Sneaking Out for Happy Hour: Yeast-Based Approaches to Explore and Modulate Immune Response and Immune Evasion
Abstract
1. Yeast as a Popular Model for Basic Research and Chemical Genetics Approaches
2. The Long-Lasting Love Affair between EBV and the Budding Yeast Saccharomyces cerevisiae
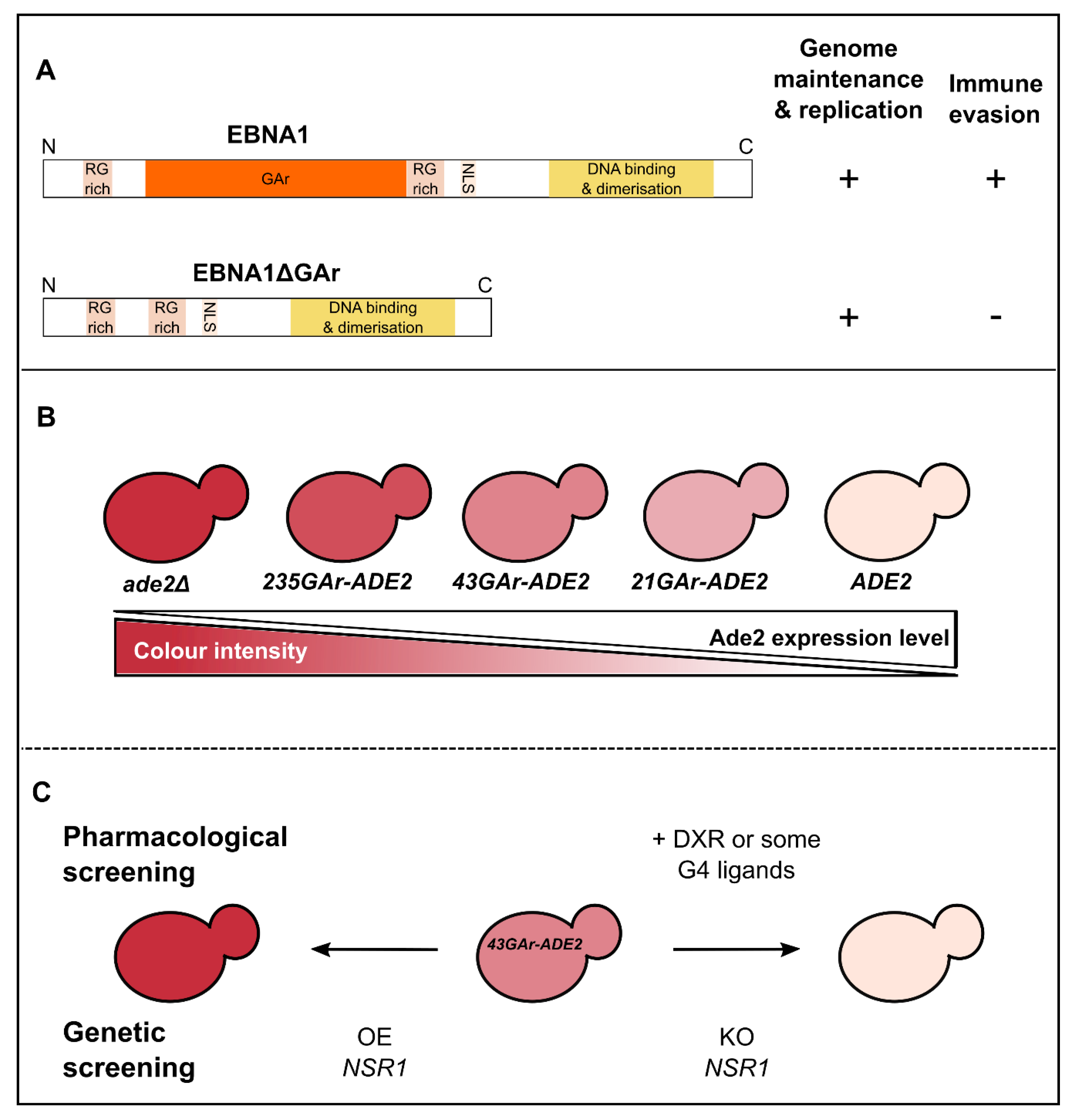
3. Use of Budding Yeast to Model and Decipher the Mechanisms at the Basis of EBV Stealthiness
4. Yeast Provided Key Insights into the Molecular Mechanisms of Gene Repression and Switching at the Basis of Immune Evasion of Some Parasites
5. Yeast as a Pathogen: Its Own Strategy to Evade the Immune System
6. Biotechnological Exploitation of Yeast to Create Vaccine or Adjuvant for Human and Animal Vaccination
7. Concluding Remarks and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Kornberg, A. DNA replication. J. Biol. Chem. 1988, 263, 1–4. [Google Scholar] [CrossRef]
- Kornberg, R. The molecular basis of eukaryotic transcription (Nobel lecture). Angew. Chem. Int. Ed. Engl. 2007, 46, 6956–6965. [Google Scholar] [CrossRef] [PubMed]
- Nurse, P.M. Nobel lecture. Cyclin dependent kinases and cell cycle control. Biosci. Rep. 2002, 22, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Schekman, R. Charting the secretory pathway in a simple eukaryote. Mol. Biol. Cell 2010, 21, 3781–3784. [Google Scholar] [CrossRef] [PubMed]
- Ohsumi, Y. Historical landmarks of autophagy research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Gutierrez, D.; Bauer, M.A.; Zimmermann, A.; Aguilera, A.; Austriaco, N.; Ayscough, K.; Balzan, R.; Bar-Nun, S.; Barrientos, A.; Belenky, P.; et al. Guidelines and recommendations on yeast cell death nomenclature. Microb. Cell 2018, 5, 4–31. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, S. Nobel yeast research. FEMS Yeast Res. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Couplan, E.; Aiyar, R.S.; Kucharczyk, R.; Kabala, A.; Ezkurdia, N.; Gagneur, J.; St Onge, R.P.; Salin, B.; Soubigou, F.; Le Cann, M.; et al. A yeast-based assay identifies drugs active against human mitochondrial disorders. Proc. Natl. Acad. Sci. USA 2011, 108, 11989–11994. [Google Scholar] [CrossRef] [PubMed]
- Lasserre, J.P.; Dautant, A.; Aiyar, R.S.; Kucharczyk, R.; Glatigny, A.; Tribouillard-Tanvier, D.; Rytka, J.; Blondel, M.; Skoczen, N.; Reynier, P.; et al. Yeast as a system for modeling mitochondrial disease mechanisms and discovering therapies. Dis. Model. Mech. 2015, 8, 509–526. [Google Scholar] [CrossRef]
- Giorgini, F.; Guidetti, P.; Nguyen, Q.; Bennett, S.C.; Muchowski, P.J. A genomic screen in yeast implicates kynurenine 3-monooxygenase as a therapeutic target for Huntington disease. Nat. Genet. 2005, 37, 526–531. [Google Scholar] [CrossRef]
- Outeiro, T.F.; Lindquist, S. Yeast cells provide insight into α-synuclein biology and pathobiology. Science 2003, 302, 1772–1775. [Google Scholar] [CrossRef] [PubMed]
- Willingham, S.; Outeiro, T.F.; DeVit, M.J.; Lindquist, S.L.; Muchowski, P.J. Yeast genes that enhance the toxicity of a mutant huntingtin fragment or α-synuclein. Science 2003, 302, 1769–1772. [Google Scholar] [CrossRef] [PubMed]
- Outeiro, T.F.; Giorgini, F. Yeast as a drug discovery platform in Huntington’s and Parkinson’s diseases. Biotechnol. J. 2006, 1, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Bach, S.; Talarek, N.; Andrieu, T.; Vierfond, J.M.; Mettey, Y.; Galons, H.; Dormont, D.; Meijer, L.; Cullin, C.; Blondel, M. Isolation of drugs active against mammalian prions using a yeast-based screening assay. Nat. Biotechnol. 2003, 21, 1075–1081. [Google Scholar] [CrossRef] [PubMed]
- Tribouillard-Tanvier, D.; Beringue, V.; Desban, N.; Gug, F.; Bach, S.; Voisset, C.; Galons, H.; Laude, H.; Vilette, D.; Blondel, M. Antihypertensive drug guanabenz is active in vivo against both yeast and mammalian prions. PLoS ONE 2008, 3, e1981. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.Y. Yeast for virus research. Microb. Cell 2017, 4, 311–330. [Google Scholar] [CrossRef] [PubMed]
- Lista, M.J.; Martins, R.P.; Angrand, G.; Quillevere, A.; Daskalogianni, C.; Voisset, C.; Teulade-Fichou, M.P.; Fahraeus, R.; Blondel, M. A yeast model for the mechanism of the Epstein-Barr virus immune evasion identifies a new therapeutic target to interfere with the virus stealthiness. Microb. Cell 2017, 4, 305–307. [Google Scholar] [CrossRef]
- Ozoya, O.O.; Sokol, L.; Dalia, S. EBV-related malignancies, outcomes and novel prevention strategies. Infect. Disord. Drug Targets 2016, 16, 4–21. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A.; Allday, M.J. The curious case of the tumour virus: 50 years of Burkitt’s lymphoma. Nat. Rev. Microbiol. 2008, 6, 913–924. [Google Scholar] [CrossRef]
- Young, L.S.; Rickinson, A.B. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 2004, 4, 757–768. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A.; Duca, K.A.; Shapiro, M. Epstein-Barr virus: A paradigm for persistent infection—For real and in virtual reality. Trends Immunol. 2008, 29, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Daskalogianni, C.; Pyndiah, S.; Apcher, S.; Mazars, A.; Manoury, B.; Ammari, N.; Nylander, K.; Voisset, C.; Blondel, M.; Fahraeus, R. Epstein-Barr virus-encoded EBNA1 and ZEBRA: Targets for therapeutic strategies against EBV-carrying cancers. J. Pathol. 2015, 235, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.I.; Fauci, A.S.; Varmus, H.; Nabel, G.J. Epstein-Barr virus: An important vaccine target for cancer prevention. Sci. Transl. Med. 2011, 3, 107fs7. [Google Scholar] [CrossRef] [PubMed]
- Lista, M.J.; Voisset, C.; Contesse, M.A.; Friocourt, G.; Daskalogianni, C.; Bihel, F.; Fahraeus, R.; Blondel, M. The long-lasting love affair between the budding yeast Saccharomyces cerevisiae and the Epstein-Barr virus. Biotechnol. J. 2015, 10, 1670–1681. [Google Scholar] [CrossRef] [PubMed]
- Henry, B.E., 2nd; Raab-Traub, N.J.; Pagano, J.S. Detection of autonomous replicating sequences (ARS) in the genome of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 1983, 80, 1096–1100. [Google Scholar] [CrossRef] [PubMed]
- Shire, K.; Ceccarelli, D.F.; Avolio-Hunter, T.M.; Frappier, L. Ebp2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 1999, 73, 2587–2595. [Google Scholar] [PubMed]
- Frappier, L. Ebna1. Curr. Top. Microbiol. Immunol. 2015, 391, 3–34. [Google Scholar] [PubMed]
- Wilson, J.B.; Manet, E.; Gruffat, H.; Busson, P.; Blondel, M.; Fahraeus, R. Ebna1: Oncogenic activity, immune evasion and biochemical functions provide targets for novel therapeutic strategies against Epstein-Barr virus-associated cancers. Cancers 2018, 10, 109. [Google Scholar] [CrossRef]
- Countryman, J.K.; Heston, L.; Gradoville, L.; Himmelfarb, H.; Serdy, S.; Miller, G. Activation of the Epstein-Barr virus bmrf1 and bzlf1 promoters by zebra in Saccharomyces cerevisiae. J. Virol. 1994, 68, 7628–7633. [Google Scholar]
- Miller, G.; Himmelfarb, H.; Heston, L.; Countryman, J.; Gradoville, L.; Baumann, R.; Chi, T.; Carey, M. Comparing regions of the Epstein-Barr virus zebra protein which function as transcriptional activating sequences in Saccharomyces cerevisiae and in B cells. J. Virol. 1993, 67, 7472–7481. [Google Scholar]
- Kuny, C.V.; Chinchilla, K.; Culbertson, M.R.; Kalejta, R.F. Cyclin-dependent kinase-like function is shared by the β- and γ- subset of the conserved herpesvirus protein kinases. PLoS Pathog. 2010, 6, e1001092. [Google Scholar] [CrossRef] [PubMed]
- Kudoh, A.; Fujita, M.; Kiyono, T.; Kuzushima, K.; Sugaya, Y.; Izuta, S.; Nishiyama, Y.; Tsurumi, T. Reactivation of lytic replication from B cells latently infected with Epstein-Barr virus occurs with high s-phase cyclin-dependent kinase activity while inhibiting cellular DNA replication. J. Virol. 2003, 77, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Blake, N. Immune evasion by gammaherpesvirus genome maintenance proteins. J. Gen. Virol. 2010, 91, 829–846. [Google Scholar] [CrossRef] [PubMed]
- Tellam, J.; Connolly, G.; Green, K.J.; Miles, J.J.; Moss, D.J.; Burrows, S.R.; Khanna, R. Endogenous presentation of cd8+ T cell epitopes from Epstein-Barr virus-encoded nuclear antigen 1. J. Exp. Med. 2004, 199, 1421–1431. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Manoury, B.; Fahraeus, R. Self-inhibition of synthesis and antigen presentation by Epstein-Barr virus-encoded EBNA1. Science 2003, 301, 1371–1374. [Google Scholar] [CrossRef] [PubMed]
- Apcher, S.; Daskalogianni, C.; Manoury, B.; Fahraeus, R. Epstein Barr virus-encoded EBNA1 interference with MHC class I antigen presentation reveals a close correlation between mRNA translation initiation and antigen presentation. PLoS Pathog. 2010, 6, e1001151. [Google Scholar] [CrossRef] [PubMed]
- Levitskaya, J.; Coram, M.; Levitsky, V.; Imreh, S.; Steigerwald-Mullen, P.M.; Klein, G.; Kurilla, M.G.; Masucci, M.G. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature 1995, 375, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Heessen, S.; Dantuma, N.P.; Tessarz, P.; Jellne, M.; Masucci, M.G. Inhibition of ubiquitin/proteasome-dependent proteolysis in Saccharomyces cerevisiae by a gly-ala repeat. FEBS Lett. 2003, 555, 397–404. [Google Scholar] [CrossRef]
- Voisset, C.; Daskalogianni, C.; Contesse, M.A.; Mazars, A.; Arbach, H.; Le Cann, M.; Soubigou, F.; Apcher, S.; Fahraeus, R.; Blondel, M. A yeast-based assay identifies drugs that interfere with immune evasion of the Epstein-Barr virus. Dis. Model. Mech. 2014, 7, 435–444. [Google Scholar] [CrossRef]
- Stotz, A.; Linder, P. The ADE2 gene from Saccharomyces cerevisiae: Sequence and new vectors. Gene 1990, 95, 91–98. [Google Scholar] [CrossRef]
- Lista, M.J.; Martins, R.P.; Billant, O.; Contesse, M.A.; Findakly, S.; Pochard, P.; Daskalogianni, C.; Beauvineau, C.; Guetta, C.; Jamin, C.; et al. Nucleolin directly mediates Epstein-Barr virus immune evasion through binding to G-quadruplexes of EBNA1 mRNA. Nat. Commun. 2017, 8, 16043. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, L.A.; Sun, H.; Hanakahi, L.A.; Maizels, N. G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D, a role for G-G pairing in immunoglobulin switch recombination. J. Biol. Chem. 1999, 274, 1066–1071. [Google Scholar] [CrossRef] [PubMed]
- Hanakahi, L.A.; Sun, H.; Maizels, N. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 1999, 274, 15908–15912. [Google Scholar] [CrossRef] [PubMed]
- Murat, P.; Zhong, J.; Lekieffre, L.; Cowieson, N.P.; Clancy, J.L.; Preiss, T.; Balasubramanian, S.; Khanna, R.; Tellam, J. G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat. Chem. Biol. 2014, 10, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, J.D.; Perreault, J.P. 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 2010, 38, 7022–7036. [Google Scholar] [CrossRef]
- Didiot, M.C.; Tian, Z.; Schaeffer, C.; Subramanian, M.; Mandel, J.L.; Moine, H. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 2008, 36, 4902–4912. [Google Scholar] [CrossRef]
- Gomez, D.; Lemarteleur, T.; Lacroix, L.; Mailliet, P.; Mergny, J.L.; Riou, J.F. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004, 32, 371–379. [Google Scholar] [CrossRef]
- Marcel, V.; Tran, P.L.; Sagne, C.; Martel-Planche, G.; Vaslin, L.; Teulade-Fichou, M.P.; Hall, J.; Mergny, J.L.; Hainaut, P.; Van Dyck, E. G-quadruplex structures in TP53 intron 3: Role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis 2011, 32, 271–278. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef]
- Song, J.; Perreault, J.P.; Topisirovic, I.; Richard, S. RNA G-quadruplexes and their potential regulatory roles in translation. Translation 2016, 4, e1244031. [Google Scholar] [CrossRef]
- Von Hacht, A.; Seifert, O.; Menger, M.; Schutze, T.; Arora, A.; Konthur, Z.; Neubauer, P.; Wagner, A.; Weise, C.; Kurreck, J. Identification and characterization of RNA guanine-quadruplex binding proteins. Nucleic Acids Res. 2014, 42, 6630–6644. [Google Scholar] [CrossRef] [PubMed]
- Prado Martins, R.; Findakly, S.; Daskalogianni, C.; Teulade-Fichou, M.P.; Blondel, M.; Fahraeus, R. In cellulo protein-mRNA interaction assay to determine the action of G-quadruplex-binding molecules. Molecules 2018, 23, 3124. [Google Scholar] [CrossRef] [PubMed]
- De Cian, A.; Delemos, E.; Mergny, J.L.; Teulade-Fichou, M.P.; Monchaud, D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007, 129, 1856–1857. [Google Scholar] [CrossRef] [PubMed]
- Reznichenko, O.; Quillevere, A.; Martins, R.P.; Loaec, N.; Kang, H.; Lista, M.J.; Beauvineau, C.; Gonzalez-Garcia, J.; Guillot, R.; Voisset, C.; et al. Novel cationic bis (acylhydrazones) as modulators of Epstein-Barr virus immune evasion acting through disruption of interaction between nucleolin and G-quadruplexes of EBNA1 mRNA. Eur. J. Med. Chem. 2019, 178, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Aznauryan, M.; Sondergaard, S.; Noer, S.L.; Schiott, B.; Birkedal, V. A direct view of the complex multi-pathway folding of telomeric G-quadruplexes. Nucleic Acids Res. 2016, 44, 11024–11032. [Google Scholar] [CrossRef] [PubMed]
- Noer, S.L.; Preus, S.; Gudnason, D.; Aznauryan, M.; Mergny, J.L.; Birkedal, V. Folding dynamics and conformational heterogeneity of human telomeric G-quadruplex structures in Na+ solutions by single molecule FRET microscopy. Nucleic Acids Res. 2016, 44, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Martins, R.P.; Malbert-Colas, L.; Lista, M.J.; Daskalogianni, C.; Apcher, S.; Pla, M.; Findakly, S.; Blondel, M.; Fahraeus, R. Nuclear processing of nascent transcripts determines synthesis of full-length proteins and antigenic peptides. Nucleic Acids Res. 2019, 47, 3086–3100. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E.; Smith, J.S.; Kozak, M.L.; Johnson, F.B. In vivo veritas: Using yeast to probe the biological functions of G-quadruplexes. Biochimie 2008, 90, 1250–1263. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.C.; Ostrowski, L.A.; Pietrobon, V.; Mekhail, K. Repetitive DNA loci and their modulation by the non-canonical nucleic acid structures R-loops and G-quadruplexes. Nucleus 2017, 8, 162–181. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.K.; Raney, K.D. Structure and function of Pif1 helicase. Biochem. Soc. Trans. 2017, 45, 1159–1171. [Google Scholar] [CrossRef] [PubMed]
- Lopes, J.; Piazza, A.; Bermejo, R.; Kriegsman, B.; Colosio, A.; Teulade-Fichou, M.P.; Foiani, M.; Nicolas, A. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011, 30, 4033–4046. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; Bochman, M.L.; Garcia, P.D.; Cejka, P.; Friedman, K.L.; Kowalczykowski, S.C.; Zakian, V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 2013, 497, 458–462. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; Capra, J.A.; Zakian, V.A. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 2011, 145, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Piazza, A.; Serero, A.; Boule, J.B.; Legoix-Ne, P.; Lopes, J.; Nicolas, A. Stimulation of gross chromosomal rearrangements by the human CEB1 and CEB25 minisatellites in Saccharomyces cerevisiae depends on G-quadruplexes or Cdc13. PLoS Genet. 2012, 8, e1003033. [Google Scholar] [CrossRef] [PubMed]
- Ribeyre, C.; Lopes, J.; Boule, J.B.; Piazza, A.; Guedin, A.; Zakian, V.A.; Mergny, J.L.; Nicolas, A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009, 5, e1000475. [Google Scholar] [CrossRef] [PubMed]
- Kirkman, L.A.; Deitsch, K.W. Antigenic variation and the generation of diversity in malaria parasites. Curr. Opin. Microbiol. 2012, 15, 456–462. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mwakalinga, S.B.; Wang, C.W.; Bengtsson, D.C.; Turner, L.; Dinko, B.; Lusingu, J.P.; Arnot, D.E.; Sutherland, C.J.; Theander, T.G.; Lavstsen, T. Expression of a type B RIFIN in Plasmodium falciparum merozoites and gametes. Malar. J. 2012, 11, 429. [Google Scholar] [CrossRef]
- Scherf, A.; Lopez-Rubio, J.J.; Riviere, L. Antigenic variation in Plasmodium falciparum. Annu. Rev. Microbiol. 2008, 62, 445–470. [Google Scholar] [CrossRef] [PubMed]
- Scherf, A.; Riviere, L.; Lopez-Rubio, J.J. Snapshot: Var gene expression in the malaria parasite. Cell 2008, 134, 190. [Google Scholar] [CrossRef][Green Version]
- Morrison, L.J.; Marcello, L.; McCulloch, R. Antigenic variation in the African trypanosome: Molecular mechanisms and phenotypic complexity. Cell Microbiol. 2009, 11, 1724–1734. [Google Scholar] [CrossRef]
- Cushion, M.T.; Stringer, J.R. Stealth and opportunism: Alternative lifestyles of species in the fungal genus Pneumocystis. Annu. Rev. Microbiol. 2010, 64, 431–452. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Fries, B.C. Antigenic and phenotypic variations in fungi. Cell Microbiol. 2009, 11, 1716–1723. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wyse, B.A.; Oshidari, R.; Jeffery, D.C.; Yankulov, K.Y. Parasite epigenetics and immune evasion: Lessons from budding yeast. Epigenet. Chromatin 2013, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Keely, S.P.; Renauld, H.; Wakefield, A.E.; Cushion, M.T.; Smulian, A.G.; Fosker, N.; Fraser, A.; Harris, D.; Murphy, L.; Price, C.; et al. Gene arrays at Pneumocystis carinii telomeres. Genetics 2005, 170, 1589–1600. [Google Scholar] [CrossRef] [PubMed]
- Ottaviani, A.; Gilson, E.; Magdinier, F. Telomeric position effect: From the yeast paradigm to human pathologies? Biochimie 2008, 90, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Taddei, A.; Gasser, S.M. Structure and function in the budding yeast nucleus. Genetics 2012, 192, 107–129. [Google Scholar] [CrossRef] [PubMed]
- Maillet, L.; Gaden, F.; Brevet, V.; Fourel, G.; Martin, S.G.; Dubrana, K.; Gasser, S.M.; Gilson, E. Ku-deficient yeast strains exhibit alternative states of silencing competence. EMBO Rep. 2001, 2, 203–210. [Google Scholar] [CrossRef][Green Version]
- Rusche, L.N.; Rine, J. Switching the mechanism of mating type switching: A domesticated transposase supplants a domesticated homing endonuclease. Genes Dev. 2010, 24, 10–14. [Google Scholar] [CrossRef][Green Version]
- Gow, N.A.R.; Latge, J.P.; Munro, C.A. The fungal cell wall: Structure, biosynthesis, and function. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Hernandez-Chavez, M.J.; Perez-Garcia, L.A.; Nino-Vega, G.A.; Mora-Montes, H.M. Fungal strategies to evade the host immune recognition. J. Fungi 2017, 3, 51. [Google Scholar] [CrossRef]
- Díaz-Jiménez, D.F.; Pérez García, L.A.; Martínez Álvarez, J.A.; Mora, H. Role of the fungal cell wall in pathogenesis and antifungal resistance. Curr. Fungal Infect. Rep. 2012, 6, 215–282. [Google Scholar] [CrossRef]
- Erwig, L.P.; Gow, N.A. Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 2016, 14, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Mata, E.; Navarro-Arias, M.J.; Perez-Garcia, L.A.; Mellado-Mojica, E.; Lopez, M.G.; Csonka, K.; Gacser, A.; Mora-Montes, H.M. Members of the Candida parapsilosis complex and Candida albicans are differentially recognized by human peripheral blood mononuclear cells. Front. Microbiol. 2015, 6, 1527. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.; Netea, M.G.; Munro, C.A.; Ferwerda, G.; Bates, S.; Mora-Montes, H.M.; Walker, L.; Jansen, T.; Jacobs, L.; Tsoni, V.; et al. Immune recognition of Candida albicans β-glucan by dectin-1. J. Infect. Dis. 2007, 196, 1565–1571. [Google Scholar] [CrossRef]
- Mora-Montes, H.M.; Bates, S.; Netea, M.G.; Castillo, L.; Brand, A.; Buurman, E.T.; Diaz-Jimenez, D.F.; Jan Kullberg, B.; Brown, A.J.; Odds, F.C.; et al. A multifunctional mannosyltransferase family in Candida albicans determines cell wall mannan structure and host-fungus interactions. J. Biol. Chem. 2010, 285, 12087–12095. [Google Scholar] [CrossRef] [PubMed]
- Mora-Montes, H.M.; Bates, S.; Netea, M.G.; Diaz-Jimenez, D.F.; Lopez-Romero, E.; Zinker, S.; Ponce-Noyola, P.; Kullberg, B.J.; Brown, A.J.; Odds, F.C.; et al. Endoplasmic reticulum α-glycosidases of Candida albicans are required for N glycosylation, cell wall integrity, and normal host-fungus interaction. Eukaryot. Cell 2007, 6, 2184–2193. [Google Scholar] [CrossRef]
- Navarro-Arias, M.J.; Defosse, T.A.; Dementhon, K.; Csonka, K.; Mellado-Mojica, E.; Dias Valerio, A.; Gonzalez-Hernandez, R.J.; Courdavault, V.; Clastre, M.; Hernandez, N.V.; et al. Disruption of protein mannosylation affects Candida guilliermondii cell wall, immune sensing, and virulence. Front. Microbiol. 2016, 7, 1951. [Google Scholar] [CrossRef]
- Perez-Garcia, L.A.; Csonka, K.; Flores-Carreon, A.; Estrada-Mata, E.; Mellado-Mojica, E.; Nemeth, T.; Lopez-Ramirez, L.A.; Toth, R.; Lopez, M.G.; Vizler, C.; et al. Role of protein glycosylation in Candida parapsilosis cell wall integrity and host interaction. Front. Microbiol. 2016, 7, 306. [Google Scholar] [CrossRef]
- West, L.; Lowman, D.W.; Mora-Montes, H.M.; Grubb, S.; Murdoch, C.; Thornhill, M.H.; Gow, N.A.; Williams, D.; Haynes, K. Differential virulence of Candida glabrata glycosylation mutants. J. Biol. Chem. 2013, 288, 22006–22018. [Google Scholar] [CrossRef]
- Wheeler, R.T.; Fink, G.R. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2006, 2, e35. [Google Scholar] [CrossRef]
- Rappleye, C.A.; Eissenberg, L.G.; Goldman, W.E. Histoplasma capsulatum α-(1,3)-glucan blocks innate immune recognition by the β-glucan receptor. Proc. Natl. Acad. Sci. USA 2007, 104, 1366–1370. [Google Scholar] [CrossRef] [PubMed]
- Garfoot, A.L.; Shen, Q.; Wuthrich, M.; Klein, B.S.; Rappleye, C.A. The Eng1 β-glucanase enhances histoplasma virulence by reducing β-glucan exposure. MBio 2016, 7, e01388-15. [Google Scholar] [CrossRef] [PubMed]
- Ballou, E.R.; Avelar, G.M.; Childers, D.S.; Mackie, J.; Bain, J.M.; Wagener, J.; Kastora, S.L.; Panea, M.D.; Hardison, S.E.; Walker, L.A.; et al. Lactate signalling regulates fungal β-glucan masking and immune evasion. Nat. Microbiol. 2016, 2, 16238. [Google Scholar] [CrossRef] [PubMed]
- Latge, J.P. Immune evasion: Face changing in the fungal opera. Nat. Microbiol. 2017, 2, 16266. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.J.P.; Gow, N.A.R.; Warris, A.; Brown, G.D. Memory in fungal pathogens promotes immune evasion, colonisation, and infection. Trends Microbiol. 2019, 27, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Garreau, H.; Hasan, R.N.; Renault, G.; Estruch, F.; Boy-Marcotte, E.; Jacquet, M. Hyperphosphorylation of Msn2p and Msn4p in response to heat shock and the diauxic shift is inhibited by cAMP in Saccharomyces cerevisiae. Microbiology 2000, 146 Pt 9, 2113–2120. [Google Scholar] [CrossRef]
- Gorner, W.; Durchschlag, E.; Martinez-Pastor, M.T.; Estruch, F.; Ammerer, G.; Hamilton, B.; Ruis, H.; Schuller, C. Nuclear localization of the c2h2 zinc finger protein Msn2p is regulated by stress and protein kinase a activity. Genes Dev. 1998, 12, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Rodaki, A.; Bohovych, I.M.; Enjalbert, B.; Young, T.; Odds, F.C.; Gow, N.A.; Brown, A.J. Glucose promotes stress resistance in the fungal pathogen Candida albicans. Mol. Biol. Cell 2009, 20, 4845–4855. [Google Scholar] [CrossRef]
- Barelle, C.J.; Priest, C.L.; Maccallum, D.M.; Gow, N.A.; Odds, F.C.; Brown, A.J. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 2006, 8, 961–971. [Google Scholar] [CrossRef]
- Borghi, M.; Renga, G.; Puccetti, M.; Oikonomou, V.; Palmieri, M.; Galosi, C.; Bartoli, A.; Romani, L. Antifungal Th immunity: Growing up in family. Front. Immunol. 2014, 5, 506. [Google Scholar] [CrossRef]
- Romani, L. Immunity to fungal infections. Nat. Rev. Immunol. 2004, 4, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, L.; Pontier-Bres, R.; Larbret, F.; Rekima, A.; Verhasselt, V.; Blin-Wakkach, C.; Czerucka, D. Saccharomyces boulardii strain CNCM I-745 modifies the mononuclear phagocytes response in the small intestine of mice following Salmonella typhimurium infection. Front. Immunol. 2019, 10, 643. [Google Scholar] [CrossRef] [PubMed]
- Blanco, J.L.; Garcia, M.E. Immune response to fungal infections. Vet. Immunol. Immunopathol. 2008, 125, 47–70. [Google Scholar] [CrossRef] [PubMed]
- Cottier, F.; Pavelka, N. Complexity and dynamics of host-fungal interactions. Immunol. Res. 2012, 53, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Zhang, L.; Xu, Z.; Zhang, J.; Jiang, Y.Y.; Cao, Y.; Yan, T. Innate immune cell response upon Candida albicans infection. Virulence 2016, 7, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [PubMed]
- Cabib, E.; Arroyo, J. How carbohydrates sculpt cells: Chemical control of morphogenesis in the yeast cell wall. Nat. Rev. Microbiol. 2013, 11, 648–655. [Google Scholar] [CrossRef]
- Paulovicova, L.; Paulovicova, E.; Farkas, P.; Cizova, A.; Bystricky, P.; Jancinova, V.; Turanek, J.; Pericolini, E.; Gabrielli, E.; Vecchiarelli, A.; et al. Bioimmunological activities of Candida glabrata cellular mannan. FEMS Yeast Res. 2019, 19, foz009. [Google Scholar] [CrossRef]
- Berner, V.K.; duPre, S.A.; Redelman, D.; Hunter, K.W. Microparticulate β-glucan vaccine conjugates phagocytized by dendritic cells activate both naive CD4 and CD8 T cells in vitro. Cell Immunol. 2015, 298, 104–114. [Google Scholar] [CrossRef]
- De Smet, R.; Demoor, T.; Verschuere, S.; Dullaers, M.; Ostroff, G.R.; Leclercq, G.; Allais, L.; Pilette, C.; Dierendonck, M.; De Geest, B.G.; et al. β-glucan microparticles are good candidates for mucosal antigen delivery in oral vaccination. J. Control. Release 2013, 172, 671–678. [Google Scholar] [CrossRef]
- Levitz, S.M.; Huang, H.; Ostroff, G.R.; Specht, C.A. Exploiting fungal cell wall components in vaccines. Semin. Immunopathol. 2015, 37, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Roohvand, F.; Shokri, M.; Abdollahpour-Alitappeh, M.; Ehsani, P. Biomedical applications of yeast-a patent view, part one: Yeasts as workhorses for the production of therapeutics and vaccines. Expert Opin. Ther. Pat. 2017, 27, 929–951. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Li, P.; Wang, F. β-glucans as potential immunoadjuvants: A review on the adjuvanticity, structure-activity relationship and receptor recognition properties. Vaccine 2018, 36, 5235–5244. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, F. Polysaccharides: Candidates of promising vaccine adjuvants. Drug Discov. Ther. 2015, 9, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.Y.; Zhang, H.; Castro-Lopez, N.; Ostroff, G.R.; Khoshlenar, P.; Abraham, A.; Cole, G.T.; Negron, A.; Forsthuber, T.; Peng, T.; et al. Glucan-chitin particles enhance Th17 response and improve protective efficacy of a multivalent antigen (rCpa1) against pulmonary Coccidioides posadasii infection. Infect. Immun. 2018, 86, e00070-18. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Askari, F.; Sahu, M.S.; Kaur, R. Candida glabrata: A lot more than meets the eye. Microorganisms 2019, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Bucarey, S.A.; Pujol, M.; Poblete, J.; Nunez, I.; Tapia, C.V.; Neira-Carrillo, A.; Martinez, J.; Bassa, O. Chitosan microparticles loaded with yeast-derived PCV2 virus-like particles elicit antigen-specific cellular immune response in mice after oral administration. Virol. J. 2014, 11, 149. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, H.; LeBert, V.; Li, M.; Lerksuthirat, T.; Galles, K.; Klein, B.; Wuthrich, M. Mannose receptor is required for optimal induction of vaccine-induced T-helper type 17 cells and resistance to Blastomyces dermatitidis infection. J. Infect. Dis. 2016, 213, 1762–1766. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, T.R.; Cowen, L.E. Insights into the host-pathogen interaction: C. Albicans manipulation of macrophage pyroptosis. Microb. Cell 2018, 5, 566–568. [Google Scholar] [CrossRef] [PubMed]
- Steger, M.; Bermejo-Jambrina, M.; Yordanov, T.; Wagener, J.; Brakhage, A.A.; Pittl, V.; Huber, L.A.; Haas, H.; Lass-Florl, C.; Posch, W.; et al. β-1,3-glucan-lacking Aspergillus fumigatus mediates an efficient antifungal immune response by activating complement and dendritic cells. Virulence 2018, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Benito-Villalvilla, C.; Soria, I.; Subiza, J.L.; Palomares, O. Novel vaccines targeting dendritic cells by coupling allergoids to mannan. Allergo J. Int. 2018, 27, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Pan, B.; Liao, G.; Zhao, Q.; Gao, Y.; Chai, X.; Zhuo, X.; Wu, Q.; Jiao, B.; Pan, W.; et al. Synthesis and immunological studies of β-1,2-mannan-peptide conjugates as antifungal vaccines. Eur. J. Med. Chem. 2019, 173, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Petrovsky, N. Comparative safety of vaccine adjuvants: A summary of current evidence and future needs. Drug Saf. 2015, 38, 1059–1074. [Google Scholar] [CrossRef] [PubMed]
- Bal, J.; Luong, N.N.; Park, J.; Song, K.D.; Jang, Y.S.; Kim, D.H. Comparative immunogenicity of preparations of yeast-derived dengue oral vaccine candidate. Microb. Cell Fact. 2018, 17, 24. [Google Scholar] [CrossRef] [PubMed]
- Patterson, R.; Eley, T.; Browne, C.; Martineau, H.M.; Werling, D. Oral application of freeze-dried yeast particles expressing the PCV2b Cap protein on their surface induce protection to subsequent PCV2b challenge in vivo. Vaccine 2015, 33, 6199–6205. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wetzel, D.; Rolf, T.; Suckow, M.; Kranz, A.; Barbian, A.; Chan, J.A.; Leitsch, J.; Weniger, M.; Jenzelewski, V.; Kouskousis, B.; et al. Establishment of a yeast-based VLP platform for antigen presentation. Microb. Cell Fact. 2018, 17, 17. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, H.J. Yeast as an expression system for producing virus-like particles: What factors do we need to consider? Lett. Appl. Microbiol. 2017, 64, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, R.K.; Ramasamy, V.; Shukla, R.; Arora, U.; Swaminathan, S.; Khanna, N. Pichia pastoris-expressed Zika virus envelope domain III on a virus-like particle platform: Design, production and immunological evaluation. Pathog. Dis. 2019, 77, ftz026. [Google Scholar] [CrossRef]
- Kim, H.J.; Lee, J.Y.; Kang, H.A.; Lee, Y.; Park, E.J.; Kim, H.J. Oral immunization with whole yeast producing viral capsid antigen provokes a stronger humoral immune response than purified viral capsid antigen. Lett. Appl. Microbiol. 2014, 58, 285–291. [Google Scholar] [CrossRef]
- Nainys, J.; Lasickiene, R.; Petraityte-Burneikiene, R.; Dabrisius, J.; Lelesius, R.; Sereika, V.; Zvirbliene, A.; Sasnauskas, K.; Gedvilaite, A. Generation in yeast of recombinant virus-like particles of porcine Circovirus type 2 capsid protein and their use for a serologic assay and development of monoclonal antibodies. BMC Biotechnol. 2014, 14, 100. [Google Scholar] [CrossRef][Green Version]
- Fernandez, E.; Toledo, J.R.; Mendez, L.; Gonzalez, N.; Parra, F.; Martin-Alonso, J.M.; Limonta, M.; Sanchez, K.; Cabrales, A.; Estrada, M.P.; et al. Conformational and thermal stability improvements for the large-scale production of yeast-derived rabbit hemorrhagic disease virus-like particles as multipurpose vaccine. PLoS ONE 2013, 8, e56417. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Shen, C.; Zhang, C.; Zhang, W.; Wang, L.; Lan, K.; Liu, Q.; Huang, Z. Yeast-produced recombinant virus-like particles of Coxsackievirus a6 elicited protective antibodies in mice. Antivir. Res. 2016, 132, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Ren, S.; Chen, X.; Ge, J.; Xu, Z.; Huang, H.; Sun, H.; Gu, Y.; Zhou, T.; Li, J.; et al. Generation of hepatitis B virus PreS2-S antigen in Hansenula polymorpha. Virol. Sin. 2014, 29, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Saraswat, S.; Athmaram, T.N.; Parida, M.; Agarwal, A.; Saha, A.; Dash, P.K. Expression and characterization of yeast derived Chikungunya virus like particles (CHIK-VLPS) and its evaluation as a potential vaccine candidate. PLoS Negl. Trop. Dis. 2016, 10, e0004782. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.; Tripathi, L.; Raut, R.; Tyagi, P.; Arora, U.; Barman, T.; Sood, R.; Galav, A.; Wahala, W.; de Silva, A.; et al. Pichia pastoris-expressed Dengue 2 envelope forms virus-like particles without pre-membrane protein and induces high titer neutralizing antibodies. PLoS ONE 2013, 8, e64595. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xiao, X.; Zhao, M.; Liu, W.; Pang, L.; Sun, X.; Cen, S.; Yang, B.B.; Huang, Y.; Sheng, W.; et al. EV71 virus-like particles produced by co-expression of capsid proteins in yeast cells elicit humoral protective response against EV71 lethal challenge. BMC Res. Notes 2016, 9, 42. [Google Scholar] [CrossRef]
- Kumar, R.; Kumar, P. Yeast-based vaccines: New perspective in vaccine development and application. FEMS Yeast Res. 2019, 19, foz007. [Google Scholar] [CrossRef]
- Palma, M.L.; Zamith-Miranda, D.; Martins, F.S.; Bozza, F.A.; Nimrichter, L.; Montero-Lomeli, M.; Marques, E.T., Jr.; Douradinha, B. Probiotic Saccharomyces cerevisiae strains as biotherapeutic tools: Is there room for improvement? Appl. Microbiol. Biotechnol. 2015, 99, 6563–6570. [Google Scholar] [CrossRef]
- Ardiani, A.; Higgins, J.P.; Hodge, J.W. Vaccines based on whole recombinant Saccharomyces cerevisiae cells. FEMS Yeast Res. 2010, 10, 1060–1069. [Google Scholar] [CrossRef]
- Habersetzer, F.; Baumert, T.F.; Stoll-Keller, F. GI-5005, a yeast vector vaccine expressing an NS3-core fusion protein for chronic HCV infection. Curr. Opin. Mol. Ther. 2009, 11, 456–462. [Google Scholar]
- Lok, A.S.; Pan, C.Q.; Han, S.H.; Trinh, H.N.; Fessel, W.J.; Rodell, T.; Massetto, B.; Lin, L.; Gaggar, A.; Subramanian, G.M.; et al. Randomized phase II study of GS-4774 as a therapeutic vaccine in virally suppressed patients with chronic hepatitis B. J. Hepatol. 2016, 65, 509–516. [Google Scholar] [CrossRef] [PubMed]
- King, T.H.; Shanley, C.A.; Guo, Z.; Bellgrau, D.; Rodell, T.; Furney, S.; Henao-Tamayo, M.; Orme, I.M. GI-19007, a novel Saccharomyces cerevisiae-based therapeutic vaccine against tuberculosis. Clin. Vaccine Immunol. 2017, 24, e00245-17. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.N.; Wang, L.; Zheng, D.Z.; Chen, S.; Shi, W.; Qiao, X.Y.; Jiang, Y.P.; Tang, L.J.; Xu, Y.G.; Li, Y.J. Oral immunization with a Lactobacillus casei-based anti-porcine epidemic diarrhoea virus (PEDV) vaccine expressing microfold cell-targeting peptide Co1 fused with the COE antigen of PEDV. J. Appl. Microbiol. 2018, 124, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Durairaj, V.; Mundt, E.; Schulze, K.; Breunig, K.D.; Behrens, S.E. Protective vaccination against infectious bursal disease virus with whole recombinant Kluyveromyces lactis yeast expressing the viral VP2 subunit. PLoS ONE 2012, 7, e42870. [Google Scholar] [CrossRef] [PubMed]
- Jacob, D.; Ruffie, C.; Dubois, M.; Combredet, C.; Amino, R.; Formaglio, P.; Gorgette, O.; Pehau-Arnaudet, G.; Guery, C.; Puijalon, O.; et al. Whole Pichia pastoris yeast expressing measles virus nucleoprotein as a production and delivery system to multimerize Plasmodium antigens. PLoS ONE 2014, 9, e86658. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bernstein, M.B.; Chakraborty, M.; Wansley, E.K.; Guo, Z.; Franzusoff, A.; Mostbock, S.; Sabzevari, H.; Schlom, J.; Hodge, J.W. Recombinant Saccharomyces cerevisiae (yeast-CEA) as a potent activator of murine dendritic cells. Vaccine 2008, 26, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Remondo, C.; Cereda, V.; Mostbock, S.; Sabzevari, H.; Franzusoff, A.; Schlom, J.; Tsang, K.Y. Human dendritic cell maturation and activation by a heat-killed recombinant yeast (Saccharomyces cerevisiae) vector encoding carcinoembryonic antigen. Vaccine 2009, 27, 987–994. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wansley, E.K.; Chakraborty, M.; Hance, K.W.; Bernstein, M.B.; Boehm, A.L.; Guo, Z.; Quick, D.; Franzusoff, A.; Greiner, J.W.; Schlom, J.; et al. Vaccination with a recombinant Saccharomyces cerevisiae expressing a tumor antigen breaks immune tolerance and elicits therapeutic antitumor responses. Clin. Cancer Res. 2008, 14, 4316–4325. [Google Scholar] [CrossRef]
- Heery, C.R.; Singh, B.H.; Rauckhorst, M.; Marte, J.L.; Donahue, R.N.; Grenga, I.; Rodell, T.C.; Dahut, W.; Arlen, P.M.; Madan, R.A.; et al. Phase I trial of a yeast-based therapeutic cancer vaccine (GI-6301) targeting the transcription factor brachyury. Cancer Immunol. Res. 2015, 3, 1248–1256. [Google Scholar] [CrossRef]
- Riemann, H.; Takao, J.; Shellman, Y.G.; Hines, W.A.; Edwards, C.K., 3rd; Franzusoff, A.; Norris, D.A.; Fujita, M. Generation of a prophylactic melanoma vaccine using whole recombinant yeast expressing MART-1. Exp. Dermatol. 2007, 16, 814–822. [Google Scholar] [CrossRef]
- Tanaka, A.; Jensen, J.D.; Prado, R.; Riemann, H.; Shellman, Y.G.; Norris, D.A.; Chin, L.; Yee, C.; Fujita, M. Whole recombinant yeast vaccine induces antitumor immunity and improves survival in a genetically engineered mouse model of melanoma. Gene Ther. 2011, 18, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Bolhassani, A.; Muller, M.; Roohvand, F.; Motevalli, F.; Agi, E.; Shokri, M.; Rad, M.M.; Hosseinzadeh, S. Whole recombinant Pichia pastoris expressing HPV16 L1 antigen is superior in inducing protection against tumor growth as compared to killed transgenic Leishmania. Hum. Vaccines Immunother. 2014, 10, 3499–3508. [Google Scholar] [CrossRef] [PubMed]
- Cherf, G.M.; Cochran, J.R. Applications of yeast surface display for protein engineering. Methods Mol. Biol. 2015, 1319, 155–175. [Google Scholar] [PubMed]
- Sun, H.; Wang, L.; Wang, T.; Zhang, J.; Liu, Q.; Chen, P.; Chen, Z.; Wang, F.; Li, H.; Xiao, Y.; et al. Display of Eimeria tenella EtMic2 protein on the surface of Saccharomyces cerevisiae as a potential oral vaccine against chicken coccidiosis. Vaccine 2014, 32, 1869–1876. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.K.; Kang, M.L.; Jung, M.H.; Cha, S.B.; Lee, W.J.; Kim, J.M.; Kim, D.H.; Yoo, H.S. Induction of protective immune responses against challenge of Actinobacillus pleuropneumoniae by oral administration with Saccharomyces cerevisiae expressing Apx toxins in pigs. Vet. Immunol. Immunopathol. 2013, 151, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Yan, L.; Zhang, X.; Yuan, L.; Fang, Q.; Zhang, Y.A.; Dai, H. Yeast surface display of capsid protein VP7 of grass carp reovirus: Fundamental investigation for the development of vaccine against hemorrhagic disease. J. Microbiol. Biotechnol. 2015, 25, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.Z.; Xu, L.M.; Liu, M.; Cao, Y.S.; LaPatra, S.E.; Yin, J.S.; Liu, H.B.; Lu, T.Y. Preliminary study of an oral vaccine against infectious hematopoietic necrosis virus using improved yeast surface display technology. Mol. Immunol. 2017, 85, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Palma, M.L.; Garcia-Bates, T.M.; Martins, F.S.; Douradinha, B. Genetically engineered probiotic Saccharomyces cerevisiae strains mature human dendritic cells and stimulate GAG-specific memory CD8(+) T cells ex vivo. Appl. Microbiol. Biotechnol. 2019, 103, 5183–5192. [Google Scholar] [CrossRef]
- Lei, H.; Jin, S.; Karlsson, E.; Schultz-Cherry, S.; Ye, K. Yeast surface-displayed H5N1 avian influenza vaccines. J. Immunol. Res. 2016, 2016, 4131324. [Google Scholar] [CrossRef]
- Epstein, M.A. The florey lecture, 1986. Vaccine prevention of virus-induced human cancers. Proc. R. Soc. Lond. B Biol. Sci. 1987, 230, 147–161. [Google Scholar]
- Wang, M.; Jiang, S.; Han, Z.; Zhao, B.; Wang, L.; Zhou, Z.; Wang, Y. Expression and immunogenic characterization of recombinant gp350 for developing a subunit vaccine against Epstein-Barr virus. Appl. Microbiol. Biotechnol. 2016, 100, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Jiang, S.; Liu, X.; Wang, Y. Expression, purification, and immunogenic characterization of Epstein-Barr virus recombinant EBNA1 protein in Pichia pastoris. Appl. Microbiol. Biotechnol. 2013, 97, 6251–6262. [Google Scholar] [CrossRef] [PubMed]
- Bill, R.M. Recombinant protein subunit vaccine synthesis in microbes: A role for yeast? J. Pharm. Pharmacol. 2015, 67, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Kwun, H.J.; da Silva, S.R.; Shah, I.M.; Blake, N.; Moore, P.S.; Chang, Y. Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mimics Epstein-Barr virus EBNA1 immune evasion through central repeat domain effects on protein processing. J. Virol. 2007, 81, 8225–8235. [Google Scholar] [CrossRef] [PubMed]
- Voisset, C.; Blondel, M. Chemobiology at happy hour: Yeast as a model for pharmacological screening. Med. Sci. 2014, 30, 1161–1168. [Google Scholar]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angrand, G.; Quillévéré, A.; Loaëc, N.; Daskalogianni, C.; Granzhan, A.; Teulade-Fichou, M.-P.; Fahraeus, R.; Prado Martins, R.; Blondel, M. Sneaking Out for Happy Hour: Yeast-Based Approaches to Explore and Modulate Immune Response and Immune Evasion. Genes 2019, 10, 667. https://doi.org/10.3390/genes10090667
Angrand G, Quillévéré A, Loaëc N, Daskalogianni C, Granzhan A, Teulade-Fichou M-P, Fahraeus R, Prado Martins R, Blondel M. Sneaking Out for Happy Hour: Yeast-Based Approaches to Explore and Modulate Immune Response and Immune Evasion. Genes. 2019; 10(9):667. https://doi.org/10.3390/genes10090667
Chicago/Turabian StyleAngrand, Gaëlle, Alicia Quillévéré, Nadège Loaëc, Chrysoula Daskalogianni, Anton Granzhan, Marie-Paule Teulade-Fichou, Robin Fahraeus, Rodrigo Prado Martins, and Marc Blondel. 2019. "Sneaking Out for Happy Hour: Yeast-Based Approaches to Explore and Modulate Immune Response and Immune Evasion" Genes 10, no. 9: 667. https://doi.org/10.3390/genes10090667
APA StyleAngrand, G., Quillévéré, A., Loaëc, N., Daskalogianni, C., Granzhan, A., Teulade-Fichou, M.-P., Fahraeus, R., Prado Martins, R., & Blondel, M. (2019). Sneaking Out for Happy Hour: Yeast-Based Approaches to Explore and Modulate Immune Response and Immune Evasion. Genes, 10(9), 667. https://doi.org/10.3390/genes10090667






