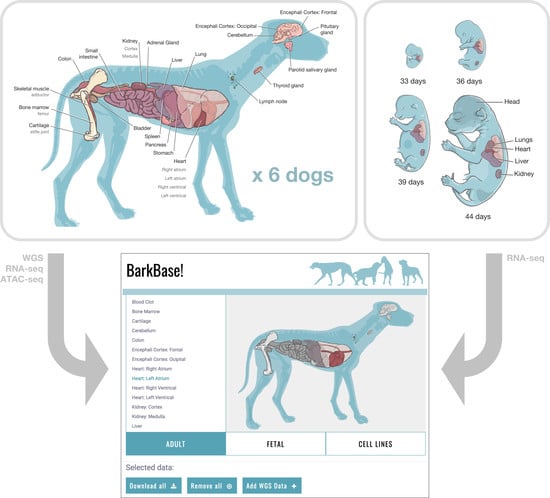
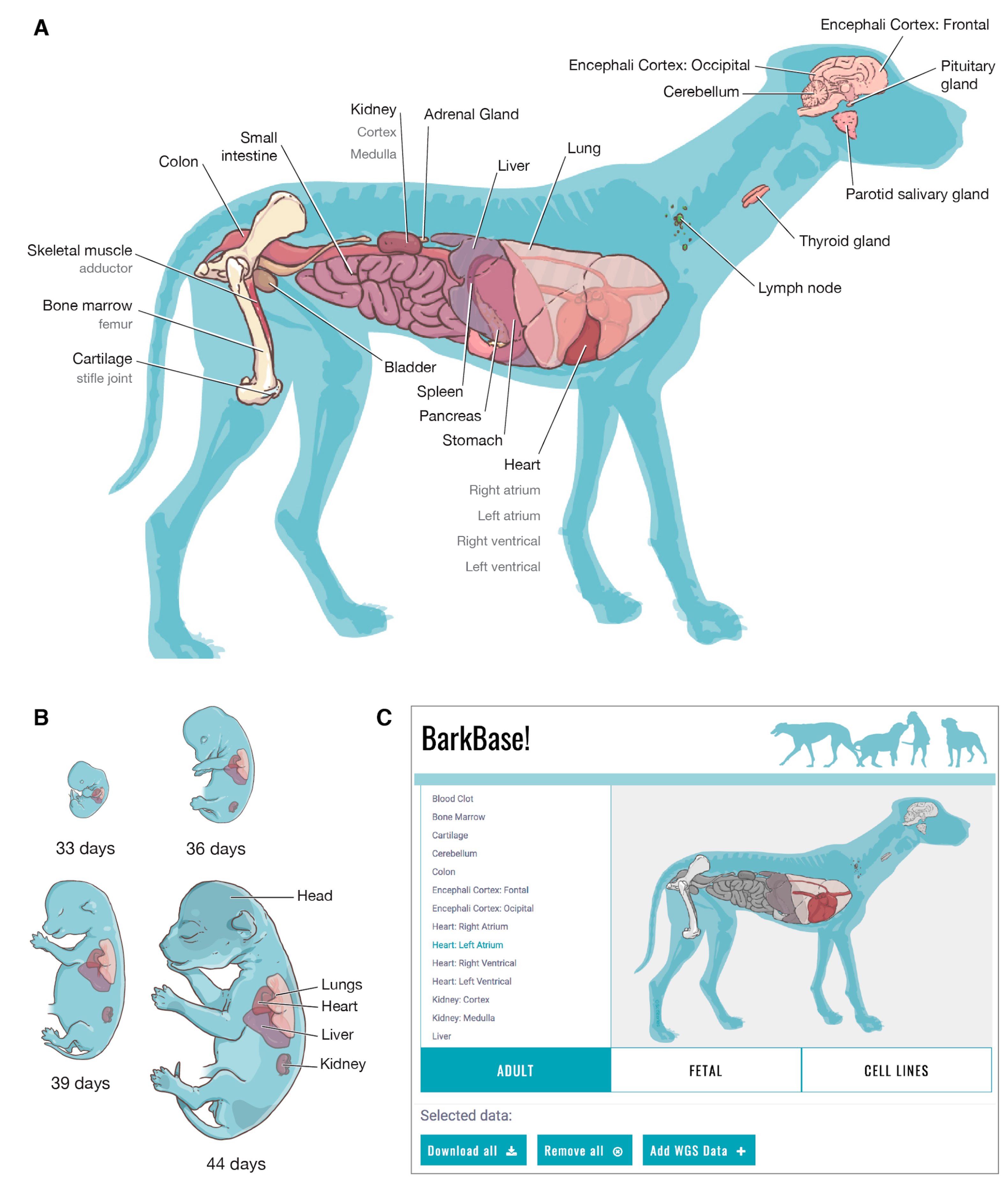
BarkBase: Epigenomic Annotation of Canine Genomes
Abstract
:1. Introduction
2. Materials and Methods
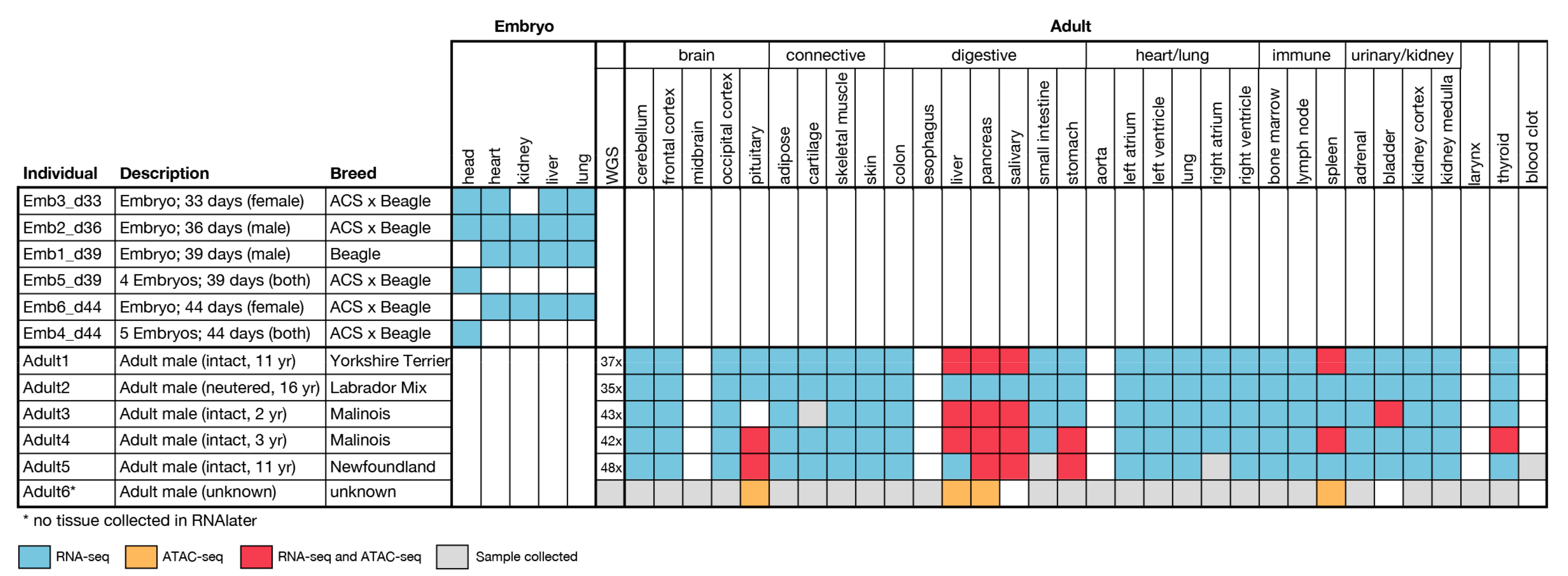
2.1. Sample Collection
2.1.1. Adult Tissue
2.1.2. Embryonic Tissue
2.2. Whole Genome Sequencing
2.2.1. Sequencing and Variant Calling
2.2.2. Relatedness
2.2.3. Annotation
2.3. RNA Sequencing, Normalization, and Analysis
2.3.1. Sequencing
2.3.2. Read Preprocessing
2.3.3. TMM Normalization to Calculate Counts Per Million (CPM)
2.3.4. Cumulative Abundance and Tissue-Specific Reads
2.3.5. Hierarchical Clustering of Samples and Adult Dogs
2.3.6. Differential Gene Expression (DGE) and Enrichment Analysis of Embryonic Data
2.3.7. Comparison of Gene Expression between Dog and Human Tissues
2.3.8. Comparison of Transcript Set to the Existing Ensembl Canine Reference Annotation
2.3.9. Comparison of Unannotated Transcripts to the RefSeq Vertebrate Mammalian Proteins
2.4. ATAC-seq
2.4.1. Sample Preparation and Sequencing
2.4.2. Processing Reads, Calling Peaks, and QC of Libraries
2.4.3. Assessing Overlap of Peaks
2.4.4. Data Sharing
3. Results
3.1. BarkBase Website
3.2. Whole Genome Sequencing
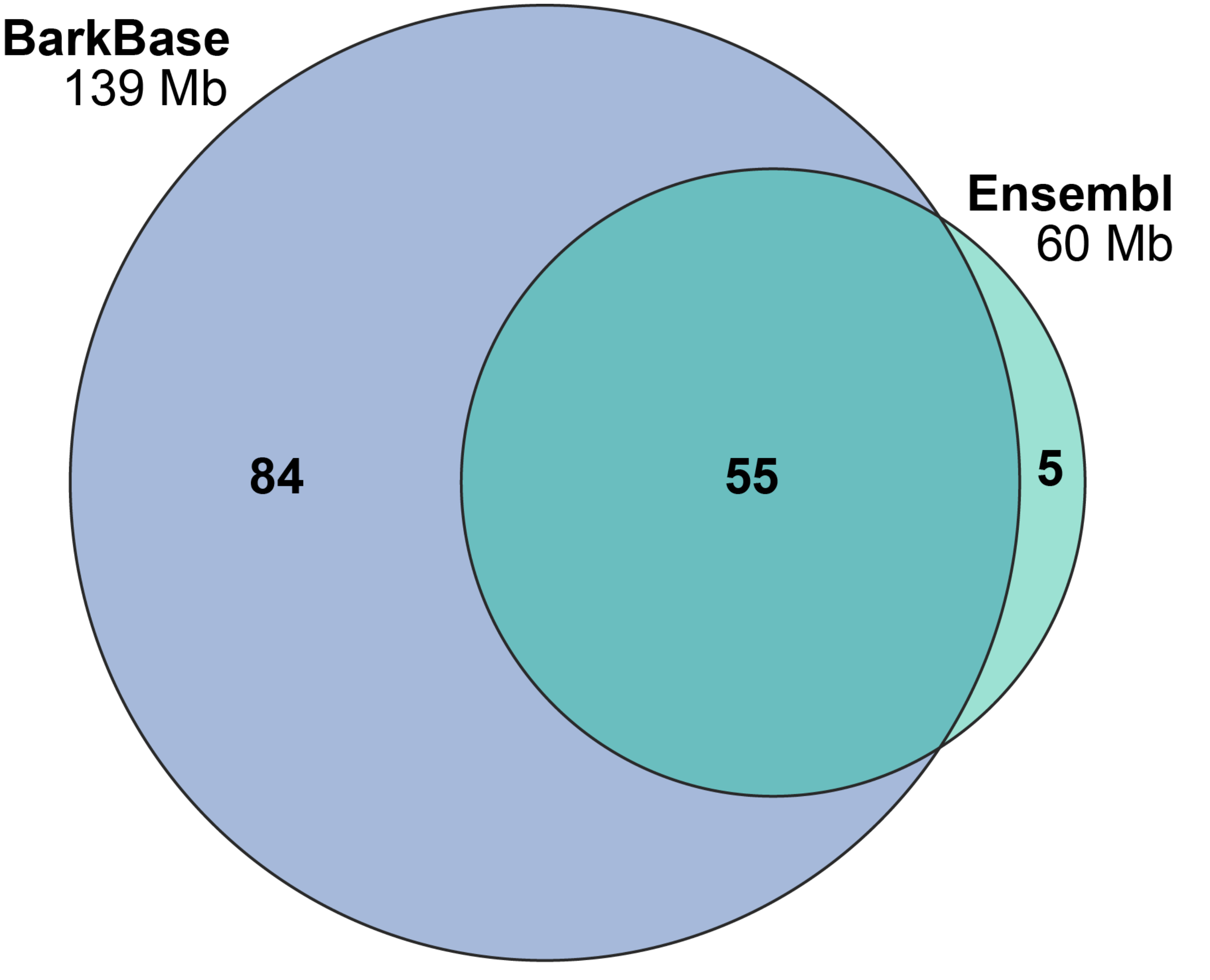
3.3. Overlap With Existing Gene Annotations
3.4. Variability Among Adult Dogs and Tissues
3.5. Comparison of Expression Profiles in Canine and Human Tissues
3.6. RNA-Sequencing of Embryonic Tissues at Multiple Time Points
3.7. ATAC-seq
3.8. Novel Genes and ATAC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schiffman, J.D.; Breen, M. Comparative oncology: What dogs and other species can teach us about humans with cancer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140231. [Google Scholar] [CrossRef] [PubMed]
- Noh, H.J.; Tang, R.; Flannick, J.; O’Dushlaine, C.; Swofford, R.; Howrigan, D.; Genereux, D.P.; Johnson, J.; van Grootheest, G.; Grünblatt, E.; et al. Integrating evolutionary and regulatory information with a multispecies approach implicates genes and pathways in obsessive-compulsive disorder. Nat. Commun. 2017, 8, 774. [Google Scholar] [CrossRef] [PubMed]
- Wilbe, M.; Jokinen, P.; Truvé, K.; Seppala, E.H.; Karlsson, E.K.; Biagi, T.; Hughes, A.; Bannasch, D.; Andersson, G.; Hansson-Hamlin, H.; et al. Genome-wide association mapping identifies multiple loci for a canine SLE-related disease complex. Nat. Genet. 2010, 42, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, E.K.; Sigurdsson, S.; Ivansson, E.; Thomas, R.; Elvers, I.; Wright, J.; Howald, C.; Tonomura, N.; Perloski, M.; Swofford, R.; et al. Genome-wide analyses implicate 33 loci in heritable dog osteosarcoma, including regulatory variants near CDKN2A/B. Genome Biol. 2013, 14, R132. [Google Scholar] [CrossRef] [PubMed]
- Schoenebeck, J.J.; Ostrander, E.A. Insights into morphology and disease from the dog genome project. Annu. Rev. Cell Dev. Biol. 2014, 30, 535–560. [Google Scholar] [CrossRef]
- Kaeberlein, M.; Creevy, K.E.; Promislow, D.E.L. The dog aging project: Translational geroscience in companion animals. Mamm. Genome 2016, 27, 279–288. [Google Scholar] [CrossRef]
- Karlsson, E.K.; Lindblad-Toh, K. Leader of the pack: Gene mapping in dogs and other model organisms. Nat. Rev. Genet. 2008, 9, 713–725. [Google Scholar] [CrossRef]
- Lindblad-Toh, K.; Wade, C.M.; Mikkelsen, T.S.; Karlsson, E.K.; Jaffe, D.B.; Kamal, M.; Clamp, M.; Chang, J.L.; Kulbokas, E.J., 3rd; Zody, M.C.; et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 2005, 438, 803–819. [Google Scholar] [CrossRef]
- Parker, H.G.; Kim, L.V.; Sutter, N.B.; Carlson, S.; Lorentzen, T.D.; Malek, T.B.; Johnson, G.S.; DeFrance, H.B.; Ostrander, E.A.; Kruglyak, L. Genetic structure of the purebred domestic dog. Science 2004, 304, 1160–1164. [Google Scholar] [CrossRef]
- Karlsson, E.K.; Baranowska, I.; Wade, C.M.; Salmon Hillbertz, N.H.C.; Zody, M.C.; Anderson, N.; Biagi, T.M.; Patterson, N.; Pielberg, G.R.; Kulbokas, E.J., 3rd; et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat. Genet. 2007, 39, 1321–1328. [Google Scholar] [CrossRef]
- Ostrander, E.A.; Wang, G.-D.; Larson, G.; vonHoldt, B.M.; Davis, B.W.; Jagannathan, V.; Hitte, C.; Wayne, R.K.; Zhang, Y.-P. Dog10K: An international sequencing effort to advance studies of canine domestication, phenotypes, and health. Natl. Sci. Rev. 2019. [Google Scholar] [CrossRef]
- Plassais, J.; Kim, J.; Davis, B.W.; Karyadi, D.M.; Hogan, A.N.; Harris, A.C.; Decker, B.; Parker, H.G.; Ostrander, E.A. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat. Commun. 2019, 10, 1489. [Google Scholar] [CrossRef] [PubMed]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Roadmap Epigenomics, Consortium; Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; et al. Integrative analysis of 111 reference human epigenomes. Nature 2015, 518, 317–330. [Google Scholar] [Green Version]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef]
- Hoffman, M.M.; Ernst, J.; Wilder, S.P.; Kundaje, A.; Harris, R.S.; Libbrecht, M.; Giardine, B.; Ellenbogen, P.M.; Bilmes, J.A.; Birney, E.; et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013, 41, 827–841. [Google Scholar] [CrossRef]
- Ernst, J.; Kheradpour, P.; Mikkelsen, T.S.; Shoresh, N.; Ward, L.D.; Epstein, C.B.; Zhang, X.; Wang, L.; Issner, R.; Coyne, M.; et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011, 473, 43–49. [Google Scholar] [CrossRef]
- Gjoneska, E.; Pfenning, A.R.; Mathys, H.; Quon, G.; Kundaje, A.; Tsai, L.-H.; Kellis, M. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature 2015, 518, 365–369. [Google Scholar] [CrossRef]
- Hoeppner, M.P.; Lundquist, A.; Pirun, M.; Meadows, J.R.S.; Zamani, N.; Johnson, J.; Sundström, G.; Cook, A.; FitzGerald, M.G.; Swofford, R.; et al. An improved canine genome and a comprehensive catalogue of coding genes and non-coding transcripts. PLoS ONE 2014, 9, e91172. [Google Scholar] [CrossRef]
- EMBL-EBI Ensembl gene annotation project (e!68): Canis lupus familiaris. Available online: https://useast.ensembl.org/info/genome/genebuild/2012_07_dog_genebuild.pdf (accessed on 1 May 2019).
- Cunningham, F.; Achuthan, P.; Akanni, W.; Allen, J.; Amode, M.R.; Armean, I.M.; Bennett, R.; Bhai, J.; Billis, K.; Boddu, S.; et al. Ensembl 2019. Nucleic Acids Res. 2019, 47, D745–D751. [Google Scholar] [CrossRef] [PubMed]
- Raney, B.J.; Dreszer, T.R.; Barber, G.P.; Clawson, H.; Fujita, P.A.; Wang, T.; Nguyen, N.; Paten, B.; Zweig, A.S.; Karolchik, D.; et al. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics 2014, 30, 1003–1005. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002. [Google Scholar] [CrossRef] [PubMed]
- Le Béguec, C.; Wucher, V.; Lagoutte, L.; Cadieu, E.; Botherel, N.; Hédan, B.; De Brito, C.; Guillory, A.-S.; André, C.; Derrien, T.; et al. Characterisation and functional predictions of canine long non-coding RNAs. Sci. Rep. 2018, 8, 13444. [Google Scholar] [CrossRef]
- Wucher, V.; Legeai, F.; Hédan, B.; Rizk, G.; Lagoutte, L.; Leeb, T.; Jagannathan, V.; Cadieu, E.; David, A.; Lohi, H.; et al. FEELnc: A tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 2017, 45, e57. [Google Scholar] [CrossRef] [PubMed]
- Haeussler, M.; Zweig, A.S.; Tyner, C.; Speir, M.L.; Rosenbloom, K.R.; Raney, B.J.; Lee, C.M.; Lee, B.T.; Hinrichs, A.S.; Gonzalez, J.N.; et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019, 47, D853–D858. [Google Scholar] [CrossRef]
- Lindblad-Toh, K.; Garber, M.; Zuk, O.; Lin, M.F.; Parker, B.J.; Washietl, S.; Kheradpour, P.; Ernst, J.; Jordan, G.; Mauceli, E.; et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 2011, 478, 476–482. [Google Scholar] [CrossRef] [Green Version]
- Rands, C.M.; Meader, S.; Ponting, C.P.; Lunter, G. 8.2% of the Human genome is constrained: Variation in rates of turnover across functional element classes in the human lineage. PLoS Genet. 2014, 10, e1004525. [Google Scholar] [CrossRef]
- Villar, D.; Berthelot, C.; Aldridge, S.; Rayner, T.F.; Lukk, M.; Pignatelli, M.; Park, T.J.; Deaville, R.; Erichsen, J.T.; Jasinska, A.J.; et al. Enhancer evolution across 20 mammalian species. Cell 2015, 160, 554–566. [Google Scholar] [CrossRef]
- Megquier, K.; Turner-Maier, J.; Swofford, R.; Kim, J.-H.; Sarver, A.L.; Wang, C.; Sakthikumar, S.; Johnson, J.; Koltookian, M.; Lewellen, M.; et al. Genomic analysis reveals shared genes and pathways in human and canine angiosarcoma. BioRxiv 2019, 570879. [Google Scholar] [CrossRef] [Green Version]
- Tonomura, N.; Elvers, I.; Thomas, R.; Megquier, K.; Turner-Maier, J.; Howald, C.; Sarver, A.L.; Swofford, R.; Frantz, A.M.; Ito, D.; et al. Genome-wide association study identifies shared risk loci common to two malignancies in golden retrievers. PLoS Genet. 2015, 11, e1004922. [Google Scholar] [CrossRef] [PubMed]
- Hendricks, W.P.D.; Zismann, V.; Sivaprakasam, K.; Legendre, C.; Poorman, K.; Tembe, W.; Perdigones, N.; Kiefer, J.; Liang, W.; DeLuca, V.; et al. Somatic inactivating PTPRJ mutations and dysregulated pathways identified in canine malignant melanoma by integrated comparative genomic analysis. PLoS Genet. 2018, 14, e1007589. [Google Scholar] [CrossRef] [PubMed]
- Ostrander, E.A.; Dreger, D.L.; Evans, J.M. Canine Cancer Genomics: Lessons for Canine and Human Health. Annu. Rev. Anim. Biosci. 2019, 7, 449–472. [Google Scholar] [CrossRef] [PubMed]
- Elvers, I.; Turner-Maier, J.; Swofford, R.; Koltookian, M.; Johnson, J.; Stewart, C.; Zhang, C.-Z.; Schumacher, S.E.; Beroukhim, R.; Rosenberg, M.; et al. Exome sequencing of lymphomas from three dog breeds reveals somatic mutation patterns reflecting genetic background. Genome Res. 2015, 25, 1634–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakthikumar, S.; Elvers, I.; Kim, J.; Arendt, M.L.; Thomas, R.; Turner-Maier, J.; Swofford, R.; Johnson, J.; Schumacher, S.E.; Alföldi, J.; et al. SETD2 Is Recurrently Mutated in Whole-Exome Sequenced Canine Osteosarcoma. Cancer Res. 2018, 78, 3421–3431. [Google Scholar] [CrossRef] [PubMed]
- Shearin, A.L.; Hedan, B.; Cadieu, E.; Erich, S.A.; Schmidt, E.V.; Faden, D.L.; Cullen, J.; Abadie, J.; Kwon, E.M.; Gröne, A.; et al. The MTAP-CDKN2A locus confers susceptibility to a naturally occurring canine cancer. Cancer Epidemiol. Biomarkers Prev. 2012, 21, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Noh, H.J.; Wang, D.; Sigurdsson, S.; Swofford, R.; Perloski, M.; Duxbury, M.; Patterson, E.E.; Albright, J.; Castelhano, M.; et al. Candidate genes and functional noncoding variants identified in a canine model of obsessive-compulsive disorder. Genome Biol. 2014, 15, R25. [Google Scholar] [CrossRef]
- Sarviaho, R.; Hakosalo, O.; Tiira, K.; Sulkama, S.; Salmela, E.; Hytönen, M.K.; Sillanpää, M.J.; Lohi, H. Two novel genomic regions associated with fearfulness in dogs overlap human neuropsychiatric loci. Transl. Psychiatry 2019, 9, 18. [Google Scholar] [CrossRef]
- Tengvall, K.; Kierczak, M.; Bergvall, K.; Olsson, M.; Frankowiack, M.; Farias, F.H.G.; Pielberg, G.; Carlborg, Ö.; Leeb, T.; Andersson, G.; et al. Genome-wide analysis in German shepherd dogs reveals association of a locus on CFA 27 with atopic dermatitis. PLoS Genet. 2013, 9, e1003475. [Google Scholar] [CrossRef]
- Vieira, N.M.; Elvers, I.; Alexander, M.S.; Moreira, Y.B.; Eran, A.; Gomes, J.P.; Marshall, J.L.; Karlsson, E.K.; Verjovski-Almeida, S.; Lindblad-Toh, K.; et al. Jagged 1 Rescues the Duchenne Muscular Dystrophy Phenotype. Cell 2015, 163, 1204–1213. [Google Scholar] [CrossRef] [Green Version]
- Hayward, J.J.; Castelhano, M.G.; Oliveira, K.C.; Corey, E.; Balkman, C.; Baxter, T.L.; Casal, M.L.; Center, S.A.; Fang, M.; Garrison, S.J.; et al. Complex disease and phenotype mapping in the domestic dog. Nat. Commun. 2016, 7, 10460. [Google Scholar] [CrossRef] [PubMed]
- Raffan, E.; Dennis, R.J.; O’Donovan, C.J.; Becker, J.M.; Scott, R.A.; Smith, S.P.; Withers, D.J.; Wood, C.J.; Conci, E.; Clements, D.N.; et al. A Deletion in the Canine POMC Gene Is Associated with Weight and Appetite in Obesity-Prone Labrador Retriever Dogs. Cell Metab. 2016, 23, 893–900. [Google Scholar] [CrossRef] [Green Version]
- Becker, D.; Minor, K.M.; Letko, A.; Ekenstedt, K.J.; Jagannathan, V.; Leeb, T.; Shelton, G.D.; Mickelson, J.R.; Drögemüller, C. A GJA9 frameshift variant is associated with polyneuropathy in Leonberger dogs. BMC Genomics 2017, 18, 662. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Faraco, J.; Li, R.; Kadotani, H.; Rogers, W.; Lin, X.; Qiu, X.; de Jong, P.J.; Nishino, S.; Mignot, E. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 1999, 98, 365–376. [Google Scholar] [CrossRef]
- Acland, G.M.; Aguirre, G.D.; Ray, J.; Zhang, Q.; Aleman, T.S.; Cideciyan, A.V.; Pearce-Kelling, S.E.; Anand, V.; Zeng, Y.; Maguire, A.M.; et al. Gene therapy restores vision in a canine model of childhood blindness. Nat. Genet. 2001, 28, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Meyers-Wallen, V.N.; Boyko, A.R.; Danko, C.G.; Grenier, J.K.; Mezey, J.G.; Hayward, J.J.; Shannon, L.M.; Gao, C.; Shafquat, A.; Rice, E.J.; et al. XX Disorder of Sex Development is associated with an insertion on chromosome 9 and downregulation of RSPO1 in dogs (Canis lupus familiaris). PLoS ONE 2017, 12, e0186331. [Google Scholar] [CrossRef]
- Meyers-Wallen, V. Canine Embryonic Atlas at Cornell University. Available online: https://www.vet.cornell.edu/canine-atlas (accessed on 1 May 2019).
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Purcell, S.; Chang, C. PLINK2 (v1.90b6.9). Available online: www.cog-genomics.org/plink/2.0/ (accessed on 1 May 2019).
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- ENCODE Consortium Current ENCODE Experiment Guidelines. Available online: https://www.encodeproject.org/about/experiment-guidelines/ (accessed on 28 May 2019).
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [Green Version]
- Law, C.W.; Alhamdoosh, M.; Su, S.; Smyth, G.K.; Ritchie, M.E. RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR. F1000Res. 2016, 5. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 28 May 2019).
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes, revisited. Trends Genet. 2013, 29, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Kinsella, R.J.; Kähäri, A.; Haider, S.; Zamora, J.; Proctor, G.; Spudich, G.; Almeida-King, J.; Staines, D.; Derwent, P.; Kerhornou, A.; et al. Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database 2011, 2011, bar030. [Google Scholar] [CrossRef]
- Jones, E.; Oliphant, T.; Peterson, P. SciPy: Open Source Scientific Tools for Python. Available online: http://www.scipy.org/ (accessed on 1 May 2019).
- GffCompare: Program for Processing GTF/GFF Files. Available online: https://ccb.jhu.edu/software/stringtie/gffcompare.shtml (accessed on 1 May 2019).
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Brown, G.R.; Hiatt, S.M.; Thibaud-Nissen, F.; Astashyn, A.; Ermolaeva, O.; Farrell, C.M.; Hart, J.; Landrum, M.J.; McGarvey, K.M.; et al. RefSeq: An update on mammalian reference sequences. Nucleic Acids Res. 2014, 42, D756–D763. [Google Scholar] [CrossRef]
- Pruitt, K.D. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2004, 33, D501–D504. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: architecture and applications. BMC Bioinf. 2009, 10, 421. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, d733–d745. [Google Scholar] [CrossRef]
- Gaspar, J.M. ATAC-seq Guidelines. Available online: https://informatics.fas.harvard.edu/atac-seq-guidelines-old-version.html (accessed on 1 May 2019).
- ataqv: A toolkit for QC and visualization of ATAC-seq results. Available online: https://github.com/ParkerLab/ataqv (accessed on 1 May 2019).
- Yu, G.; Wang, L.-G.; He, Q.-Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef]
- Aken, B.L.; Ayling, S.; Barrell, D.; Clarke, L.; Curwen, V.; Fairley, S.; Fernandez Banet, J.; Billis, K.; García Girón, C.; Hourlier, T.; et al. The Ensembl gene annotation system. Database 2016. [Google Scholar] [CrossRef]
- Roadmap Epigenomics Project Visual Browser. Available online: http://www.roadmapepigenomics.org/data/visualbrowser/adult (accessed on 1 May 2019).
- Leinonen, R.; Sugawara, H.; Shumway, M. International Nucleotide Sequence Database Collaboration The sequence read archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef]
- Kang, N.; Koo, J. Olfactory receptors in non-chemosensory tissues. BMB Rep. 2012, 45, 612–622. [Google Scholar] [CrossRef] [Green Version]
- Li, F. Taste perception: From the tongue to the testis. Mol. Hum. Reprod. 2013, 19, 349–360. [Google Scholar] [CrossRef]
- Mooney, M.; Bond, J.; Monks, N.; Eugster, E.; Cherba, D.; Berlinski, P.; Kamerling, S.; Marotti, K.; Simpson, H.; Rusk, T.; et al. Comparative RNA-Seq and microarray analysis of gene expression changes in B-cell lymphomas of Canis familiaris. PLoS ONE 2013, 8, e61088. [Google Scholar] [CrossRef]
- Maeda, S.; Tomiyasu, H.; Tsuboi, M.; Inoue, A.; Ishihara, G.; Uchikai, T.; Chambers, J.K.; Uchida, K.; Yonezawa, T.; Matsuki, N. Comprehensive gene expression analysis of canine invasive urothelial bladder carcinoma by RNA-Seq. BMC Cancer 2018, 18, 472. [Google Scholar] [CrossRef]
- Gorden, B.H.; Kim, J.-H.; Sarver, A.L.; Frantz, A.M.; Breen, M.; Lindblad-Toh, K.; O’Brien, T.D.; Sharkey, L.C.; Modiano, J.F.; Dickerson, E.B. Identification of three molecular and functional subtypes in canine hemangiosarcoma through gene expression profiling and progenitor cell characterization. Am. J. Pathol. 2014, 184, 985–995. [Google Scholar] [CrossRef]
- Scott, M.C.; Temiz, N.A.; Sarver, A.E.; LaRue, R.S.; Rathe, S.K.; Varshney, J.; Wolf, N.K.; Moriarity, B.S.; O’Brien, T.D.; Spector, L.G.; et al. Comparative Transcriptome Analysis Quantifies Immune Cell Transcript Levels, Metastatic Progression, and Survival in Osteosarcoma. Cancer Res. 2018, 78, 326–337. [Google Scholar] [CrossRef]
- Schurch, N.J.; Schofield, P.; Gierliński, M.; Cole, C.; Sherstnev, A.; Singh, V.; Wrobel, N.; Gharbi, K.; Simpson, G.G.; Owen-Hughes, T.; et al. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 2016, 22, 839–851. [Google Scholar] [CrossRef] [Green Version]
- Ramsköld, D.; Wang, E.T.; Burge, C.B.; Sandberg, R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 2009, 5, e1000598. [Google Scholar] [CrossRef]
- Danielsson, A.; Pontén, F.; Fagerberg, L.; Hallström, B.M.; Schwenk, J.M.; Uhlén, M.; Korsgren, O.; Lindskog, C. The Human Pancreas Proteome Defined by Transcriptomics and Antibody-Based Profiling. PLoS ONE 2014, 9, e115421. [Google Scholar] [CrossRef]
- Melé, M.; Ferreira, P.G.; Reverter, F.; DeLuca, D.S.; Monlong, J.; Sammeth, M.; Young, T.R.; Goldmann, J.M.; Pervouchine, D.D.; Sullivan, T.J.; et al. Human genomics. The human transcriptome across tissues and individuals. Science 2015, 348, 660–665. [Google Scholar] [CrossRef]
- Wright, F.A.; Sullivan, P.F.; Brooks, A.I.; Zou, F.; Sun, W.; Xia, K.; Madar, V.; Jansen, R.; Chung, W.; Zhou, Y.-H.; et al. Heritability and genomics of gene expression in peripheral blood. Nat. Genet. 2014, 46, 430–437. [Google Scholar] [CrossRef]
- Zeisel, A.; Muñoz-Manchado, A.B.; Codeluppi, S.; Lönnerberg, P.; La Manno, G.; Juréus, A.; Marques, S.; Munguba, H.; He, L.; Betsholtz, C.; et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 2015, 347, 1138–1142. [Google Scholar] [CrossRef]
- GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef] [Green Version]
- McKenzie, A.T.; Wang, M.; Hauberg, M.E.; Fullard, J.F.; Kozlenkov, A.; Keenan, A.; Hurd, Y.L.; Dracheva, S.; Casaccia, P.; Roussos, P.; et al. Brain Cell Type Specific Gene Expression and Co-expression Network Architectures. Sci. Rep. 2018, 8, 8868. [Google Scholar] [CrossRef]
- Rackley, C.R.; Stripp, B.R. Building and maintaining the epithelium of the lung. J. Clin. Investig. 2012, 122, 2724–2730. [Google Scholar] [CrossRef] [Green Version]
- Mallo, M. Reassessing the Role of Hox Genes during Vertebrate Development and Evolution. Trends Genet. 2018, 34, 209–217. [Google Scholar] [CrossRef] [Green Version]
- Di-Poï, N.; Koch, U.; Radtke, F.; Duboule, D. Additive and global functions of HoxA cluster genes in mesoderm derivatives. Dev. Biol. 2010, 341, 488–498. [Google Scholar] [CrossRef] [Green Version]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinf. 2016, 54, 1.30.1–1.30.33. [Google Scholar]
- Buenrostro, J.D.; Wu, B.; Chang, H.Y.; Greenleaf, W.J. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol. 2015, 109, 21–29. [Google Scholar] [PubMed]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, A.M.; Wang, Z.; Schug, J.; Naji, A.; Kaestner, K.H. Integration of ATAC-seq and RNA-seq identifies human alpha cell and beta cell signature genes. Mol. Metab. 2016, 5, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantile, M.; Franco, R.; Tschan, A.; Baumhoer, D.; Zlobec, I.; Schiavo, G.; Forte, I.; Bihl, M.; Liguori, G.; Botti, G.; et al. HOX D13 expression across 79 tumor tissue types. Int. J. Cancer 2009, 125, 1532–1541. [Google Scholar] [CrossRef] [Green Version]
- Song, I.-S.; Oh, N.S.; Kim, H.T.; Ha, G.-H.; Jeong, S.-Y.; Kim, J.-M.; Kim, D.-I.; Yoo, H.-S.; Kim, C.-H.; et al. Human ZNF312b Promotes the Progression of Gastric Cancer by Transcriptional Activation of the K-ras Gene. Cancer Res. 2009, 69, 3131–3139. [Google Scholar] [CrossRef]
- Inoue, K.; Tsubamoto, H.; Isono-Nakata, R.; Sakata, K.; Nakagomi, N. Itraconazole treatment of primary malignant melanoma of the vagina evaluated using positron emission tomography and tissue cDNA microarray: A case report. BMC Cancer 2018, 18. [Google Scholar] [CrossRef]
- Wang, M.; Li, X.; Zhang, J.; Yang, Q.; Chen, W.; Jin, W.; Huang, Y.-R.; Yang, R.; Gao, W.-Q. AHNAK2 is a Novel Prognostic Marker and Oncogenic Protein for Clear Cell Renal Cell Carcinoma. Theranostics 2017, 7, 1100–1113. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [Green Version]
- Mattick, J.S.; Rinn, J.L. Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 2015, 22, 5–7. [Google Scholar] [CrossRef]
- Schoenfelder, S.; Fraser, P. Long-range enhancer–promoter contacts in gene expression control. Nat. Rev. Genet. 2019, 1. [Google Scholar] [CrossRef]
- Viñuela, A.; Brown, A.A.; Buil, A.; Tsai, P.-C.; Davies, M.N.; Bell, J.T.; Dermitzakis, E.T.; Spector, T.D.; Small, K.S. Age-dependent changes in mean and variance of gene expression across tissues in a twin cohort. Hum. Mol. Genet. 2018, 27, 732–741. [Google Scholar] [CrossRef]
- Chaker, L.; Cappola, A.R.; Mooijaart, S.P.; Peeters, R.P. Clinical aspects of thyroid function during ageing. Lancet Diabetes Endocrinol. 2018, 6, 733–742. [Google Scholar] [CrossRef]
- Diagnostic Center for Population & Animal Health. Canine Thyroid Registry; Michigan State University: East Lansing, MI, USA, 2010. [Google Scholar]
- Consortium, G. GTEx Consortium Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef]
- Werber, M.; Wittler, L.; Timmermann, B.; Grote, P.; Herrmann, B.G. The tissue-specific transcriptomic landscape of the mid-gestational mouse embryo. Development 2014, 141, 2325–2330. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-J.; Shirahige, K.; Ohsugi, M.; Nakai, K. DBTMEE: A database of transcriptome in mouse early embryos. Nucleic Acids Res. 2015, 43, D771–D776. [Google Scholar] [CrossRef]
- Sahakyan, A.; Plath, K. Transcriptome Encyclopedia of Early Human Development. Cell 2016, 165, 777–779. [Google Scholar] [CrossRef] [Green Version]






| Category | Diseases or Functions | p | No. of Genes | Genes | |
|---|---|---|---|---|---|
| head | Organismal Injury and Abnormalities | Hypertrophy | 9.8 × 10−10 | 15 | TRIM55, CXCL12, TNNI3K, TNNT2, ADGRG1, GATA6, JPH2, INHBA, RPS6KA2, NR3C1, SLC25A4, CAST, RRAD, TRIM63, IL33 |
| Visceromegaly | 1.6 × 10−9 | 18 | TRIM55, CXCL12, TNNI3K, TNNT2, NEXN, ADGRG1, GATA6, JPH2, INHBA, TBX20, NR3C1, SLC25A4, CAST, RRAD, SSTR2, BIK, TRIM63, IL33 | ||
| Cardiovascular Disease, Cardiovascular System Development and Function, Organ Morphology, Organismal Development, Organismal Injury and Abnormalities | Enlargement of heart | 3.7 × 10−9 | 15 | TRIM55, CXCL12, TNNI3K, TNNT2, NEXN, ADGRG1, GATA6, JPH2, INHBA, TBX20, SLC25A4, CAST, RRAD, TRIM63, IL33 | |
| Abnormal morphology of heart | 5.0 × 10−9 | 17 | TRIM55, CXCL12, TNNI3K, TNNT2, NEXN, ADGRG1, GATA6, JPH2, DHRS3, INHBA, TBX20, RPS6KA2, SLC25A4, CAST, RRAD, TRIM63, IL33 | ||
| Muscular hypertrophy | 5.7 × 10−9 | 10 | INHBA, TRIM55, RPS6KA2, CAST, RRAD, ADGRG1, GATA6, JPH2, TRIM63, IL33 | ||
| Hypertrophy of heart | 1.6 × 10−7 | 11 | CXCL12, INHBA, TRIM55, TNNI3K, SLC25A4, TNNT2, RRAD, ADGRG1, GATA6, TRIM63, IL33 | ||
| Cardiovascular System Development and Function | Morphology of cardiovascular system | 6.4 × 10−9 | 19 | CXCL12, TRIM55, TNNI3K, TNNT2, PLA2G7, NEXN, ADGRG1, GATA6, JPH2, DHRS3, INHBA, TBX20, RPS6KA2, SLC25A4, RRAD, CAST, SSTR2, TRIM63, IL33 | |
| Cardiovascular Disease, Cardiovascular System Development and Function | Abnormal morphology of cardiovascular system | 8.2 × 10−9 | 18 | TRIM55, CXCL12, TNNI3K, TNNT2, NEXN, ADGRG1, GATA6, JPH2, DHRS3, INHBA, TBX20, RPS6KA2, SLC25A4, CAST, RRAD, SSTR2, TRIM63, IL33 | |
| Organismal Development, Organismal Injury and Abnormalities | Abnormal morphology of thoracic cavity | 2.9 × 10−8 | 18 | TRIM55, CXCL12, TNNI3K, TNNT2, NEXN, ADGRG1, GATA6, JPH2, DHRS3, INHBA, TBX20, RPS6KA2, NR3C1, SLC25A4, CAST, RRAD, TRIM63, IL33 | |
| Organismal Development | Abnormal morphology of body cavity | 8.0 × 10−8 | 22 | TRIM55, RBMS1, CXCL12, TNNI3K, TNNT2, MAPK8IP2, NEXN, ADGRG1, GATA6, JPH2, DHRS3, INHBA, TBX20, RPS6KA2, NR3C1, SLC25A4, RRAD, CAST, BIK, SSTR2, TRIM63, IL33 | |
| heart | Skeletal and Muscular System Development and Function | Morphogenesis of embryonic skeleton | 4.4 × 10−12 | 7 | HOXB8, HOXA6, HOXA3, HOXA7, HOXA4, HOXA2, HOXA5 |
| Morphology of axial skeleton | 1.1 × 10−8 | 8 | HSD11B2, HOXB8, HOXA3, HOXA6, HOXB9, HOXA4, HOXA5, mir-196 | ||
| Fusion of bone | 1.6 × 10−8 | 6 | HOXA6, HOXA3, HOXB9, HOXA7, HOXA4, HOXA5 | ||
| Morphology of skeleton | 1.8 × 10−8 | 9 | HSD11B2, HOXB8, HOXA6, HOXA3, HOXB9, HOXA4, HOXA2, mir-196, HOXA5 | ||
| Embryonic Development, Organismal Development | Patterning of rostrocaudal axis | 1.2 × 10−11 | 8 | HOXB8, HOXA6, HOXA3, HOXB9, HOXA7, HOXA4, HOXA2, HOXA5 | |
| Organismal Development | Abnormal morphology of body cavity | 9.2 × 10−9 | 17 | TRIM55, HSD11B2, MYH7, SMYD1, TNNC1, TNNI3K, HOXA3, HOXB9, ATP2A2, HOXA5, PDZK1, TBX20, SLC25A4, HOXA7, HOXA2, TRIM63, SGPP2 | |
| Cardiovascular System Development and Function, Organ Development, Organ Morphology, Skeletal and Muscular System Development and Function | Contraction of cardiac muscle | 3.9 × 10−8 | 6 | MYH7, TNNC1, TNNI3K, ATP2A2, TRIM63, SRL | |
| Organ Morphology, Skeletal and Muscular System Development and Function | Quantity of rib | 6.0 × 10−8 | 5 | HOXA6, HOXB9, HOXA4, HOXA5, mir-196 | |
| Cancer, Skeletal and Muscular Disorders, Tissue Morphology | Transformation of vertebrae | 7.1 × 10−8 | 5 | HOXA6, HOXB9, HOXA4, HOXA5, mir-196 | |
| Organismal Development, Organismal Injury and Abnormalities | Abnormal morphology of thoracic cavity | 8.8 × 10−8 | 13 | TRIM55, MYH7, SMYD1, TNNC1, TNNI3K, HOXA3, HOXB9, ATP2A2, HOXA5, TBX20, SLC25A4, HOXA7, TRIM63 | |
| kidney | Cell Cycle | Cell division of neural stem cells | 9.1 × 10−5 | 1 | EMX2 |
| Embryonic Development, Nervous System Development and Function, Organ Development, Organismal Development, Tissue Development | Development of hippocampal fissure | 9.1 × 10−5 | 1 | EMX2 | |
| Nervous System Development and Function, Organ Morphology, Organismal Development | Size of primary visual cortex | 9.1 × 10−5 | 1 | EMX2 | |
| Nervous System Development and Function, Neurological Disease, Organ Morphology, Organismal Development, Organismal Injury and Abnormalities | Abnormal morphology of medial ganglionic eminences | 1.8 × 10−4 | 1 | EMX2 | |
| Developmental Disorder, Embryonic Development, Tissue Morphology | Degeneration of Wolffian duct | 1.8 × 10−4 | 1 | EMX2 | |
| liver | Cancer, Gastrointestinal Disease, Hepatic System Disease, Organismal Injury and Abnormalities | Hepatitis B virus-related hepatocellular carcinoma | 9.2 × 10−5 | 3 | TF, ALDOB, RGN |
| Cell-To-Cell Signaling and Interaction, Renal and Urological System Development and Function | Activation of kidney cells | 9.8 × 10−5 | 2 | TF, MST1 | |
| Organismal Injury and Abnormalities | Organ Degeneration | 9.8 × 10−5 | 8 | EFEMP1, GSTZ1, TF, GRID2, RP2, RGN, ZNF408, mir-22 | |
| Developmental Disorder, Hereditary Disorder, Metabolic Disease, Organismal Injury and Abnormalities | Hyperphenylalaninemia | 2.3 × 10−4 | 2 | GCH1, DNAJC12 | |
| Lipid Metabolism, Small Molecule Biochemistry | Metabolism of acylglycerol | 3.2 × 10−4 | 4 | ACSL5, SLC22A4, RGN, F2 | |
| lung | Cancer,Organismal Injury and Abnormalities | Epithelial neoplasm | 0.0 | 6961 | many |
| Non-hematological solid tumor | 0.0 | 7039 | many | ||
| Nonhematologic malignant neoplasm | 0.0 | 7021 | many | ||
| Carcinoma | 0.0 | 6949 | many | ||
| Tumorigenesis of tissue | 0.0 | 6969 | many |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Megquier, K.; Genereux, D.P.; Hekman, J.; Swofford, R.; Turner-Maier, J.; Johnson, J.; Alonso, J.; Li, X.; Morrill, K.; Anguish, L.J.; et al. BarkBase: Epigenomic Annotation of Canine Genomes. Genes 2019, 10, 433. https://doi.org/10.3390/genes10060433
Megquier K, Genereux DP, Hekman J, Swofford R, Turner-Maier J, Johnson J, Alonso J, Li X, Morrill K, Anguish LJ, et al. BarkBase: Epigenomic Annotation of Canine Genomes. Genes. 2019; 10(6):433. https://doi.org/10.3390/genes10060433
Chicago/Turabian StyleMegquier, Kate, Diane P. Genereux, Jessica Hekman, Ross Swofford, Jason Turner-Maier, Jeremy Johnson, Jacob Alonso, Xue Li, Kathleen Morrill, Lynne J. Anguish, and et al. 2019. "BarkBase: Epigenomic Annotation of Canine Genomes" Genes 10, no. 6: 433. https://doi.org/10.3390/genes10060433
APA StyleMegquier, K., Genereux, D. P., Hekman, J., Swofford, R., Turner-Maier, J., Johnson, J., Alonso, J., Li, X., Morrill, K., Anguish, L. J., Koltookian, M., Logan, B., Sharp, C. R., Ferrer, L., Lindblad-Toh, K., Meyers-Wallen, V. N., Hoffman, A., & Karlsson, E. K. (2019). BarkBase: Epigenomic Annotation of Canine Genomes. Genes, 10(6), 433. https://doi.org/10.3390/genes10060433