNA subset shared by human chromosomes 13 and 21. J. Mol. Evol. 1993, 37, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, A.L.; Kolvraa, S.; Jones, C.; Bak, A.L. A subfamily of alphoid repetitive DNA shared by the NOR-bearing human chromosomes 14 and 22. Genomics 1988, 3, 100–109. [Google Scholar] [CrossRef]
- Jorgensen, A.L.; Laursen, H.B.; Jones, C.; Bak, A.L. Evolutionarily different alphoid repeat DNA on homologous chromosomes in human and chimpanzee. Proc. Natl. Acad. Sci. USA 1992, 89, 3310–3314. [Google Scholar] [CrossRef]
- Sabourin, M.; Nitiss, J.L.; Nitiss, K.C.; Tatebayashi, K.; Ikeda, H.; Osheroff, N. Yeast recombination pathways triggered by topoisomerase II-mediated DNA breaks. Nucleic Acids Res. 2003, 31, 4373–4384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salser, W.; Bowen, S.; Browne, D.; el-Adli, F.; Fedoroff, N.; Fry, K.; Heindell, H.; Paddock, G.; Poon, R.; Wallace, B.; et al. Investigation of the organization of mammalian chromosomes at the DNA sequence level. Fed. Proc. 1976, 35, 23–35. [Google Scholar]
- Cacheux, L.; Ponger, L.; Gerbault-Seureau, M.; Loll, F.; Gey, D.; Richard, F.A.; Escude, C. The targeted sequencing of α satellite DNA in Cercopithecus pogonias provides new insight into the diversity and dynamics of centromeric repeats in old world monkeys. Genome Biol. Evol. 2018, 10, 1837–1851. [Google Scholar] [CrossRef] [PubMed]
- da Silva, E.L.; Busso, A.F.; Parise-Maltempi, P.P. Characterization and genome organization of a repetitive element associated with the nucleolus organizer region in Leporinus elongatus (Anostomidae: Characiformes). Cytogenet. Genome Res. 2013, 139, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Mestrovic, N.; Plohl, M.; Mravinac, B.; Ugarkovic, D. Evolution of satellite DNAs from the genus Palorus--experimental evidence for the “library” hypothesis. Mol. Biol. Evol. 1998, 15, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.R.; Zhang, W.; Langdon, T.; Jin, W.; Yan, H.; Cheng, Z.; Jiang, J. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc. Natl. Acad. Sci. USA 2005, 102, 11793–11798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faravelli, M.; Moralli, D.; Bertoni, L.; Attolini, C.; Chernova, O.; Raimondi, E.; Giulotto, E. Two extended arrays of a satellite DNA sequence at the centromere and at the short-arm telomere of Chinese hamster chromosome 5. Cytogenet. Cell Genet. 1998, 83, 281–286. [Google Scholar] [CrossRef]
- Gong, Z.; Wu, Y.; Koblizkova, A.; Torres, G.A.; Wang, K.; Iovene, M.; Neumann, P.; Zhang, W.; Novak, P.; Buell, C.R.; et al. Repeatless and repeat-based centromeres in potato: Implications for centromere evolution. Plant Cell 2012, 24, 3559–3574. [Google Scholar] [CrossRef] [PubMed]
- Bulazel, K.; Ferreri, G.C.; Eldridge, M.D.; O’Neill, R.J. Species-specific shifts in centromere sequence composition are coincident with breakpoint reuse in karyotypically divergent lineages. Genome Biol. 2007, 8, R170. [Google Scholar] [CrossRef] [PubMed]
- Chaves, R.; Adega, F.; Heslop-Harrison, J.S.; Guedes-Pinto, H.; Wienberg, J. Complex satellite DNA reshuffling in the polymorphic t(1;29) Robertsonian translocation and evolutionarily derived chromosomes in cattle. Chromosome Res. 2003, 11, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Chaves, R.; Guedes-Pinto, H.; Heslop-Harrison, J.; Schwarzacher, T. The species and chromosomal distribution of the centromeric α-satellite I sequence from sheep in the tribe Caprini and other Bovidae. Cytogenet. Cell Genet. 2000, 91, 62–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malik, H.S. The centromere-drive hypothesis: A simple basis for centromere complexity. Prog. Mol. Subcell. Biol. 2009, 48, 33–52. [Google Scholar] [CrossRef]
- Iwata-Otsubo, A.; Dawicki-McKenna, J.M.; Akera, T.; Falk, S.J.; Chmatal, L.; Yang, K.; Sullivan, B.A.; Schultz, R.M.; Lampson, M.A.; Black, B.E. Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr. Biol. 2017, 27, 2365–2373.e8. [Google Scholar] [CrossRef]
- Drpic, D.; Almeida, A.C.; Aguiar, P.; Renda, F.; Damas, J.; Lewin, H.A.; Larkin, D.M.; Khodjakov, A.; Maiato, H. Chromosome segregation is biased by kinetochore size. Curr. Biol. 2018, 28, 1344–1356. [Google Scholar] [CrossRef]
- Zwick, M.E.; Salstrom, J.L.; Langley, C.H. Genetic variation in rates of nondisjunction: Association of two naturally occurring polymorphisms in the chromokinesin nod with increased rates of nondisjunction in Drosophila melanogaster. Genetics 1999, 152, 1605–1614. [Google Scholar] [PubMed]
- Hirsch, C.D.; Wu, Y.; Yan, H.; Jiang, J. Lineage-specific adaptive evolution of the centromeric protein CENH3 in diploid and allotetraploid Oryza species. Mol. Biol. Evol. 2009, 26, 2877–2885. [Google Scholar] [CrossRef] [PubMed]
- Schueler, M.G.; Swanson, W.; Thomas, P.J.; Green, E.D. Adaptive evolution of foundation kinetochore proteins in primates. Mol. Biol. Evol. 2010, 27, 1585–1597. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Masuelli, R.; Tyagi, A.P.; Comai, L.; Henikoff, S. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 2002, 14, 1053–1066. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Bryson, T.D.; Henikoff, S. Adaptive evolution of centromere proteins in plants and animals. J. Biol. 2004, 3, 18. [Google Scholar] [CrossRef] [PubMed]
- Zedek, F.; Bures, P. Evidence for centromere drive in the holocentric chromosomes of Caenorhabditis. PLoS ONE 2012, 7, e30496. [Google Scholar] [CrossRef] [PubMed]
- Chmatal, L.; Gabriel, S.I.; Mitsainas, G.P.; Martinez-Vargas, J.; Ventura, J.; Searle, J.B.; Schultz, R.M.; Lampson, M.A. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr. Biol. 2014, 24, 2295–2300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardo-Manuel de Villena, F.; Sapienza, C. Female meiosis drives karyotypic evolution in mammals. Genetics 2001, 159, 1179–1189. [Google Scholar] [PubMed]
- Rosin, L.F.; Mellone, B.G. Centromeres drive a hard bargain. Trends Genet. 2017, 33, 101–117. [Google Scholar] [CrossRef] [PubMed]
- Scott, K.C.; Sullivan, B.A. Neocentromeres: A place for everything and everything in its place. Trends Genet. 2014, 30, 66–74. [Google Scholar] [CrossRef]
- Garsed, D.W.; Marshall, O.J.; Corbin, V.D.; Hsu, A.; Di Stefano, L.; Schroder, J.; Li, J.; Feng, Z.P.; Kim, B.W.; Kowarsky, M.; et al. The architecture and evolution of cancer neochromosomes. Cancer Cell 2014, 26, 653–667. [Google Scholar] [CrossRef] [PubMed]
- Schubert, I. What is behind “centromere repositioning”? Chromosoma 2018, 127, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, M.M.; Vilkomerson, H. On the anaphase movement of chromosomes. Proc. Natl. Acad. Sci. USA 1942, 28, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Fritz, B.; Hasson, D.; Abrusan, G.; Cheung, F.; Yoda, K.; Radlwimmer, B.; Ladurner, A.G.; Warburton, P.E. Co-localization of CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol. 2007, 8, R148. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Mahmood, R.; Li, S.; Cheung, F.; Yoda, K.; Warburton, P.E. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum. Mol. Genet. 2003, 12, 2711–2721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voullaire, L.E.; Slater, H.R.; Petrovic, V.; Choo, K.H. A functional marker centromere with no detectable α-satellite, satellite III, or CENP-B protein: Activation of a latent centromere? Am. J. Hum. Genet. 1993, 52, 1153–1163. [Google Scholar]
- du Sart, D.; Cancilla, M.R.; Earle, E.; Mao, J.I.; Saffery, R.; Tainton, K.M.; Kalitsis, P.; Martyn, J.; Barry, A.E.; Choo, K.H. A functional neo-centromere formed through activation of a latent human centromere and consisting of non-α-satellite DNA. Nat. Genet. 1997, 16, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Amor, D.J.; Bentley, K.; Ryan, J.; Perry, J.; Wong, L.; Slater, H.; Choo, K.H. Human centromere repositioning “in progress”. Proc. Natl. Acad. Sci. USA 2004, 101, 6542–6547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barry, A.E.; Howman, E.V.; Cancilla, M.R.; Saffery, R.; Choo, K.H. Sequence analysis of an 80 kb human neocentromere. Hum. Mol. Genet. 1999, 8, 217–227. [Google Scholar] [CrossRef] [Green Version]
- Amor, D.J.; Choo, K.H.A. Neocentromeres: Role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 2002, 71, 695–714. [Google Scholar] [CrossRef]
- Warburton, P.E. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004, 12, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Eldridge, M.D.; Close, R.L. Radiation of chromosome shuffles. Curr. Opin. Genet. Dev. 1993, 3, 915–922. [Google Scholar] [CrossRef]
- Suja, J.A.; Camacho, J.P.M.; Cabrero, J.; Rufas, J.S. Analysis of a centric shift in the S11 chromosome of Aiolopus strepens (Orthoptera: Acrididae). Genetica 1986, 70, 211–216. [Google Scholar] [CrossRef]
- Ventura, M.; Archidiacono, N.; Rocchi, M. Centromere emergence in evolution. Genome Res. 2001, 11, 595–599. [Google Scholar] [CrossRef]
- Iannuzzi, L.; Di Meo, G.P.; Perucatti, A.; Incarnato, D.; Schibler, L.; Cribiu, E.P. Comparative FISH mapping of bovid X chromosomes reveals homologies and divergences between the subfamilies bovinae and caprinae. Cytogenet. Cell Genet. 2000, 89, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wu, Y.; Zhang, W.; Dawe, R.K.; Jiang, J. Maize centromeres expand and adopt a uniform size in the genetic background of oat. Genome Res. 2014, 24, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Rothfels, K.H.; Mason, G.F. Achiasmate meiosis and centromere shift in Eusimulium aureum (Diptera-Simuliidae). Chromosoma 1975, 51, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Carbone, L.; Nergadze, S.G.; Magnani, E.; Misceo, D.; Francesca Cardone, M.; Roberto, R.; Bertoni, L.; Attolini, C.; Francesca Piras, M.; de Jong, P.; et al. Evolutionary movement of centromeres in horse, donkey, and zebra. Genomics 2006, 87, 777–782. [Google Scholar] [CrossRef] [Green Version]
- Klein, S.J.; O’Neill, R.J. Transposable elements: Genome innovation, chromosome diversity, and centromere conflict. Chromosome Res. 2018, 26, 5–23. [Google Scholar] [CrossRef]
- Nergadze, S.G.; Piras, F.M.; Gamba, R.; Corbo, M.; Cerutti, F.; McCarter, J.G.W.; Cappelletti, E.; Gozzo, F.; Harman, R.M.; Antczak, D.F.; et al. Birth, evolution, and transmission of satellite-free mammalian centromeric domains. Genome Res. 2018, 28, 789–799. [Google Scholar] [CrossRef]
- Piras, F.M.; Nergadze, S.G.; Magnani, E.; Bertoni, L.; Attolini, C.; Khoriauli, L.; Raimondi, E.; Giulotto, E. Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet. 2010, 6, e1000845. [Google Scholar] [CrossRef] [PubMed]
- Birchler, J.A.; Presting, G.G. Retrotransposon insertion targeting: A mechanism for homogenization of centromere sequences on nonhomologous chromosomes. Genes Dev. 2012, 26, 638–640. [Google Scholar] [CrossRef] [PubMed]
- Rocchi, M.; Archidiacono, N.; Schempp, W.; Capozzi, O.; Stanyon, R. Centromere repositioning in mammals. Heredity 2012, 108, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Rocchi, M.; Stanyon, R.; Archidiacono, N. Evolutionary new centromeres in primates. Prog. Mol. Subcell. Biol. 2009, 48, 103–152. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, R.J.; Eldridge, M.D.; Metcalfe, C.J. Centromere dynamics and chromosome evolution in marsupials. J. Hered. 2004, 95, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Wade, C.; Giulotto, E.; Sigurdsson, S.; Zoli, M.; Gnerre, S.; Imsland, F.; Lear, T.; Adelson, D.; Bailey, E.; Bellone, R.; et al. Genome sequence, comparative analysis and population genetics of the domestic horse (Equus caballus). Science 2009, 326, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Purgato, S.; Belloni, E.; Piras, F.M.; Zoli, M.; Badiale, C.; Cerutti, F.; Mazzagatti, A.; Perini, G.; Della Valle, G.; Nergadze, S.G.; et al. Centromere sliding on a mammalian chromosome. Chromosoma 2015, 124, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.N.; O’Meally, D.; Chen, Z.; Etherington, G.J.; Ho, S.Y.W.; Nash, W.J.; Grueber, C.E.; Cheng, Y.; Whittington, C.M.; Dennison, S.; et al. Adaptation and conservation insights from the koala genome. Nat. Genet. 2018, 50, 1102–1111. [Google Scholar] [CrossRef]
- Carbone, L.; Harris, R.A.; Gnerre, S.; Veeramah, K.R.; Lorente-Galdos, B.; Huddleston, J.; Meyer, T.J.; Herrero, J.; Roos, C.; Aken, B.; et al. Gibbon genome and the fast karyotype evolution of small apes. Nature 2014, 513, 195–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagaki, K.; Cheng, Z.; Ouyang, S.; Talbert, P.B.; Kim, M.; Jones, K.M.; Henikoff, S.; Buell, C.R.; Jiang, J. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 2004, 36, 138–145. [Google Scholar] [CrossRef] [Green Version]
- Renfree, M.B.; Papenfuss, A.T.; Deakin, J.E.; Lindsay, J.; Heider, T.; Belov, K.; Rens, W.; Waters, P.D.; Pharo, E.A.; Shaw, G.; et al. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 2011, 12, R81. [Google Scholar] [CrossRef] [PubMed]
- Carone, D.; Longo, M.; Ferreri, G.; Hall, L.; Harris, M.; Shook, N.; Bulazel, K.; Carone, B.; Obergfell, C.; O’Neill, M.; et al. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma 2009, 118, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, K.; Baum, M.; Carbon, J. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc. Natl. Acad. Sci. USA 2004, 101, 11374–11379. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.K.; Baum, M.; Carbon, J. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Mol. Genet. Genom. 2007, 278, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Harrington, J.J.; Van Bokkelen, G.; Mays, R.W.; Gustashaw, K.; Willard, H.F. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 1997, 15, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Ebersole, T.A.; Ross, A.; Clark, E.; McGill, N.; Schindelhauer, D.; Cooke, H.; Grimes, B. Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum. Mol. Genet. 2000, 9, 1623–1631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doolittle, W.F.; Sapienza, C. Selfish genes, the phenotype paradigm and genome evolution. Nature 1980, 284, 601–603. [Google Scholar] [CrossRef]
- Orgel, L.E.; Crick, F.H.C. Selfish DNA: the ultimate parasite. Nature 1980, 284, 604–607. [Google Scholar] [CrossRef]
- Ravindran, S. Barbara McClintock and the discovery of jumping genes. Proc. Natl. Acad. Sci. USA 2012. [Google Scholar] [CrossRef]
- Wessler, S.R. Transposable elements and the evolution of eukaryotic genomes. Proc. Natl. Acad. Sci. USA 2006, 103, 17600–17601. [Google Scholar] [CrossRef] [Green Version]
- Craig, N.L.; Craigie, R.; Gellert, M.; Lambowitz, A.M. Mobile DNA II; American Society for Microbiology Press: Washington, DC, USA, 2002. [Google Scholar]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sotero-Caio, C.G.; Platt, R.N.; Suh, A.; Ray, D.A. Evolution and diversity of transposable elements in vertebrate genomes. Genome Biol. Evol. 2017, 9, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Mills, R.E.; Bennett, E.A.; Iskow, R.C.; Devine, S.E. Which transposable elements are active in the human genome? Trends Genet. 2007, 23, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, V.; Cheng, Y.; Ma, Z.; Li, D.; Xing, X.; Edge, P.; Snyder, M.P.; Wang, T. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014, 24, 1963–1976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wittkopp, P.J.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2011, 13, 59–69. [Google Scholar] [CrossRef]
- Jacques, P.E.; Jeyakani, J.; Bourque, G. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet. 2013, 9, e1003504. [Google Scholar] [CrossRef]
- Bourque, G.; Leong, B.; Vega, V.B.; Chen, X.; Lee, Y.L.; Srinivasan, K.G.; Chew, J.L.; Ruan, Y.; Wei, C.L.; Ng, H.H.; et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008, 18, 1752–1762. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Zeng, J.; Lowe, C.B.; Sellers, R.G.; Salama, S.R.; Yang, M.; Burgess, S.M.; Brachmann, R.K.; Haussler, D. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl. Acad. Sci. USA 2007, 104, 18613–18618. [Google Scholar] [CrossRef] [Green Version]
- Roman, A.C.; Benitez, D.A.; Carvajal-Gonzalez, J.M.; Fernandez-Salguero, P.M. Genome-wide B1 retrotransposon binds the transcription factors dioxin receptor and Slug and regulates gene expression in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 1632–1637. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Chen, G.; Wu, G.; Zhang, X.; McDermott, J.; Chen, X.; Xu, C.; Jiang, Q.; Chen, Z.; Zeng, Y.; et al. Widespread roles of enhancer-like transposable elements in cell identity and long-range genomic interactions. Genome Res. 2018. [Google Scholar] [CrossRef]
- Makarevitch, I.; Waters, A.J.; West, P.T.; Stitzer, M.; Hirsch, C.N.; Ross-Ibarra, J.; Springer, N.M. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 2015, 11, e1004915. [Google Scholar] [CrossRef]
- Feschotte, C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008, 9, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Ariumi, Y. Guardian of the human genome: host defense mechanisms against LINE-1 retrotransposition. Front. Chem. 2016, 4, 28. [Google Scholar] [CrossRef] [PubMed]
- Schueler, M.G.; Higgins, A.W.; Rudd, M.K.; Gustashaw, K.; Willard, H.F. Genomic and genetic definition of a functional human centromere. Science 2001, 294, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, A.E.; Shepelev, V.A.; Tumeneva, I.G.; Alexandrov, A.A.; Yurov, Y.B.; Alexandrov, I.A. Interspersed repeats are found predominantly in the “old” α satellite families. Genomics 2003, 82, 619–627. [Google Scholar] [CrossRef]
- Rosenbloom, K.R.; Armstrong, J.; Barber, G.P.; Casper, J.; Clawson, H.; Diekhans, M.; Dreszer, T.R.; Fujita, P.A.; Guruvadoo, L.; Haeussler, M.; et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015, 43, D670–D681. [Google Scholar] [CrossRef]
- Burrack, L.S.; Berman, J. Neocentromeres and epigenetically inherited features of centromeres. Chromosome Res. 2012, 20, 607–619. [Google Scholar] [CrossRef] [Green Version]
- Chueh, A.C.; Northrop, E.L.; Brettingham-Moore, K.H.; Choo, K.H.; Wong, L.H. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 2009, 5, e1000354. [Google Scholar] [CrossRef]
- Carbone, L.; Harris, R.A.; Mootnick, A.R.; Milosavljevic, A.; Martin, D.I.; Rocchi, M.; Capozzi, O.; Archidiacono, N.; Konkel, M.K.; Walker, J.A.; et al. Centromere remodeling in Hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons. Genome Biol. Evol. 2012, 4, 648–658. [Google Scholar] [CrossRef] [PubMed]
- Tolomeo, D.; Capozzi, O.; Stanyon, R.R.; Archidiacono, N.; D’Addabbo, P.; Catacchio, C.R.; Purgato, S.; Perini, G.; Schempp, W.; Huddleston, J.; et al. Epigenetic origin of evolutionary novel centromeres. Sci. Rep. 2017, 7, 41980. [Google Scholar] [CrossRef] [Green Version]
- Kato, M.; Takashima, K.; Kakutani, T. Epigenetic control of CACTA transposon mobility in Arabidopsis thaliana. Genetics 2004, 168, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Bourc’his, D.; Bestor, T.H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 2004, 431, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Metcalfe, C.J.; Bulazel, K.V.; Ferreri, G.C.; Schroeder-Reiter, E.; Wanner, G.; Rens, W.; Obergfell, C.; Eldridge, M.D.; O’Neill, R.J. Genomic instability within centromeres of interspecific marsupial hybrids. Genetics 2007, 177, 2507–2517. [Google Scholar] [CrossRef]
- O’Neill, R.J.; O’Neill, M.J.; Graves, J.A. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 1998, 393, 68–72. [Google Scholar] [CrossRef] [PubMed]
- May, B.P.; Lippman, Z.B.; Fang, Y.; Spector, D.L.; Martienssen, R.A. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet. 2005, 1, e79. [Google Scholar] [CrossRef] [PubMed]
- Ferreri, G.C.; Brown, J.D.; Obergfell, C.; Jue, N.; Finn, C.E.; O’Neill, M.J.; O’Neill, R.J. Recent amplification of the kangaroo endogenous retrovirus, KERV, limited to the centromere. J. Virol. 2011, 85, 4761–4771. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Galindo, R.; Kaplan, M.H.; He, S.; Contreras-Galindo, A.C.; Gonzalez-Hernandez, M.J.; Kappes, F.; Dube, D.; Chan, S.M.; Robinson, D.; Meng, F.; et al. HIV infection reveals widespread expansion of novel centromeric human endogenous retroviruses. Genome Res. 2013, 23, 1505–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Z.J.; Murata, M. A centromeric tandem repeat family originating from a part of Ty3/gypsy-retroelement in wheat and its relatives. Genetics 2003, 164, 665–672. [Google Scholar] [PubMed]
- Kapitonov, V.V.; Jurka, J. Molecular paleontology of transposable elements from Arabidopsis thaliana. Genetica 1999, 107, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Heikkinen, E.; Launonen, V.; Muller, E.; Bachmann, L. The pvB370 BamHI satellite DNA family of the Drosophila virilis group and its evolutionary relation to mobile dispersed genetic pDv elements. J. Mol. Evol. 1995, 41, 604–614. [Google Scholar] [CrossRef] [PubMed]
- Kapitonov, V.V.; Holmquist, G.P.; Jurka, J. L1 repeat is a basic unit of heterochromatin satellites in cetaceans. Mol. Biol. Evol. 1998, 15, 611–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGurk, M.P.; Barbash, D.A. Double insertion of transposable elements provides a substrate for the evolution of satellite DNA. Genome Res. 2018, 28, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Dias, G.B.; Svartman, M.; Delprat, A.; Ruiz, A.; Kuhn, G.C. Tetris is a foldback transposon that provided the building blocks for an emerging satellite DNA of Drosophila virilis. Genome Biol. Evol. 2014, 6, 1302–1313. [Google Scholar] [CrossRef] [PubMed]
- Mestrovic, N.; Mravinac, B.; Pavlek, M.; Vojvoda-Zeljko, T.; Satovic, E.; Plohl, M. Structural and functional liaisons between transposable elements and satellite DNAs. Chromosome Res. 2015, 23, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Liang, P. Transposable elements are a significant contributor to tandem repeats in the human genome. Comp. Funct. Genom. 2012, 2012, 947089. [Google Scholar] [CrossRef]
- Satovic, E.; Vojvoda Zeljko, T.; Luchetti, A.; Mantovani, B.; Plohl, M. Adjacent sequences disclose potential for intra-genomic dispersal of satellite DNA repeats and suggest a complex network with transposable elements. BMC Genom. 2016, 17, 997. [Google Scholar] [CrossRef]
- McLaughlin, R.N., Jr.; Malik, H.S. Genetic conflicts: The usual suspects and beyond. J. Exp. Biol. 2017, 220, 6–17. [Google Scholar] [CrossRef]
- Yoder, J.A.; Walsh, C.P.; Bestor, T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997, 13, 335–340. [Google Scholar] [CrossRef]
- Fedoroff, N.V. Presidential address. Transposable elements, epigenetics, and genome evolution. Science 2012, 338, 758–767. [Google Scholar] [CrossRef]
- Symer, D.E.; Connelly, C.; Szak, S.T.; Caputo, E.M.; Cost, G.J.; Parmigiani, G.; Boeke, J.D. Human l1 retrotransposition is associated with genetic instability in vivo. Cell 2002, 110, 327–338. [Google Scholar] [CrossRef]
- Gilbert, N.; Lutz, S.; Morrish, T.A.; Moran, J.V. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 2005, 25, 7780–7795. [Google Scholar] [CrossRef]
- Kazazian, H.H., Jr.; Moran, J.V. Mobile DNA in health and disease. N. Engl. J. Med. 2017, 377, 361–370. [Google Scholar] [CrossRef]
- Beck, C.R.; Garcia-Perez, J.L.; Badge, R.M.; Moran, J.V. LINE-1 elements in structural variation and disease. Annu. Rev. Genom. Hum. Genet. 2011, 12, 187–215. [Google Scholar] [CrossRef]
- Divashuk, M.G.; Khuat, T.M.; Kroupin, P.Y.; Kirov, I.V.; Romanov, D.V.; Kiseleva, A.V.; Khrustaleva, L.I.; Alexeev, D.G.; Zelenin, A.S.; Klimushina, M.V.; et al. Variation in copy number of Ty3/Gypsy centromeric retrotransposons in the genomes of Thinopyrum intermedium and its diploid progenitors. PLoS ONE 2016, 11, e0154241. [Google Scholar] [CrossRef]
- Han, J.; Masonbrink, R.E.; Shan, W.; Song, F.; Zhang, J.; Yu, W.; Wang, K.; Wu, Y.; Tang, H.; Wendel, J.F.; et al. Rapid proliferation and nucleolar organizer targeting centromeric retrotransposons in cotton. Plant J. 2016, 88, 992–1005. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, H.; Zhang, T.; Zeng, Z.; Zhang, P.; Zhu, B.; Han, Y.; Braz, G.T.; Casler, M.D.; Schmutz, J.; et al. Amplification and adaptation of centromeric repeats in polyploid switchgrass species. New Phytol. 2018, 218, 1645–1657. [Google Scholar] [CrossRef]
- Pardue, M.L.; Gall, J.G. Chromosomal localization of mouse satellite DNA. Science 1970, 168, 1356–1358. [Google Scholar] [CrossRef]
- Jones, K.W. Chromosomal and nuclear location of mouse satellite DNA in individual cells. Nature 1970, 225, 912–915. [Google Scholar] [CrossRef] [PubMed]
- Yunis, J.J.; Yasmineh, W.G. Heterochromatin, satellite DNA, and cell function. Structural DNA of eucaryotes may support and protect genes and aid in speciation. Science 1971, 174, 1200–1209. [Google Scholar] [CrossRef] [PubMed]
- Britten, R.J.; Kohne, D.E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science 1968, 161, 529–540. [Google Scholar] [CrossRef]
- Hall, L.E.; Mitchell, S.E.; O’Neill, R.J. Pericentric and centromeric transcription: A perfect balance required. Chromosome Res. 2012, 20, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Henikoff, S. Transcribing centromeres: noncoding RNAs and kinetochore assembly. Trends Genet. 2018, 34, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Braselton, J.P. Ribonucleoprotein staining of Allium cepa kinetochores. Cytobiologie 1975, 12, 148–151. [Google Scholar]
- Rieder, C.L. Ribonucleoprotein staining of centrioles and kinetochores in newt lung cell spindles. J. Cell Biol. 1979, 80, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Blower, M.D.; Sullivan, B.A.; Karpen, G.H. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2002, 2, 319–330. [Google Scholar] [CrossRef]
- Sullivan, B.A.; Karpen, G.H. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 2004, 11, 1076–1083. [Google Scholar] [CrossRef] [Green Version]
- Bergmann, J.H.; Rodriguez, M.G.; Martins, N.M.; Kimura, H.; Kelly, D.A.; Masumoto, H.; Larionov, V.; Jansen, L.E.; Earnshaw, W.C. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011, 30, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.S.; Stralfors, A.; Castillo, A.G.; Durand-Dubief, M.; Ekwall, K.; Allshire, R.C. Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J. Biol. Chem. 2011, 286, 23600–23607. [Google Scholar] [CrossRef] [PubMed]
- Chan, F.L.; Marshall, O.J.; Saffery, R.; Kim, B.W.; Earle, E.; Choo, K.H.; Wong, L.H. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc. Natl. Acad. Sci. USA 2012, 109, 1979–1984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosic, S.; Kohler, F.; Erhardt, S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Biol. 2014, 207, 335–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.C.; Bowers, S.; Lipinszki, Z.; Palladino, J.; Trusiak, S.; Bettini, E.; Rosin, L.; Przewloka, M.R.; Glover, D.M.; O’Neill, R.J.; et al. Establishment of centromeric chromatin by the CENP-A assembly factor CAL1 requires FACT-mediated transcription. Dev. Cell 2015, 34, 73–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohkuni, K.; Kitagawa, K. Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Curr. Biol. 2011, 21, 1695–1703. [Google Scholar] [CrossRef]
- Chan, F.L.; Wong, L.H. Transcription in the maintenance of centromere chromatin identity. Nucleic Acids Res. 2012, 40, 11178–11188. [Google Scholar] [CrossRef]
- McNulty, S.M.; Sullivan, L.L.; Sullivan, B.A. Human centromeres produce chromosome-specific and array-specific α satellite transcripts that are complexed with CENP-A and CENP-C. Dev. Cell 2017, 42, 226–240. [Google Scholar] [CrossRef]
- Topp, C.N.; Zhong, C.X.; Dawe, R.K. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl. Acad. Sci. USA 2004, 101, 15986–15991. [Google Scholar] [CrossRef] [Green Version]
- Carone, D.M.; Zhang, C.; Hall, L.E.; Obergfell, C.; Carone, B.R.; O’Neill, M.J.; O’Neill, R.J. Hypermorphic expression of centromeric retroelement-encoded small RNAs impairs CENP-A loading. Chromosome Res. 2013, 21, 49–62. [Google Scholar] [CrossRef]
- Saffery, R.; Sumer, H.; Hassan, S.; Wong, L.H.; Craig, J.M.; Todokoro, K.; Anderson, M.; Stafford, A.; Choo, K.H. Transcription within a functional human centromere. Mol. Cell 2003, 12, 509–516. [Google Scholar] [CrossRef]
- Wong, L.H.; Brettingham-Moore, K.H.; Chan, L.; Quach, J.M.; Anderson, M.A.; Northrop, E.L.; Hannan, R.; Saffery, R.; Shaw, M.L.; Williams, E.; et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007, 17, 1146–1160. [Google Scholar] [CrossRef] [Green Version]
- Ugarkovic, D. Functional elements residing within satellite DNAs. EMBO Rep. 2005, 6, 1035–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumann, P.; Yan, H.; Jiang, J. The centromeric retrotransposons of rice are transcribed and differentially processed by RNA interference. Genetics 2007, 176, 749–761. [Google Scholar] [CrossRef] [PubMed]
- Scott, K.C.; White, C.V.; Willard, H.F. An RNA polymerase III-dependent heterochromatin barrier at fission yeast centromere 1. PLoS ONE 2007, 2, e1099. [Google Scholar] [CrossRef]
- Cardinale, S.; Bergmann, J.H.; Kelly, D.; Nakano, M.; Valdivia, M.M.; Kimura, H.; Masumoto, H.; Larionov, V.; Earnshaw, W.C. Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol. Biol. Cell 2009, 20, 4194–4204. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Cardinale, S.; Noskov, V.N.; Gassmann, R.; Vagnarelli, P.; Kandels-Lewis, S.; Larionov, V.; Earnshaw, W.C.; Masumoto, H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell 2008, 14, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, J.H.; Jakubsche, J.N.; Martins, N.M.; Kagansky, A.; Nakano, M.; Kimura, H.; Kelly, D.A.; Turner, B.M.; Masumoto, H.; Larionov, V.; et al. Epigenetic engineering: Histone H3K9 acetylation is compatible with kinetochore structure and function. J. Cell Sci. 2012, 125, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Ting, D.T.; Lipson, D.; Paul, S.; Brannigan, B.W.; Akhavanfard, S.; Coffman, E.J.; Contino, G.; Deshpande, V.; Iafrate, A.J.; Letovsky, S.; et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 2011, 331, 593–596. [Google Scholar] [CrossRef]
- Hill, A.; Bloom, K. Genetic manipulation of centromere function. Mol. Cell. Biol. 1987, 7, 2397–2405. [Google Scholar] [CrossRef]
- Bouzinba-Segard, H.; Guais, A.; Francastel, C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc. Natl. Acad. Sci. USA 2006, 103, 8709–8714. [Google Scholar] [CrossRef] [Green Version]
- Ferri, F.; Bouzinba-Segard, H.; Velasco, G.; Hube, F.; Francastel, C. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 2009, 37, 5071–5080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, D.Y.L.; Moralli, D.; Khoja, S.; Monaco, Z.L. Noncoding centromeric RNA expression impairs chromosome stability in human and murine stem cells. Dis. Mark. 2017, 2017, 7506976. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Topp, C.N.; Dawe, R.K. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 2010, 6, e1000835. [Google Scholar] [CrossRef]
- Sandmann, M.; Talbert, P.; Demidov, D.; Kuhlmann, M.; Rutten, T.; Conrad, U.; Lermontova, I. Targeting of Arabidopsis KNL2 to centromeres depends on the conserved CENPC-k motif in its C terminus. Plant Cell 2017, 29, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Dawe, R.K. RNA interference, transposons, and the centromere. Plant Cell 2003, 15, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; Choo, K.H. Evolutionary dynamics of transposable elements at the centromere. Trends Genet. 2004, 20, 611–616. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, R.J.; Carone, D.M. The role of ncRNA in centromeres: A lesson from marsupials. Prog. Mol. Subcell. Biol. 2009, 48, 77–101. [Google Scholar] [CrossRef] [PubMed]
- Kasinathan, S.; Henikoff, S. Non-B-form DNA is enriched at centromeres. Mol. Biol. Evol. 2018, 35, 949–962. [Google Scholar] [CrossRef]
- Bobkov, G.O.M.; Gilbert, N.; Heun, P. Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. J. Cell Biol. 2018, 217, 1957–1972. [Google Scholar] [CrossRef] [Green Version]
- Quenet, D.; Dalal, Y. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. Elife 2014, 3, e03254, Erratum in 2018, 7, e41593. [Google Scholar] [CrossRef]
- Chueh, A.C.; Wong, L.H.; Wong, N.; Choo, K.H. Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere. Hum. Mol. Genet. 2005, 14, 85–93. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The behaviour of successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl. Acad. Sci. USA 1939, 25, 405–416. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The stability of broken ends of chromosomes in Zea Mays. Genetics 1941, 26, 234–282. [Google Scholar] [PubMed]
- McClintock, B. The production of homozygous deficient tissues with mutant characteristics by means of the aberrant mitotic behavior of ring-shaped chromosomes. Genetics 1938, 23, 315–376. [Google Scholar] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hartley, G.; O’Neill, R.J. Centromere Repeats: Hidden Gems of the Genome. Genes 2019, 10, 223. https://doi.org/10.3390/genes10030223
Hartley G, O’Neill RJ. Centromere Repeats: Hidden Gems of the Genome. Genes. 2019; 10(3):223. https://doi.org/10.3390/genes10030223
Chicago/Turabian StyleHartley, Gabrielle, and Rachel J. O’Neill. 2019. "Centromere Repeats: Hidden Gems of the Genome" Genes 10, no. 3: 223. https://doi.org/10.3390/genes10030223
APA StyleHartley, G., & O’Neill, R. J. (2019). Centromere Repeats: Hidden Gems of the Genome. Genes, 10(3), 223. https://doi.org/10.3390/genes10030223



