Prevalence, Type, and Molecular Spectrum of NF1 Mutations in Patients with Neurofibromatosis Type 1 and Congenital Heart Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. Molecular Studies
2.3. Statistical Analysis
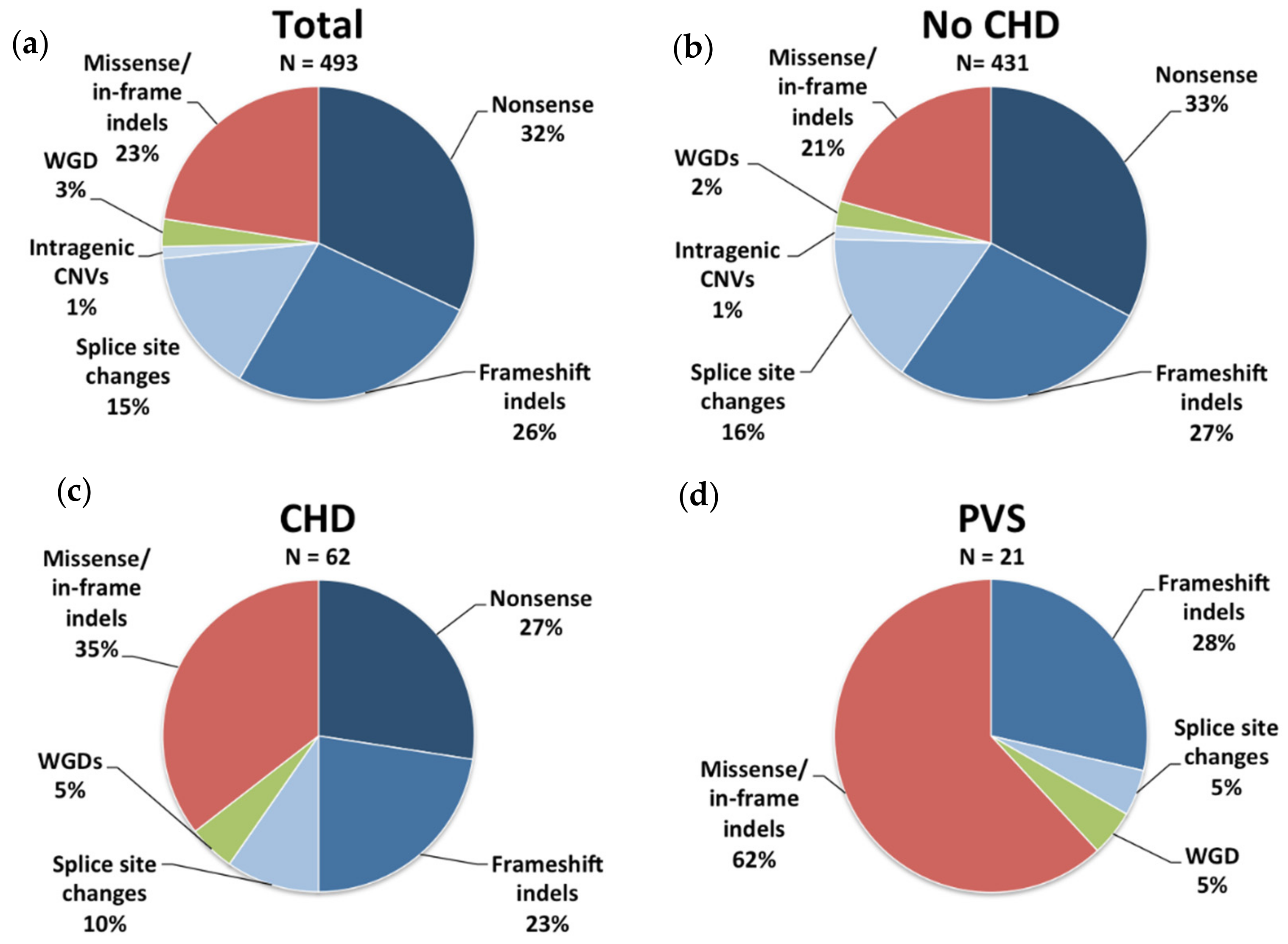
3. Results
3.1. Congenital Heart Disease
3.2. Pulmonary Valve Stenosis
4. Discussion
5. Conclusions
Supplementary Materials
Supplementary File 1Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Uusitalo, E.; Leppävirta, J.; Koffert, A.; Suominen, S.; Vahtera, J.; Vahlberg, T.; Pöyhönen, M.; Peltonen, J.; Peltonen, S. Incidence and mortality of neurofibromatosis: A total population study in Finland. J. Investig. Dermatol. 2015, 135, 904–906. [Google Scholar] [CrossRef] [PubMed]
- Huson, S.M.; Hughes, R. The Neurofibromatoses: A Clinical and Pathogenetic Overview; Chapman and Hall: London, UK, 1994. [Google Scholar]
- Stumpf, D.; Alksne, J.; Annegers, J.; Brown, S.; Conneally, P.; Housman, D.; Leppert, M.; Miller, J.; Moss, M.; Pileggi, A.; et al. Neurofibromatosis. Conference statement. National Institutes of Health Consensus Development Conference. Arch. Neurol. 1988, 45, 575–578. [Google Scholar]
- Cawthon, R.M.; Weiss, R.; Xu, G.F.; Viskochil, D.; Culver, M.; Stevens, J.; Robertson, M.; Dunn, D.; Gesteland, R.; O’Connell, P.; et al. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell 1990, 62, 193–201. [Google Scholar] [CrossRef]
- Viskochil, D.; Buchberg, A.M.; Xu, G.; Cawthon, R.M.; Stevens, J.; Wolff, R.K.; Culver, M.; Carey, J.C.; Copeland, N.G.; Jenkins, N.A.; et al. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell 1990, 62, 187–192. [Google Scholar] [CrossRef]
- Martin, G.A.; Viskochil, D.; Bollag, G.; McCabe, P.C.; Crosier, W.J.; Haubruck, H.; Conroy, L.; Clark, R.; O’Connell, P.; Cawthon, R.M.; et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell 1990, 63, 843–849. [Google Scholar] [CrossRef]
- De Luca, A.; Schirinzi, A.; Buccino, A.; Bottillo, I.; Sinibaldi, L.; Torrente, I.; Ciavarella, A.; Dottorini, T.; Porciello, R.; Giustini, S.; et al. Novel and recurrent mutations in the NF1 gene in Italian patients with neurofibromatosis type 1. Hum. Mutat. 2004, 23, 629. [Google Scholar] [CrossRef] [PubMed]
- Messiaen, L.; Wimmer, K. NF1 Mutational Spectrum. In Neurofibromatoses; Kaufmann, D., Ed.; Karger Publishers: Basel, Switzerland, 2008; Volume 16, pp. 63–77. [Google Scholar]
- Kehrer-Sawatzki, H.; Mautner, V.F.; Cooper, D.N. Emerging genotype-phenotype relationships in patients with large NF1 deletions. Hum. Genet. 2017, 136, 349–376. [Google Scholar] [CrossRef]
- Nguyen, R.; Mir, T.S.; Kluwe, L.; Jett, K.; Kentsch, M.; Mueller, G.; Kehrer-Sawatzki, H.; Friedman, J.M.; Mautner, V.F. Cardiac characterization of 16 patients with large NF1 gene deletions. Clin. Genet. 2013, 84, 344–349. [Google Scholar] [CrossRef]
- Upadhyaya, M.; Huson, S.M.; Davies, M.; Thomas, N.; Chuzhanova, N.; Giovannini, S.; Evans, D.G.; Howard, E.; Kerr, B.; Griffiths, S.; et al. An absence of cutaneous neurofibromas associated with a 3-bp inframe deletion in exon 17 of the NF1 gene (c.2970-2972 delAAT): Evidence of a clinically significant NF1 genotype-phenotype correlation. Am. J. Hum. Genet. 2007, 80, 140–151. [Google Scholar] [CrossRef]
- Koczkowska, M.; Callens, T.; Gomes, A.; Sharp, A.; Chen, Y.; Hicks, A.D.; Aylsworth, A.S.; Azizi, A.A.; Basel, D.G.; Bellus, G.; et al. Expanding the clinical phenotype of individuals with a 3-bp in-frame deletion of the NF1 gene (c.2970_2972del): An update of genotype-phenotype correlation. Genet. Med. 2019, 21, 867–876. [Google Scholar] [CrossRef]
- Pinna, V.; Lanari, V.; Daniele, P.; Consoli, F.; Agolini, E.; Margiotti, K.; Bottillo, I.; Torrente, I.; Bruselles, A.; Fusilli, C.; et al. p.Arg1809Cys substitution in neurofibromin is associated with a distinctive NF1 phenotype without neurofibromas. Eur. J. Hum. Genet. 2015, 23, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Rojnueangnit, K.; Xie, J.; Gomes, A.; Sharp, A.; Callens, T.; Chen, Y.; Liu, Y.; Cochran, M.; Abbott, M.A.; Atkin, J.; et al. High Incidence of Noonan Syndrome Features Including Short Stature and Pulmonic Stenosis in Patients carrying NF1 Missense Mutations Affecting p.Arg1809: Genotype-Phenotype Correlation. Hum. Mutat. 2015, 36, 1052–1063. [Google Scholar] [CrossRef] [PubMed]
- Aoki, Y.; Niihori, T.; Inoue, S.; Matsubara, Y. Recent advances in RASopathies. J. Hum. Genet. 2016, 61, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.E.; Allanson, J.E.; Tartaglia, M.; Gelb, B.D. Noonan syndrome. Lancet. 2013, 381, 333–342. [Google Scholar] [CrossRef]
- Koczkowska, M.; Chen, Y.; Callens, T.; Gomes, A.; Sharp, A.; Johnson, S.; Hsiao, M.C.; Chen, Z.; Balasubramanian, M.; Barnett, C.P.; et al. Genotype-Phenotype Correlation in NF1: Evidence for a More Severe Phenotype Associated with Missense Mutations Affecting NF1 Codons 844-848. Am. J. Hum. Genet. 2018, 102, 69–87. [Google Scholar] [CrossRef] [PubMed]
- Trevisson, E.; Morbidoni, V.; Forzan, M.; Daolio, C.; Fumini, V.; Parrozzani, R.; Cassina, M.; Midena, E.; Salviati, L.; Clementi, M. The Arg1038Gly missense variant in the NF1 gene causes a mild phenotype without neurofibromas. Mol. Genet. Genomic Med. 2019, 7, e616. [Google Scholar] [CrossRef] [PubMed]
- Brems, H.; Chmara, M.; Sahbatou, M.; Denayer, E.; Taniguchi, K.; Kato, R.; Somers, R.; Messiaen, L.; De Schepper, S.; Fryns, J.P.; et al. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat. Genet. 2007, 39, 1120–1126. [Google Scholar] [CrossRef] [PubMed]
- Messiaen, L.; Yao, S.; Brems, H.; Callens, T.; Sathienkijkanchai, A.; Denayer, E.; Spencer, E.; Arn, P.; Babovic-Vuksanovic, D.; Bay, C.; et al. Clinical and mutational spectrum of neurofibromatosis type 1-like syndrome. JAMA 2009, 302, 2111–2118. [Google Scholar] [CrossRef]
- Carey, J.C. Neurofibromatosis-Noonan syndrome. Am. J. Med. Genet. 1998, 75, 263–264. [Google Scholar] [CrossRef]
- De Luca, A.; Bottillo, I.; Sarkozy, A.; Carta, C.; Neri, C.; Bellacchio, E.; Schirinzi, A.; Conti, E.; Zampino, G.; Battaglia, A.; et al. NF1 gene mutations represent the major molecular event underlying neurofibromatosis-Noonan syndrome. Am. J. Hum. Genet. 2005, 77, 1092–1101. [Google Scholar] [CrossRef]
- Bertola, D.R.; Pereira, A.C.; Passetti, F.; de Oliveira, P.S.; Messiaen, L.; Gelb, B.D.; Kim, C.A.; Krieger, J.E. Neurofibromatosis-Noonan syndrome: Molecular evidence of the concurrence of both disorders in a patient. Am. J. Med. Genet. A 2005, 136, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Nyström, A.M.; Ekvall, S.; Strömberg, B.; Holmström, G.; Thuresson, A.C.; Annerén, G.; Bondeson, M.L. A severe form of Noonan syndrome and autosomal dominant café-au-lait spots—Evidence for different genetic origins. Acta Paediatr. 2009, 98, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Thiel, C.; Wilken, M.; Zenker, M.; Sticht, H.; Fahsold, R.; Gusek-Schneider, G.C.; Rauch, A. Independent NF1 and PTPN11 mutations in a family with neurofibromatosis-Noonan syndrome. Am. J. Med. Genet. A 2009, 149, 1263–1267. [Google Scholar] [CrossRef] [PubMed]
- Prada, C.E.; Zarate, Y.A.; Hagenbuch, S.; Lovell, A.; Schorry, E.K.; Hopkin, R.J. Lethal presentation of neurofibromatosis and Noonan syndrome. Am. J. Med. Genet. A 2011, 155, 1360–1366. [Google Scholar] [CrossRef] [PubMed]
- Pasmant, E.; Amiel, J.; Rodriguez, D.; Vidaud, M.; Vidaud, D.; Parfait, B. Two independent de novo mutations as a cause for neurofibromatosis type 1 and Noonan syndrome in a single family. Am. J. Med. Genet. A 2012, 158, 2290–2291. [Google Scholar] [CrossRef]
- Baquedano Lobera, I.; Izquierdo Álvarez, S.; Oliván Del Cacho, M.J. Rasopathies case report: Concurrence of two pathogenic variations de novo in NF1 and KRAS genes in a patient. BMC Pediatr. 2019, 19, 92. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, D.A.; Viskochil, D.H.; Rope, A.F.; Carey, J.C. Clinical and molecular aspects of an informative family with neurofibromatosis type 1 and Noonan phenotype. Clin. Genet. 2006, 69, 246–253. [Google Scholar] [CrossRef]
- Ben-Shachar, S.; Constantini, S.; Hallevi, H.; Sach, E.K.; Upadhyaya, M.; Evans, G.D.; Huson, S.M. Increased rate of missense/in-frame mutations in individuals with NF1-related pulmonary stenosis: A novel genotype-phenotype correlation. Eur. J. Hum. Genet. 2013, 21, 535–539. [Google Scholar] [CrossRef]
- Watson, G.H. Pulmonary stenosis, café-au-lait spots, and dull intelligence. Arch. Dis. Child. 1967, 42, 303–307. [Google Scholar] [CrossRef]
- Tassabehji, M.; Strachan, T.; Sharland, M.; Colley, A.; Donnai, D.; Harris, R.; Thakker, N. Tandem duplication within a neurofibromatosis type 1 (NF1) gene exon in a family with features of Watson syndrome and Noonan syndrome. Am. J. Hum. Genet. 1993, 53, 90–95. [Google Scholar]
- Lin, A.E.; Birch, P.H.; Korf, B.R.; Tenconi, R.; Niimura, M.; Poyhonen, M.; Armfield Uhas, K.; Sigorini, M.; Virdis, R.; Romano, C.; et al. Cardiovascular malformations and other cardiovascular abnormalities in neurofibromatosis 1. Am. J. Med. Genet. 2000, 95, 108–117. [Google Scholar] [CrossRef]
- Friedman, J.M.; Arbiser, J.; Epstein, J.A.; Gutmann, D.H.; Huot, S.J.; Lin, A.E.; McManus, B.; Korf, B.R. Cardiovascular disease in neurofibromatosis 1: Report of the NF1 Cardiovascular Task Force. Genet. Med. 2002, 4, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Tedesco, M.A.; Di Salvo, G.; Natale, F.; Pergola, V.; Calabrese, E.; Grassia, C.; Ratti, G.; Iarussi, D.; Iacono, A.; Calabrò, R.; et al. The heart in neurofibromatosis type 1: An echocardiographic study. Am. Heart. J. 2002, 143, 883–888. [Google Scholar] [CrossRef] [PubMed]
- Ferese, R.; Bonetti, M.; Consoli, F.; Guida, V.; Sarkozy, A.; Lepri, F.R.; Versacci, P.; Gambardella, S.; Calcagni, G.; Margiotti, K.; et al. Heterozygous missense mutations in NFATC1 are associated with atrioventricular septal defect. Hum. Mutat. 2018, 39, 1428–1441. [Google Scholar] [CrossRef] [PubMed]
- De Luca, A.; Bottillo, I.; Dasdia, M.C.; Morella, A.; Lanari, V.; Bernardini, L.; Divona, L.; Giustini, S.; Sinibaldi, L.; Novelli, A.; et al. Deletions of NF1 gene and exons detected by multiplex ligation-dependent probe amplification. J. Med. Genet. 2007, 44, 800–808. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Altman, D.G. Practical Statistics for Medical Research; Chapman and Hall: London, UK, 1991. [Google Scholar]
- Pros, E.; Larriba, S.; López, E.; Ravella, A.; Gili, M.L.; Kruyer, H.; Valls, J.; Serra, E.; Lázaro, C. NF1 mutation rather than individual genetic variability is the main determinant of the NF1-transcriptional profile of mutations affecting splicing. Hum. Mutat. 2006, 27, 1104–1114. [Google Scholar] [CrossRef]
- Colley, A.; Donnai, D.; Evans, D.G. Neurofibromatosis/Noonan phenotype: A variable feature of type 1 neurofibromatosis. Clin. Genet. 1996, 49, 59–64. [Google Scholar] [CrossRef]
- İncecik, F.; Hergüner, Ö.M.; Alınç Erdem, S.; Altunbaşak, Ş. Neurofibromatosis type 1 and cardiac manifestations. Turk. Kardiyol. Dern. Ars. 2015, 43, 714–716. [Google Scholar] [CrossRef][Green Version]
- Banerjee, A.; Kohl, T.; Silverman, N.H. Echocardiographic evaluation of congenital mitral valve anomalies in children. Am. J. Cardiol. 1995, 76, 1284–1291. [Google Scholar] [CrossRef]
- Lakkis, M.M.; Epstein, J.A. Neurofibromin modulation of ras activity is required for normal endocardial-mesenchymal transformation in the developing heart. Development 1998, 125, 4359–4367. [Google Scholar]
- Friedman, J.M.; Birch, P.; Greene, C. National Neurofibromatosis Foundation International Database. Am. J. Med. Genet. 1993, 45, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ismat, F.A.; Wang, T.; Lu, M.M.; Antonucci, N.; Epstein, J.A. Cardiomyocyte-specific loss of neurofibromin promotes cardiac hypertrophy and dysfunction. Circ. Res. 2009, 105, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Baralle, D.; Mattocks, C.; Kalidas, K.; Elmslie, F.; Whittaker, J.; Lees, M.; Ragge, N.; Patton, M.A.; Winter, R.M.; ffrench-Constant, C. Different mutations in the NF1 gene are associated with Neurofibromatosis-Noonan syndrome (NFNS). Am. J. Med. Genet. A 2003, 119, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hüffmeier, U.; Zenker, M.; Hoyer, J.; Fahsold, R.; Rauch, A. A variable combination of features of Noonan syndrome and neurofibromatosis type I are caused by mutations in the NF1 gene. Am. J. Med. Genet. A 2006, 140, 2749–2756. [Google Scholar] [CrossRef] [PubMed]
- Nyström, A.M.; Ekvall, S.; Allanson, J.; Edeby, C.; Elinder, M.; Holmström, G.; Bondeson, M.L.; Annerén, G. Noonan syndrome and neurofibromatosis type I in a family with a novel mutation in NF1. Clin. Genet. 2009, 76, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Ekvall, S.; Sjörs, K.; Jonzon, A.; Vihinen, M.; Annerén, G.; Bondeson, M.L. Novel association of neurofibromatosis type 1-causing mutations in families with neurofibromatosis-Noonan syndrome. Am. J. Med. Genet. A 2014, 164, 579–587. [Google Scholar] [CrossRef]
- Wu, R.; Legius, E.; Robberecht, W.; Dumoulin, M.; Cassiman, J.J.; Fryns, J.P. Neurofibromatosis type I gene mutation in a patient with features of LEOPARD syndrome. Hum. Mutat. 1996, 8, 51–56. [Google Scholar] [CrossRef]

| CHDs (n = 62/493, 12.6%) | CHD Feature, n (%) |
|---|---|
| Pulmonary valve stenosis a (PVS)/dysplasia (PVD) (total) | 23/62 (37.1) |
| • PVS b | 19/23 (82.6) |
| • PVS plus atrial septal defects (ASD) | 2/23 (8.7) |
| • PVD | 1/23 (4.3) |
| • PVD plus aortic valve dysplasia | 1/23 (4.3) |
| Mitral valve anomalies (total) | 20/62 (32.3) |
| • Mitral valve prolapse (MVP) | 5/20 (25) |
| • Mitral valve insufficiency (MVI) | 4/20 (20.0) |
| • MVP and tricuspid vale insufficiency (TVI) | 3/20 (15.0) |
| • MVP and MVI | 2/20 (10.0) |
| • Mitral valve dysplasia (MVD) | 1/20 (5.0) |
| • Mild MVD plus bicuspid aortic valve | 1/20 (5.0) |
| • MVI and arching of the posterior mitral leaflet | 1/20 (5.0) |
| • MVI and mild septal hypertrophic cardiomyopathy | 1/20 (5.0) |
| • Mitral valve thickening | 1/20 (5.0) |
| • Mild arching mitral valve | 1/20 (5.0) |
| Polyvalvular dysplasia c | 1/62 (1.6) |
| Mild aortic valve insufficiency with normal tricuspid aortic valve | 1/62 (1.6) |
| Patent ductus arteriosus (PDA) d | 2/62 (3.2) |
| ASD e | 2/62 (3.2) |
| Ventricular septal defects (VSD) f | 2/62 (3.2) |
| Patent foramen ovale (PFO) | 6/62 (9.7) |
| Transposition of the great arteries | 1/62 (1.6) |
| Dextrocardia h | 1/62 (1.6) |
| Tetralogy of Fallot | 1/62 (1.6) |
| Hypertrophy of the left ventricle | 1/62 (1.6) |
| Lipomatous hypertrophy of the interatrial septum | 1/62 (1.6) |
| CHDs (n = 62/493, 12.6%) | CHD feature, n (%) |
| CHD (n = 62) | No CHD (n = 431) | Chi-Square Significance | |
|---|---|---|---|
| Sex | 20 M, 42 F | 198 M, 233 F | |
| Age at observation | 12 y 7 m, max = 54 y, min = 4 m | 22 y 10 m, max = 64 y, min = 4 m | |
| CaLS | 61/61 (100.0%) | 276/277 (99.6%) | p > 0.05 |
| Freckling | 51/60 (83.6%) | 205/269 (76.2%) | p > 0.05 |
| Cutaneous NF a | 5/9 (55.6%) | 107/143 (74.8%) | p > 0.05 |
| Subcutaneous NF a | 4/7 (57.1%) | 78/133 (58.6%) | p > 0.05 |
| Plexiform NF b | 4/29 (13.8%) | 51/168 (30.4%) | p > 0.05 |
| Lish nodules | 23/50 (46.0%) | 89/206 (43.2%) | p > 0.05 |
| Optic glioma | 8/41 (19.5%) | 29/180 (16.1%) | p > 0.05 |
| Skeletal dysplasia | 0/52 (0.0%) | 19/252 (7.5%) | p > 0.05 c |
| Scoliosis | 20/60 (33.3%) | 70/270 (25.9%) | p > 0.05 |
| Learning disabilities | 23/56 (41.1%) | 62/225 (27.6%) | p > 0.05 |
| Intellectual disability | 6/59 (10.2%) | 23/229 (10.0%) | p > 0.05 |
| Macrocephaly | 9/19 (47.4%) | 26/110 (23.6%) | p > 0.05 |
| NS facial features | 37/55 (64.9%) | 48/250 (19.2%) | p < 0.00001 |
| Thoracic anomalies | 4/27 (14.8%) | 13/250 (5.2%) | p > 0.05 |
| Urogenital anomalies | 0/24 (0.0%) | 3/209 (1.4%) c | p > 0.05 d |
| Short stature | 22/57 (38.6%) | 42/150 (28.0%) | p > 0.05 |
| Hypertension | 2/54 (3.7%) | 14/256 (5.5%) | p > 0.05 |
| Vasculopathy | 2/40 (5.0%) | 7/46 (15.2%) | p > 0.05 |
| Other neoplasms | 7/48 (14.6%) | 31/194 (16.0%) | p > 0.05 |
| CHDs (n = 62/493, 12.6%) | Intragenic NF1 Mutations | WGD, n = 3 (%) | |
|---|---|---|---|
| In-Frame Mutations, n = 22 (%) | Out-of-Frame Mutations, n = 37 (%) | ||
| Pulmonary valve stenosis a (PVS)/dysplasia (PVD) (total) | 14/22 (63.6) * | 8/37 (21.6) * | 1/3 (33.3) |
| • PVS | 13/14 (92.9) | 5/8 (62.5) b | 1/1 (100) |
| • PVS plus ASD | 2/8 (25.0) | ||
| • PVD | 1/8 (12.5) | ||
| • PVD plus aortic valve dysplasia | 1/14 (7.1) | ||
| Mitral valve anomalies (total) | 3/22 (13.6) ** | 17/37 (45.9) ** | 0/3 (0.0%) |
| • Mitral valve prolapse (MVP) | 1/3 (33.3) | 4/17 (23.5) | |
| • Mitral valve insufficiency (MVI) | 1/3 (33.3) | 4/17 (23.5) | |
| • MVP and tricuspid valve insufficiency (TVI) | 1/17 (5.9) | ||
| • MVP and MVI | 1/3 (33.3) | 2/17 (11.8) | |
| • Mitral valve dysplasia (MVD) | 1/17 (5.9) | ||
| • Mild MVD plus bicuspid aortic valve | 1/17 (5.9) | ||
| • MVI and arching of the posterior mitral leaflet | 1/17 (5.9) | ||
| • MVI and mild septal hypertrophic cardiomyopathy | 1/17 (5.9) | ||
| • Mitral valve thickening | 1/17 (5.9) | ||
| • Mild arching mitral valve | 1/17 (5.9) | ||
| Polyvalvular dysplasia | 1/3 (33.3) c | ||
| Mild aortic valve insufficiency with normal tricuspid aortic valve | 1/37 (2.7) | ||
| Patent ductus arteriosus | 2/37 (5.4) d | ||
| Atrial septal defects (ASD) | 1/22 (4.5) | 1/37 (2.7) e | |
| Ventricular septal defects | 2/22 (9.1) f | ||
| Patent foramen ovale (PFO) | 1/22 (4.5) | 4/37 (10.8) | 1/3 (33.3) |
| Transposition of the great arteries | 1/37 (2.7) | ||
| Dextrocardia | 1/37 (2.7) h | ||
| Tetralogy of Fallot | 1/37 (2.7) | ||
| Hypertrophy of the left ventricle | 1/37 (2.7) | ||
| Lipomatous hypertrophy of the interatrial septum | 1/22 (4.5) | ||
| PVS (n = 21) | No PVS (n = 472) | Chi-Square Values | |
|---|---|---|---|
| Sex | 6 M, 15 F | 212 M, 260 F | |
| Age at diagnosis | 9 y 6 m, max = 33 year, min = 2 year | 21 y 11 m, max = 64 year, min = 4 min | |
| CaLS | 21/21 (100.0%) | 316/317 (99.7%) | p > 0.05 |
| Freckling | 17/21 (81.0%) | 239/308 (77.6%) | p > 0.05 |
| Cutaneous NF a | 1/1 (100.0%) | 110/150 (73.3%) | p > 0.05 c |
| Subcutaneous NF a | 1/1 (100.0%) | 81/139 (58.3%) | p > 0.05 c |
| Plexiform NF b | 0/9 (0.0%) | 53/186 (28.5%) | p > 0.05 c |
| Lish nodules | 4/17 (23.5%) | 108/240 (45.0%) | p > 0.05 |
| Optic Glioma | 3/13 (23.1%) | 34/208 (16.3%) | p > 0.05 |
| Skeletal dysplasia | 0/16 (0.0%) | 19/289 (6.6%) | p > 0.05 c |
| Scoliosis | 6/20 (30.0%) | 84/310 (27.1%) | p > 0.05 |
| Learning disability | 11/20 (55.0%) | 74/261 (28.4%) | p > 0.05 |
| Intellectual disability | 1/21 (4.8%) | 28/267 (10.5%) | p > 0.05 c |
| Macrocephaly | 4/8 (50.0%) | 31/121 (25.6%) | p > 0.05 |
| NS facial features | 19/20 (95.0%) | 66/285 (23.2%) | p = 0.000018 |
| Thoracic anomalies | 2/13 (15.4%) | 14/264 (5.3%) | p > 0.05 |
| Urogenital anomalies | 0/12 (0.0%) | 3/221 (1.4%) | p > 0.05 c |
| Short stature | 10/18 (55.6%) | 54/189 (28.6%) | p > 0.05 |
| Hypertension | 0/18 (0.0%) | 17/292 (5.8%) | p > 0.05 c |
| Vasculopathy | 0/11 (0.0%) | 9/75 (12.0%) | p > 0.05 c |
| Other neoplasms | 3/19 (15.8%) | 36/223 (16.1%) | p > 0.05 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinna, V.; Daniele, P.; Calcagni, G.; Mariniello, L.; Criscione, R.; Giardina, C.; Lepri, F.R.; Hozhabri, H.; Alberico, A.; Cavone, S.; et al. Prevalence, Type, and Molecular Spectrum of NF1 Mutations in Patients with Neurofibromatosis Type 1 and Congenital Heart Disease. Genes 2019, 10, 675. https://doi.org/10.3390/genes10090675
Pinna V, Daniele P, Calcagni G, Mariniello L, Criscione R, Giardina C, Lepri FR, Hozhabri H, Alberico A, Cavone S, et al. Prevalence, Type, and Molecular Spectrum of NF1 Mutations in Patients with Neurofibromatosis Type 1 and Congenital Heart Disease. Genes. 2019; 10(9):675. https://doi.org/10.3390/genes10090675
Chicago/Turabian StylePinna, Valentina, Paola Daniele, Giulio Calcagni, Lucio Mariniello, Roberta Criscione, Chiara Giardina, Francesca Romana Lepri, Hossein Hozhabri, Angela Alberico, Stefania Cavone, and et al. 2019. "Prevalence, Type, and Molecular Spectrum of NF1 Mutations in Patients with Neurofibromatosis Type 1 and Congenital Heart Disease" Genes 10, no. 9: 675. https://doi.org/10.3390/genes10090675
APA StylePinna, V., Daniele, P., Calcagni, G., Mariniello, L., Criscione, R., Giardina, C., Lepri, F. R., Hozhabri, H., Alberico, A., Cavone, S., Morella, A. T., Mandile, R., Annunziata, F., Di Giosaffatte, N., D’Asdia, M. C., Versacci, P., Capolino, R., Strisciuglio, P., Giustini, S., ... De Luca, A. (2019). Prevalence, Type, and Molecular Spectrum of NF1 Mutations in Patients with Neurofibromatosis Type 1 and Congenital Heart Disease. Genes, 10(9), 675. https://doi.org/10.3390/genes10090675





