Abstract
Antioxidants play an important role in animal and plant life owing to their involvement in complex metabolic and signaling mechanisms, hence uncovering the genetic basis associated with antioxidant activity is very important for the development of improved varieties. Here, a total of 182 common bean (Phaseolus vulgaris) landraces and six commercial cultivars collected from 19 provinces of Turkey were evaluated for seed antioxidant activity under four environments and two locations. Antioxidant activity was measured using ABTS radical scavenging capacity and mean antioxidant activity in common bean landraces was 20.03 µmol TE/g. Analysis of variance reflected that genotype by environment interaction was statistically non-significant and heritability analysis showed higher heritability of antioxidant activity. Variations in seed color were observed, and a higher antioxidant activity was present in seeds having colored seed as compared to those having white seeds. A negative correlation was found between white-colored seeds and antioxidant activity. A total of 7900 DArTseq markers were used to explore the population structure that grouped the studied germplasm into two sub-populations on the basis of their geographical origins and trolox equivalent antioxidant capacity contents. Mean linkage disequilibrium (LD) was 54%, and mean LD decay was 1.15 Mb. Mixed linear model i.e., the Q + K model demonstrated that four DArTseq markers had significant association (p < 0.01) for antioxidant activity. Three of these markers were present on chromosome Pv07, while the fourth marker was located on chromosome Pv03. Among the identified markers, DArT-3369938 marker showed maximum (14.61%) variation. A total of four putative candidate genes were predicted from sequences reflecting homology to identified DArTseq markers. This is a pioneering study involving the identification of association for antioxidant activity in common bean seeds. We envisage that this study will be very helpful for global common bean breeding community in order to develop cultivars with higher antioxidant activity.
1. Introduction
The world is facing a great threat of climate change, and extreme events throughout the world are being observed. Besides climate change, rapid growth in the world population is a global issue, and concerns have been raised to produce a sufficient quantity of food in terms of calories and nutrients [1]. A report issued by the food and agriculture organization (FAO) [2] stated that the daily average per capita calories available to the world population were 2789 kcal in the year 2000, but it is estimated that 3130 kcal will be required on a daily basis during the year 2050. In this context, grain legumes are key commodities having great potential to mitigate world food shortages and provide well balanced and cheap food [3].
The common bean (Phaseolus vulgaris L.) is one of the most important proteinaceous crops of Latin America. The common bean serves a source of food for more than 300 million people [4]. Although the common bean is an underrated crop, the presence of higher quantity of proteins, carbohydrates, vitamins, and minerals makes it a rational food of choice, particularly for poor people all around the world [5]. Global common bean production was 23 million tons in 2010 and 26 million tons in 2016 [6]. The common bean reflects a great level of genotypic and phenotypic diversity, and domestication of this crop resulted in two distinct gene pools—the Mesoamerican gene pool from Central America and the Andean gene pool from the Andes Mountains in South America [7]. Turkey is not the center of origin of the common bean, and it is known that this crop was introduced to Turkey from Europe in the 17th century [8]. However, hundreds of landraces, especially those having bigger seed size due to local consumer preference, are found in Turkey. The common bean is a very popular food commodity for Turkish people and is an important part of their daily diet [9]. An increase in the yield of the common bean has been observed in Turkey during the last two decades; 212,758 tons were produced in the year 2010, and 651,094 tons in the year 2016 [6], making up a share of about 2.75% of the world production. To date, 200 fresh bean and 39 dry bean cultivars have been registered in Turkey (Variety Registration and Seed Certification Center; www.tarimorman.gov.tr). According to Ozturk et al. [10], the observed gradual rise in common bean production is due to the development of improved cultivars reflecting better adaptation to environmental stresses.
Advancements in the molecular markers and sequencing technologies have boosted breeding activities [11]. Among major techniques, genome-wide association studies (GWAS) and quantitative trait loci (QTL) mapping are the two main approaches used to elucidate the genetic basis for traits of interest. QTL mapping or bi-parental mapping is mainly used for the investigation of QTLs associated with traits of interest. However, in addition to its advantages, QTL mapping also has several drawbacks, like low recombination, being time-consuming, and the population specificity of the identified QTLs [11]. GWAS overcomes all the drawbacks of QTL mapping, and the identified markers through this approach can be used for any population [12]. GWAS has been successfully carried out for the common bean in order to elucidate marker-trait association for agronomic traits [13], seed weight [14], disease resistance [15], cooking properties [16], anthracnose resistance [17], and drought tolerance [18]. These studies resulted in the identification of some candidate genes that can be helpful in the development of new genetic stocks for breeding perspectives in the common bean.
Antioxidants are an important class of chemicals present in our food and play an essential role in reducing the oxidative stress of the physiological system [19]. Antioxidants play a significant role against various disorders like cancer, aging, cardiovascular disease, cataracts, immune system decline, and brain dysfunction. Antioxidants minimize the chances of these diseases by preventing the production of free radicals [20]. Sudhir et al. [21] stated that higher antioxidants in food minimize the incidence of cardio and cerebrovascular diseases. Oxygen under various conditions may have serious deleterious effects on the body [22]. Oxygen is a highly reactive atom with the ability to become part of potentially damaging molecules known as “free radicals.” These free radicals have the ability to attack healthy body cells, leading to various diseases like cardiovascular disease, cancer, brain dysfunction, and a decline in the immune system [20,21,22,23]. Antioxidants can electrons to stabilize these free radicals and prevent their detrimental effects. Antioxidants protect the body from both endogenous and exogenous molecules [24]. Sikora et al. [25] stated that food containing a higher concentration of antioxidants can significantly affect the increase of the reactive antioxidant potential of the organism and decrease the risk of some diseases caused by the production of free radicals. Antioxidants can also be endogenous or exogenous; exogenous antioxidants enter the body when we eat antioxidant-rich food, while endogenous antioxidants are solely produced by the body. Exogenous antioxidants are found in plant and animal sources, and those of plant origin are of great interest because of their presence in relatively higher quantities [26,27].
The role of antioxidants against various stress conditions in plants has been well documented [28]. Plant species containing a higher level of antioxidant contents, either constitutive or induced, reflect a higher resistance to environmental stresses [29]. According to Silvana et al. [30], the extent of oxidative damage to plant cells when exposed to abiotic stresses is mainly governed by the capacity of their antioxidant defense system. As antioxidant activity is a key trait, its manipulation can be a relevant target for common bean breeding community to develop new varieties having improved antioxidant activity. This in turn will be helpful for improving human health. GWAS has been used previously for the identification of genes associated with antioxidant activity and total flavonoid contents in barley [31] and rice [32] in order to investigate polyphenol contents and antioxidant activity. Despite a good understanding of the role of antioxidants for human health, the genetic basis for antioxidant activity has not been elucidated for most of crops. The common bean is a good source of antioxidants [29], and Cardador-Martiänez et al. [33] stated that bean extract from the hull contains a significant level of antioxidants. Weidner et al. [34] explored the antioxidant levels in different colored common bean genotypes and observed a good amount of antioxidant capacity. The common bean serves as a source of food for a large population of the world, so variations in the common bean for antioxidant activities can be used to develop cultivars with better antioxidant content. Therefore, an effort is made through this study to explore the variations for antioxidant activity and antioxidant activity–associated markers in Turkish common bean germplasm.
2. Materials and Methods
2.1. Plant Material
A total of 182 common bean landraces and six commercial cultivars (Akman, Goynuk, Karacaşehir, Onceler, Goksun, and Akdag) were used as plant material in this study. Germplasm was collected from 19 geographical regions of Turkey (Table S1; Figure 1) and maintained at Bolu Abant Izzet Baysal University (BAIBU). Commercial cultivars used in this study were developed through a single plant selection and have been used as standard cultivars in previous studies [35,36].
Figure 1.
Collection provinces of Turkish common bean germplasm.
2.2. Field Experiment and Phenotypic Evaluation
Field experiments were conducted during 2017 and 2018 at two experimental sites—BAIBU (40°44′45″ N/31°37′44″ E, altitude; 752 m) and Cumhuriyet University Sivas (39°42′16.69″ K/37°01′57.48″ D, altitude; 1293 m). The experiment was laid out according to augmented block design, which has been found to be more effective, precise, time-saving, and trustworthy compared to other experimental designs [37]. Augmented block design contains more than one control cultivar that are taken as replicated treatments, and these cultivars are repeated in each block. These control cultivars are used to standardize the data to calculate adjusted means. Evaluated means are later used for various statistical analyses [37,38]. In this study, seeds were planted in elementary plots, each consisting of a 2 m row with a 50 cm inter-row and 10 cm intra-row spacing. Studied germplasm was sown according to augmented block design in eight blocks. Six commercial cultivars (Akman, Goynuk, Karacaşehir, Onceler, Goksun, and Akdag) were included in each block as a control group. Sowing was done on 27 April 2017 and 16 April 2018, respectivelyat the BAIBU, while the trial in Sivas location was sown on 15 May 2017 and 19 May 32018. Di-ammonium phosphate (DAP) (130 kg/ha) and ammonium sulfate (51 kg/ha) were used as a source of fertilizer, while standard agriculture practices were performed at both locations. Harvesting was done at the time of 90% pods per plant maturity.
2.3. DNA Extraction and Genotyping for DArTseq Markers
DNA was extracted from two-week-old seedlings according to CTAB protocol of Doyle and Doyle [39] with some modifications [40]. Agarose gel electrophoresis (0.8%) was used to check the purity of isolated DNA and quantification was performed through DS-11 FX series spectrophotometer/fluorometer (Denovix, Wilmington, DE, USA). High-quality DNA was further diluted and a final concentration of 50 ng μL-1 was prepared. For genotyping-by-sequencing (GBS), the prepared DNA samples were processed at Diversity Array Technology Pty, Ltd., Bruce, Australia (http://www.diversityarrays.com/). DArTseq analysis was carried out as previously described [9].
2.4. Extraction and Determination of Total Antioxidant Activity
Antioxidant activity was measured using ABTS radical scavenging capacity. For the investigation of antioxidant activity, TEAC extract was firstly prepared to calculate trolox equivalent antioxidant capacity (TEAC). For this purpose, dry samples were extracted with acetone, water, and acetic acid solution (70/29.5/0.5), respectively for ten days. For TEAC analysis, 7 mM ABTS (2,2′-Azino-bis 3-ethylbenzothiazoline-6-sulfonic acid) was mixed with 2.45 mM potassium sulfate for 12–16 h as suggested by Saracoglu et al. [41]. This solution was then mixed with 20 mM sodium acetate (pH 4.5) buffer solution. Finally, 30 μL of extract was mixed with 2.97 mL of prepared buffer and incubated for 10 min and measured on a spectrophotometer at a wavelength of 734 nm to 0.700 ± 0.01. Absorbance values obtained were calculated with Trolox (10–100 µmol/L) and presented as µmol TE/g.
2.5. Statistical Analysis
2.5.1. Phenotypic Data Analysis
An online software for augmented block design developed by Rathore et al. [42] was used to derive statistical inferences. The analysis was performed in two steps. First, analysis of variance (ANOVA) was investigated within the environments, and adjusted means were derived. Second, these evaluated means were used to run ANOVA across the environments, solving the appropriate mixed model equation to contemporarily account for both genotype and genotype × environment interaction effects. The Fligner–Killeen test was also computed before combined analysis to evaluate the homogeneity of variance. Environmental effects were taken as random, and genotypic effects were taken fixed in the computed mixed model equation, as suggested by Gomez and Gomez [43]. For the calculation of heritability, appropriate variance components were derived from the linear mixed model equation fitted attributing genotype and environment random effects as reported by Habyarimana [44]. The model was fitted with a restricted maximum likelihood using the R software [45]. The boxplot and frequency distribution of antioxidant activity were investigated using GenStat software [46]. Pearson’s correlation coefficient among various colored seeds and antioxidant activity was investigated using the statistical software XLSTAT (www.xlstat.com).
2.5.2. Population Structure and GWAS Analysis
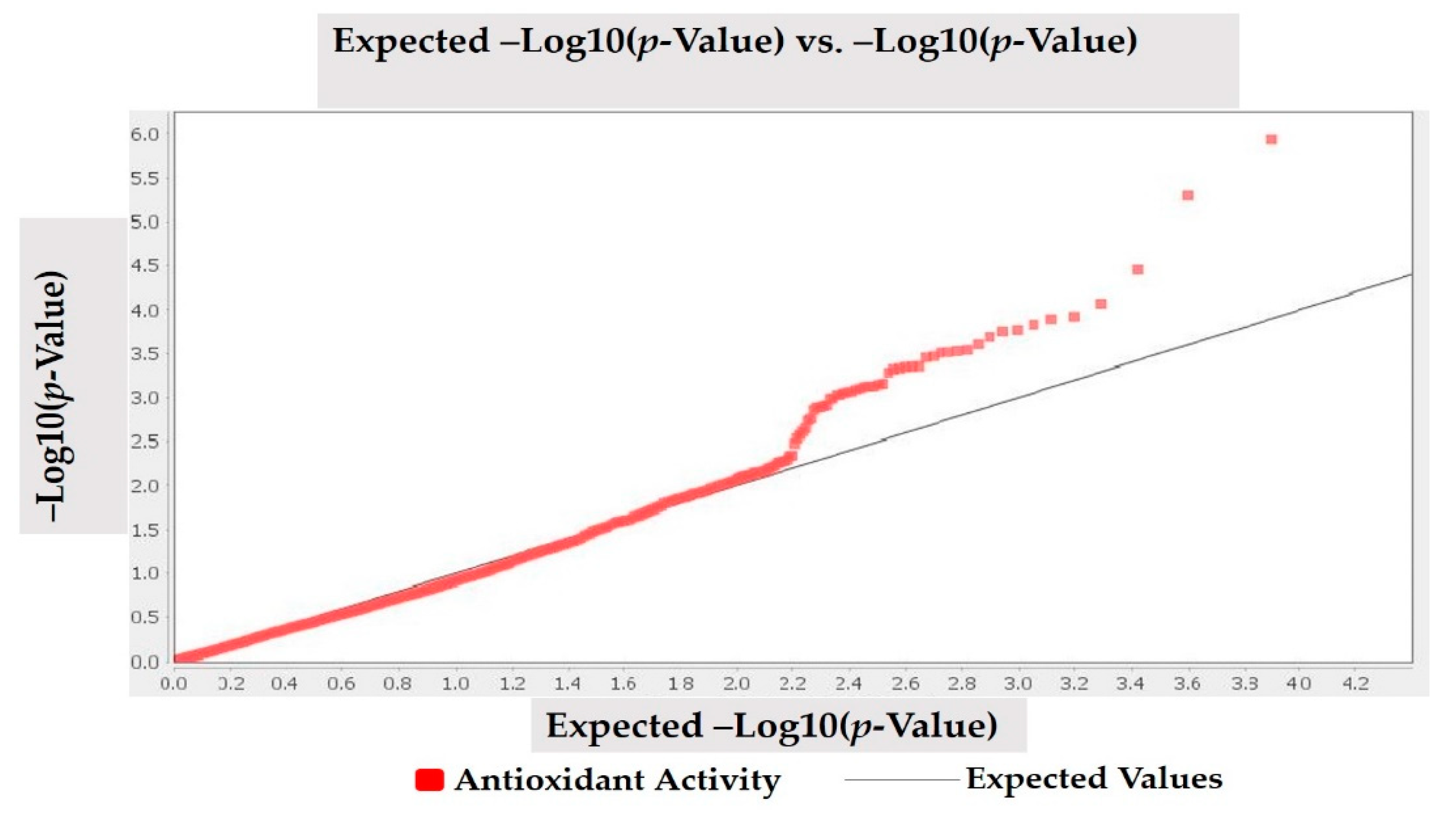
Population structure was carried as described by Nadeem et al. [9]. A mixed linear model (MLM, Q + K) approach was used to investigate the marker-trait association. Q-metrics (Q) and kinship (K) were used to correct the population and family structure during association analysis. The kinship matrix was investigated by following the methodology reported by Bradbury et al. [47] through TASSEL 5.0.5 (https://tassel.bitbucket.io) software. Linkage disequilibrium (LD) was estimated for all DArTseq markers with r2 based on allele frequencies [48,49]. Pairwise LD values of polymorphic sites were plotted on both the X- and Y-axis to generate LD heat map. To calculate LD decay, the pairwise correlations among linked markers in significant LD (p < 0.001) were used. The threshold for LD decay was considered below r2 = 0.1. Results of association analysis reflected that p value (marker) shows whether a marker is associated with a trait, and R2 (marker) defines the proportion of phenotypic variation accounted for by a significant marker [49,50]. Both Bonferroni and FDR thresholds were used, while DArTseq markers having FDR and Bonferroni p = 0.01 thresholds were taken significantly associated with antioxidant activity. Statistically significant markers for antioxidant activity were visualized through the Manhattan plot in R 3.4.1 statistical software (http://www.r-project.org/) by using qq-man R Package [51]. Quantile-Quantile (q-q) plot was drawn to visualized important p-value distributions (expected vs. observed p-values on a –log10 scale) through TASSEL 5.2.50 (https://tassel.bitbucket.io) software.
2.5.3. Putative Candidate Gene Analysis
Sequences of DArTseq markers associated with the antioxidant activity were used to BLAST-search against common bean genome in Phytozome V.12.1 (http://phytozome.jgi.doe.gov/pz/portal.html), the legume information system (LIS: https://legumeinfo.org/), and the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/) databases to detect putative genes homologous to these sequences.
3. Results
3.1. Phenotypic Diversity Evaluation
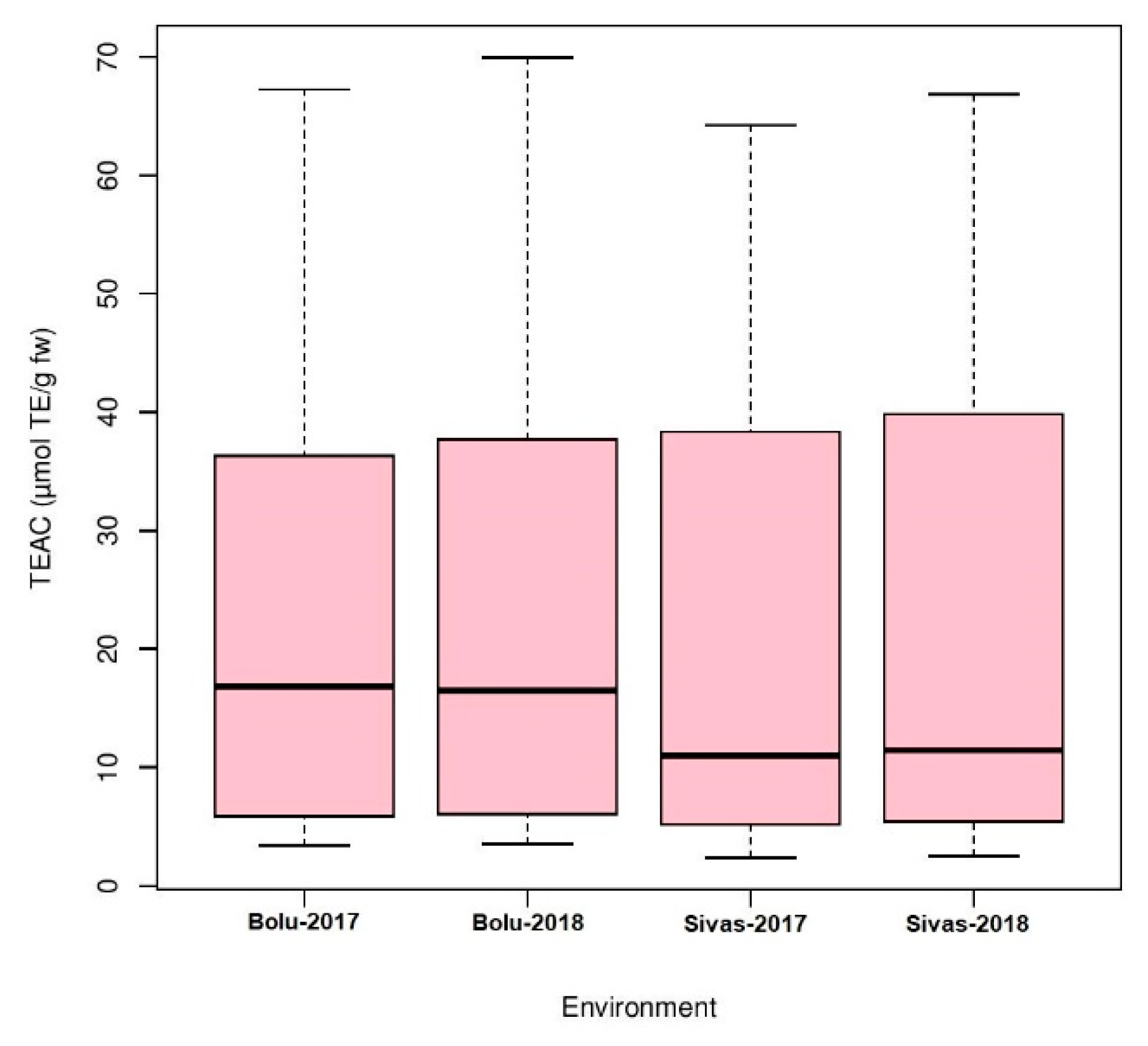
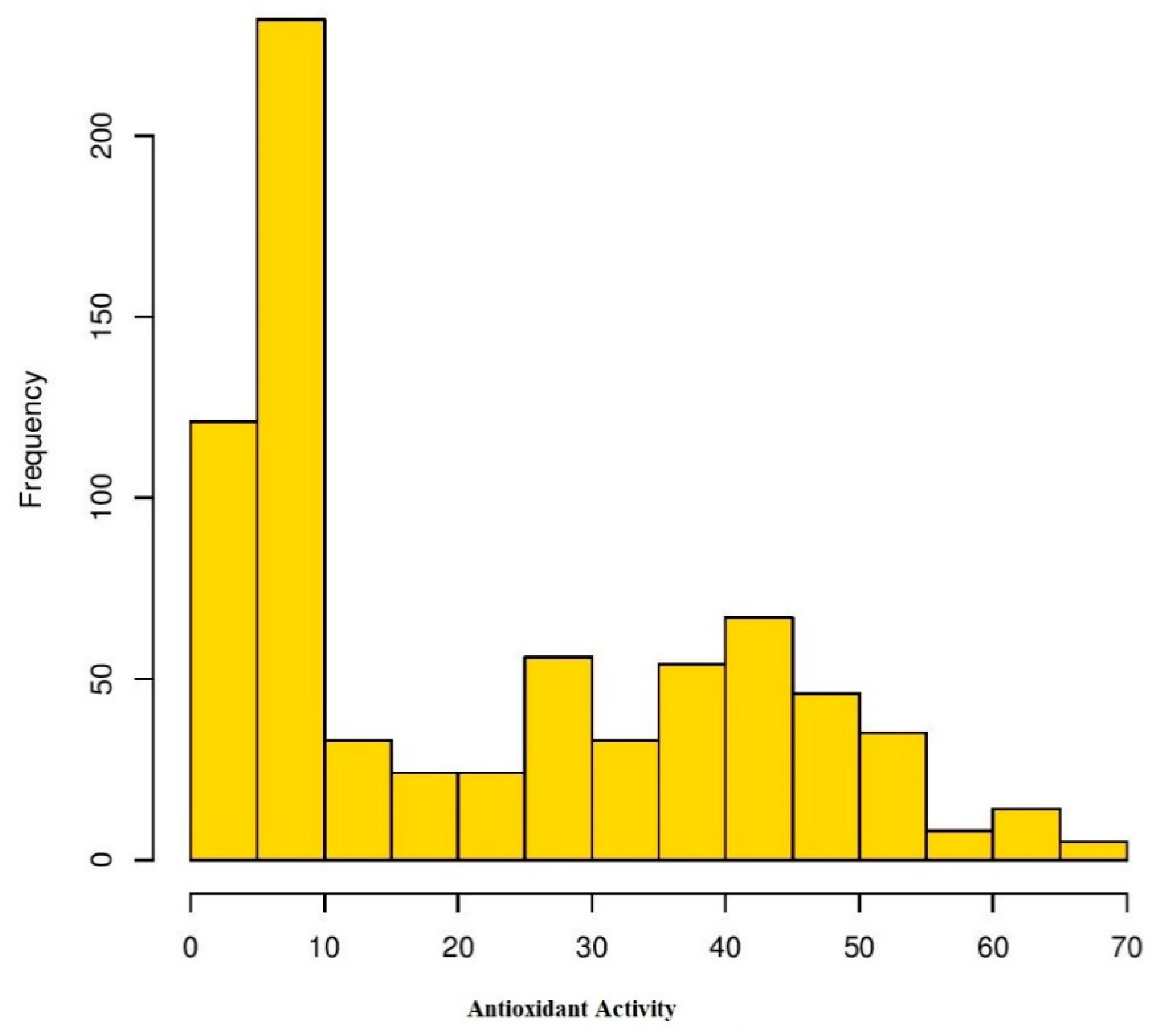
Analysis of variance (ANOVA) was performed for four environments to understand the effects of genotype, environment, and the interaction of both (GEI) for antioxidant activity in Turkish common bean germplasm (Table 1). Genotypic effects were found to be statistically significant at (p < 0.05; F value 11.9), while GEI was statistically non-significant for antioxidants activity in this study. Heritability analysis revealed the existence of higher heritability (0.92) (Table 1). Antioxidant activity was evaluated as TEAC for each environment for both locations (Table S2). At BAIBU in 2017, the mean TEAC for common bean landraces was 19.78 ranging 1.8 (Hakkari-65) to 67.2 (Balikesir-19) (Table S3). At the same location in 2018, 20.25 was the mean TEAC, while 1.9 (Hakkari-65) and 69.9 (Balikesir-19) were the minimum and maximum TEAC, respectively. At the Sivas location in 2017, mean TEAC was 19.65, while minimum and maximum TEAC reflected by Hakkari-65 and Bitlis-22 landraces were 2.4 and 64.3, respectively. Mean TEAC was 20.43 at Sivas location during 2018, while minimum and maximum TEAC reflected by Hakkari-65 and Bitlis-22 landraces were 2.5 and 66.8, respectively. Overall mean TEAC in Turkish common bean landraces was 20.03, ranging from 32.15 (Hakkari-65) to 60.48 (Balikesir-19). Mean TEAC in common bean cultivars at Bolu in 2017 and 2018 were 13.9 and 14.3, respectively (Table S3). Mean TEAC in common bean cultivars at Sivas in 2017 and 2018 were 15.7 and 16.3, respectively, while overall mean TEAC in common bean cultivars during the whole period of study was 15.1. Boxplot analysis also confirmed the results of ANOVA by revealing that there are no significant environmental effects on the seed antioxidant contents (Figure 2). Frequency distributions for TEAC in common bean are presented in Figure 3, which shows that most of the landraces have less than 10 µmol TE/g TEAC. Mean antioxidant activity in the colored seeds and white seeds were 30.04 and 16.22 µmol TE/g TEAC, respectively. Correlation analysis between antioxidant activity and seed caot colors was also performed, which revealed a negative correlation of antioxidant activity with white seeds (Table 2).

Table 1.
Summary of analysis of variance and estimates of heritability in Turkish common bean germplasm.
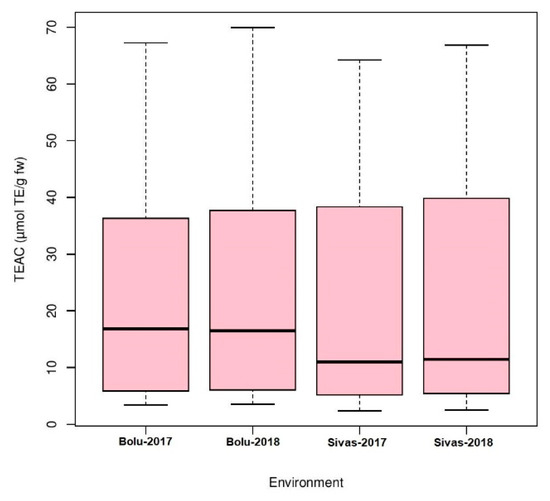
Figure 2.
Boxplot analysis of antioxidant activity in Turkish common bean germplasm under four environments.
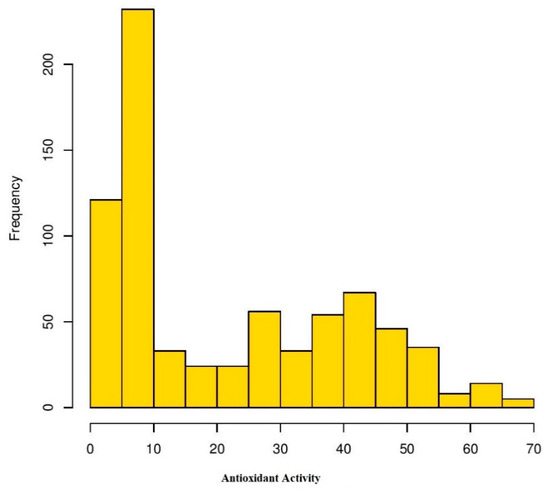
Figure 3.
Frequency distribution of antioxidant activity in Turkish common bean germplasm under four environments.

Table 2.
Pearson’s correlation coefficient between various seeds colors and antioxidant activity.
3.2. Population Structure and LD
Whole-genome DArTseq profiling of Turkish common bean germplasm resulted in a total of 15,608 DArTseq markers. This marker dataset was filtered to retain 7900 high-quality markers with less than 5% missing data, PIC value > 0.10, call rate > 0.60, and 100% reproducibility. The Bayesian clustering model implemented in STRUCTURE software grouped studied germplasm into two groups; population A (red) containing 117 landraces (59.57%) and population B (green) 71 landraces (37.76%) (Figure S1). The 7900 highly informative DArTseq markers were used for LD and GWAS analysis. Based on each chromosome, the number of mapped markers ranged from 931 (Chr. 2) to 591 (Chr. 6) (Table 3). The mean distance between markers on whole-genome was 15.54 Mb with a range of 13.15 (Chr 7) to 18.99 Mbps (Chr. 2). Mean r2 was 0.47, and 54% was the mean significant LD (Table 3). Mean LD decay for whole-genome was 1.15 Mbps which ranged from 1.05 Mbps for chromosome 9 to 1.3 for chromosome 5. Nonlinear trend line of LD measure r2 vs. the physical map distance decayed with almost 1.3 Mbs to r2 value of 0.5 (Figure S2). Linkage disequilibrium measured r2 plotted vs. p-value between pairs of DArTseq markers revealed LD decay on LG6 (Figure S3).

Table 3.
Distribution and linkage disequilibrium of DArTseq markers on different chromosome of common bean.
3.3. Marker Trait Association for Antioxidant Activity
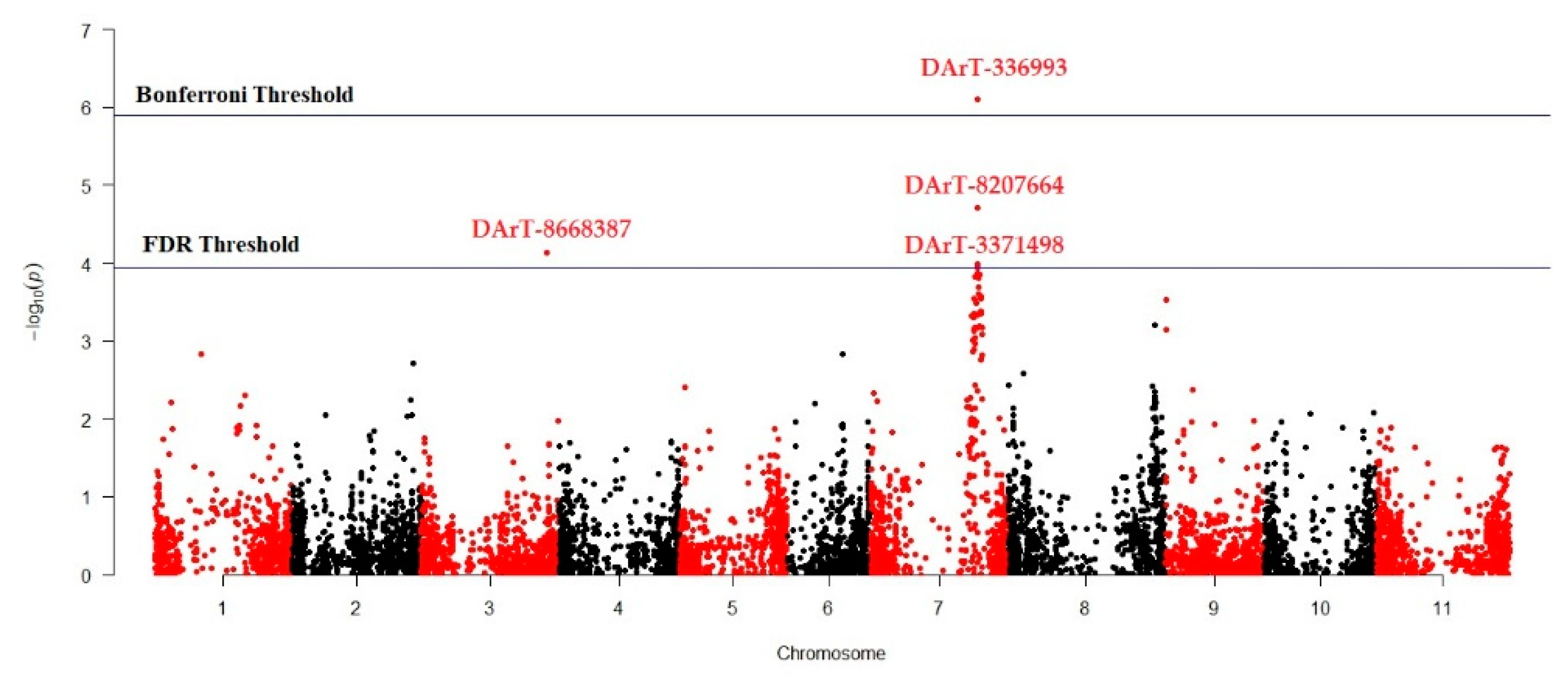
MLM (Q + K) model was applied to investigate marker-trait association for antioxidant activity in studied Turkish common bean germplasm. MLM based on Q + K model showed that out of 7900 DArTseq markers, only four markers (DArT-3369938, DArT-8668387, DArT-8207664, and DArT-3371498) reflected statistically significant association for antioxidant activity (Table 4; Figure 4). The phenotypic variation percentage (R2) reflected by each marker ranged between 0.094% to 0.156% for DArT-3371498 and DArT-3369938, respectively. The p-value of each marker reflecting significant level of association between marker and antioxidant activity is presented in the form of Manhattan plots (Figure 4). Manhattan plots showed that markers associated with antioxidant activity are present on chromosomes 3 and 7. Quantile-quantile (q-q) plot (Figure 5) reflected the goodness of fit of the best model by implementing p-value distributions (expected vs. observed p-values on a -log10 scale).

Table 4.
Significant marker-trait associations identified for antioxidant activity in Turkish common bean germplasm.
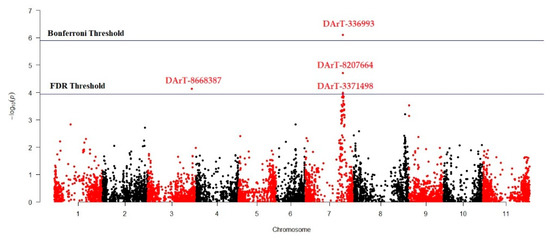
Figure 4.
Manhattan plot reflecting marker trait association for antioxidant activity in common bean germplasm.
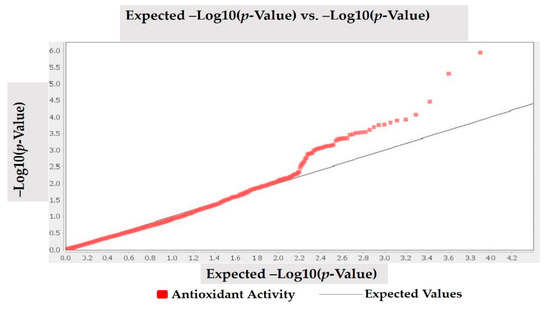
Figure 5.
Quantile-quantile (q-q) plots for antioxidant activity in Turkish common bean germplasm.
3.4. Putative Candidate Genes for Antioxidant Activity
Sequences of all four identified linked markers were used as queries to BLAST-search against Phytozome (V.12.1), LIS, and NCBI databases to detect predicted genes homologous to these sequences. A zinc-finger protein family gene (Phvul.009G180200.2) located on Ch Pv07 was retrieved in BLAST using DArT-3369938 marker as a querry (Table 4). The search for the homologous candidate gene against DArT-8207664 marker resulted in the retrieval of Phvul.007G281200.1 on chromosome Pv07. The gene encodes a methylene tetrahydrofolate reductase also known as the MTHFR gene, and folic acid production is predicted function of this gene. A search against the query marker DArT-8668387 resulted in the retrieval of Phvul.003G239700 located on chromosome Pv03. This gene encodes for a UDP-glycosyltransferase superfamily protein. Phvul.007G171700.1 was found against DArT-3371498. The gene located only 20 kb upstream of DArT-3371498 belongs to the Pentatricopeptide repeat family (PPR_2).
4. Discussion
In this study, a total of 182 Turkish common bean landraces and six commercial cultivars were used to identify marker trait association for antioxidant activity. Results of phenotypic characterization for antioxidant activity showed a great level of diversity ranging from 2.15 to 60.48 with the mean TEAC of 20.03 µmol TE/g (Table S3). The observed range and average TEAC in the studied germplasm was much higher than reported by Huber et al. [52], Weidner et al. [34], and Bojilov et al. [53]. Antioxidant activity during 2018 at both locations was recorded as higher compared to 2017. García-Díaz et al. [54] also observed higher antioxidant activity in common bean seeds in one year as compared to the other year of study. These differences in the antioxidant activity of common seeds grown in different years have been previously explained to be influenced by environmental variations, crop management practices, and genetic differences [55]. Frequency distribution reflected that a good number of landraces acquired TEAC contents similar to the results of earlier studies [34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53]. The ANOVA was performed within and across the four environments to evaluate the genotypic effect and explore the GEI for antioxidant capacity. The ANOVA confirmed that statistically significant variations in TEAC are due to the genetic background of landraces, while GEI reflected a non-significant effect for antioxidant capacity. These results were further strengthened by the results of heritability analysis. Higher heritability (0.92) in this study reflected that this trait is mainly under the control of genetic forces, while the environmental forces have very little effect. García-Díaz et al. [54] investigated the effect of cropping season and genotype on antioxidant activity in the common bean and revealed that variations are mainly due to genotypic effect. Di Silvestro et al. [56] investigated how genotype and environment effect antioxidant activity in wheat under multiyear experiments. A lesser effect of the environmental forces was observed in their results supports our observations of the common bean. A similar study on corn concluded that most of the variations for studied traits are contributed by the genotypes as compared to environmental forces [57].
We also observed variations in seed color and recorded that white color (58% of the accessions) was prevalent in our selected population. Seed color in the common bean is associated with bioactive compounds, such as anthocyanin and tannins which are found in higher quantity in colored seeds as compared to white colored seeds [58]. Earlier studies explored the relationship between the seed color and antioxidant activity in common bean and concluded the role of seed color in higher antioxidant activity [58,59]. In this study, mean antioxidant activity was found much higher in colored seeds as compared to the white ones. These results are in line with the previous studies stating the existence of higher antioxidant activity in colored seeds as compared to white seeds [58,59,60]. Pearson’s correlation coefficient showed a highly significant correlation between colored seeds (brown, beige) and antioxidant activity. On the other hand, there was a negative correlation between white seeds and antioxidant activity. Earlier studies revealed the presence of higher anthocyanin in the seed coat of the colored seed, which ultimately contributes toward a higher antioxidant activity [58,59].
4.1. DArTseq Markers Analysis and LD
A total of 7900 markers were employed in this study to investigate population structure and marker-trait association for antioxidant activity. The number of mapped markers ranged from 931 (Chr. 2) to 591 (Chr. 6) (Table 3). The mean distance between markers on the whole nuclear genome was 15.54 Mb with a range of 13.15 (Chr. 7) to 18.99 Mbps (Chr. 2). The number of markers used in this study was much higher than earlier studies used for GWAS in various plant [61,62], which in turn leads toward a more efficient genome-wide screening. Using marker information, the structure algorithm divided whole germplasm into two heterotic populations on the basis of their 100-seeds weight, TEAC contents, and collection point. Population A clustered a total 117 landraces, while 71 landraces clustered in population B. Akdag, Onceler, and Goynuk cultivars grouped in population A and Akman, Goksun, and Karacasehir reflected much similarity with population B. Landraces clustered in population A mainly contained higher 100-seed weight and relatively more TEAC contents, while landraces having lower to moderate TEAC contents with lower 100-seed weight clustered into population B. Geographic locations also played a role in the clustering and landraces from same provinces were grouped together. Earlier studies have confirmed the clustering of common bean germplasm based on plant height, 100-seed weight, and collection point. These previous reports strengthen the findings of our study [63].
Linkage disequilibrium is a nonrandom association of alleles at particular loci in a sampled population genome [64]. In this study, mean r2 was 0.47 while the mean significant LD was 54% (Table 3). The maximum and minimum LD was 59.8% and 45.89% reflected by chromosomes 11 and 9, respectively. Mean LD decay was 1.15 Mb for the whole genome and ranged from 1.05 Mbps for chromosome 7 to 1.3 Mbps for chromosome 5. A nonlinear trend line between r2 vs. distance bp showed that LD decayed at 1.3 Mbs and r2 value was 0.5 (Figure S2). When an LD decay plot is constructed, usually the distance point is looked for where the LD value (r2) decreases below 0.1 or half of D’ strength (D’ = 0.5) based on curve of nonlinear logarithmic trend line [65,66]. This shows a rough estimation of extent of LD, however, highly significant threshold LD values (r2 ≥ 0.2) are used for more precise LD calculation. Our results are in a disagreement with the findings of Campa et al. [67], who stated that chromosome 9 shows maximum LD. Linkage disequilibrium measured r2 plotted vs. p-value between pairs of DArTseq markers the studied germplasm showed a relatively bigger LD decay on LG6 (Figure S3). Mean r2 found in this study was slightly lower as compared to the findings by Ates et al. [68]. The higher LD in their study may be due to human selection, which leads in particular usage, combining distinct variations through a long history of cultivation and multiple nationwide expeditions.
4.2. Marker Trait Association and Putative Candidate Genes for Antioxidant Activity
We used GWAS for the whole-genome screening of the selected germplasm. This approach has rapidly gained the attention of breeders to identify genomic regions for various traits of interest that can be used for marker-assisted breeding [69]. For the investigation of marker–trait association, the MLM (Q + K) approach was used during GWAS analysis to eliminate possible spurious associations [68]. In this study a total of four markers (DArT-3369938, DArT-8668387, DArT-8207664, and DArT-3371498) were significantly associated with seed antioxidant contents (Table 4; Figure 4). DArT-3371498 accounted for 15.6% of the total variation and was the most statistically significant marker, while DArT-3371498 accounted for the least variation among these four identified markers. The Manhattan plot showed that DArT-3369938, DArT-8207664, and DArT-3371498 are located on chromosome 7, while DArT-8668387 reflected association with chromosome 3. As these markers reflected significant association for antioxidant activity in the common bean, these markers can be used for marker-assisted breeding to develop cultivars with better antioxidant activity.
Located only 100 kb upstream of DArT-3369938, on Pv09, Phvul.009G180200.2 is putative candidate gene to explain the phenotypic variation associated with this marker. This gene encodes for zinc-finger domain–containing protein. Zinc-finger proteins (ZFPs) constitute large protein families and have an important role in various plant developmental stages and to biotic and abiotic stress [70]. Hichri et al. [71] stated that this protein family contains transcription factors that control various aspects of plant development and shown a pivotal role in abiotic stress tolerance. Cao et al. [72] found that ZFPs have resistant to biotic stress such as rice blast fungus infection. Cheuk and Houde [73] explored the role of ZFPs in abiotic stress and investigated 53 Q-type C2H2 zinc-finger proteins (TaZFPs) from Triticum aestivum, and stated that these TaZFPs have positive response to high light (44/53), H2O2 (37/53), drought (37/53), and flooding (31/53). Zang et al. [74] confirmed that this protein is involved in abiotic stress by investigating abiotic stress in Arabidopsis and concluded that ZFPs increase the salt and osmotic tolerance in this plant through a series of physiological processes. Zhang et al. [75] identified ZFPs in rice and concluded that overexpression of ZFPs in their study elevated the activities of antioxidant enzymes and enhanced the tolerance of rice plants against water and oxidative stresses.
The BLAST search against DArT-8668387 retrieved a gene (Phvul.003G239700) encoding UDP-Glycosyltransferase superfamily protein; a multifunctional protein family in plants. Glycosylation is one of the final steps involved in the biosynthesis of triterpenoid for the production of various plant defensive compounds like phenolic, glucosinolates, salicylates, and anthocyanins [76]. UDP-glycosyltransferases (UGTs) are one of the biggest gene families in the plant kingdom and play an important role in transferring sugar, controlling various metabolic processes, and play an effective role against various biotic and abiotic stresses [77,78]. Various studies have explored the biological role of the putative UGTs against different abiotic stresses. Li et al. [77] comprehensively explained the role and functioning of a homolog of this protein in enhanced plant tolerance to low temperatures, drought, and salt stresses in Arabidopsis. Sun et al. [79] found that ectopic expression of UGT85A5 in tobacco resulted in enhanced salt stress tolerance. Rehman et al. [78] performed a comparative genomic and transcriptomic analyses study for brassica species and Arabidopsis and found the role of UDP-Glycosyltransferase in biotic and abiotic stresses.
Phvul.007G281200.1 resulted as the putative candidate gene for DArT-8207664 marker that encodes for the methylene tetrahydrofolate reductase MTHFR gene. MTHFR enzyme is involved in the metabolism of folate and homocysteine (Hcy). This gene is linked with folate metabolism which plays a positive effects on oxidative stress by its protecting action against cell death and increased folic concentrations stabilize MTHFR enzyme [80]. Earlier studies have found the synergetic effect of folic acid on growth, yield, and yield quality of many plant species. Folic acid can play an effective role by catching the free radicals or active oxygen produced during the stress conditions and help the plant defense against environmental stresses [81]. Phvul.007G171700.1 encodes for PPR containing plant protein (RNA-binding proteins) having a key role in post-transcription via RNA editing, cleavage, splicing, stability, or translation in plastids or mitochondria [82]. Earlier studies revealed that PPR proteins might be target genes of some miRNAs regulating abiotic stress responses [83]. Zsigmond et al. [84] revealed that PPR proteins are involved in oxidative respiration that contributes to abiotic stress tolerance in Arabidopsis. Recently, Chen et al. [85] aimed to investigate the functioning of PPR gene family in rice and explored the role of this family under stress condition. They investigated the expression pattern of this gene family and concluded that PPR proteins have crucial roles in response to different abiotic stresses in rice.
As is obvious from the previously discussed evidence, our diverse association panel reflected a wide range of antioxidant activity in common bean seeds. These genetic variations can be utilized for various common bean improvement programs. The role of antioxidants in human health has been universally accepted, and earlier studies revealed that higher antioxidant levels in food can prevent various disorders [20,21,22]. Germplasm characterization is an important way to elucidate novel variations which can be used for the breeding perspective of any crop [86,87,88]. Therefore, there is a need to identify the genetic basis for association with antioxidant activity. The common bean serves as a source of food for millions of people all over the world, and thus, the intake of beans with higher antioxidant levels will reduce the risk of various disorders. We identified four DArTseq markers (DArT-3369938, DArT-8668387, DArT-8207664, and DArT-3371498) with significant association for antioxidant activity in Turkish common bean germplasm. These DArTseq marker can be used for the marker-assisted breeding of the common bean in order to develop cultivars having improved antioxidant activity.
5. Conclusions
A wide range of phenotypic variations were observed for antioxidant activity in Turkish common bean germplasm. Heritability analysis revealed that antioxidant activity is controlled by genotypic effect. A negative correlation was observed between antioxidant activity and white seeds. This is a pioneer study revealing the marker trait association for antioxidant activity in Turkish common bean seeds. A total of four DArTseq markers showed statistically significant association for seed antioxidant activity, and these markers can be used in the future for the development of common bean cultivars with better antioxidant contents.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/1/36/s1, Figure S1: Population structure of Turkish common bean germplasm revealed by DArTseq markers, Figure S2: Scatter plot reflecting association between linkage disequilibrium (r2) and distance bp, Figure S3: Linkage disequilibrium (LD) measured r2 plotted vs. P-value between pairs of DArTseq markers in Turkish common bean germplasm Table S1: Passport data of Turkish common bean germplasm used in this study, Table S2: Antioxidant activity (µmol TE/g fw) in four environments for Turkish common bean germplasm, Table S3: Comparison of mean, maximum and minimum for antioxidant activity (µmol TE/g fw) between Turkish common bean landraces and commercial cultivars under four environments.
Author Contributions
Conceptualization, F.S.B.; methodology, F.S.B. and S.E.; software, M.A.N. (Muhammad Azhar Nadeem), E.H. and X.L.; validation, T.K. and M.A.N. (Muhammad Azhar Nadeem); formal analysis, M.A.N. (Muhammad Azhar Nadeem), E.H., X.L., and M.S.; investigation, T.K. and M.G.; resources, F.S.B., G.C., and B.-M.J.; data curation, M.A.N. (Muhammad Azhar Nadeem), O.S., and T.K.; writing—original draft preparation, M.A.N. (Muhammad Azhar Nadeem); writing—review and editing, M.A.N. (Muhammad Amjad Nawaz), R.H., F.A., and S.E.; visualization, F.S.B.; supervision, F.S.B.; project administration, F.S.B; funding acquisition, F.S.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Scientific and Technological Research Council of Turkey TUBİTAK under research grant number (TOVAG-2015O630 to FSB).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Long, S.P.; Marshall-Colon, A.; Zhu, X.G. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 2015, 161, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization. 2012. Available online: http://www.faostat.fao.org/site/339/default.aspx> (accessed on 5 September 2018).
- Jensen, E.S.; Peoples, M.B.; Boddey, R.M.; Gresshoff, P.M.; Hauggaard-Nielsen, H.; Alves, B.J.; Morrison, M.J. Legumes for mitigation of climate change and the provision of feedstock for biofuels and biorefineries. A review. Agron. Sustain. Dev. 2012, 32, 329–364. [Google Scholar] [CrossRef]
- Petry, N.; Boy, E.; Wirth, J.; Hurrell, R. The potential of the common bean (Phaseolus vulgaris) as a vehicle for iron biofortification. Nutrients 2015, 7, 1144–1173. [Google Scholar] [CrossRef] [PubMed]
- Celmeli, T.; Sari, H.; Canci, H.; Sari, D.; Adak, A.; Eker, T.; Toker, C. The nutritional content of common bean (Phaseolus vulgaris L.) landraces in comparison to modern varieties. Agronomy 2018, 8, 166. [Google Scholar] [CrossRef]
- Food and Agriculture Organization. FAO Statistical Yearbook. 2016. Available online: http://www.fao.org/docrep/017/i3138e/i3138e.pdf (accessed on 5 September 2018).
- Bitocchi, E.; Bellucci, E.; Giardini, A.; Rau, D.; Rodriguez, M.; Biagetti, E.; Santilocchi, R.; Spagnoletti Zeuli, P.; Gioia, T.; Logozzo, G.; et al. Molecular analysis of the parallel domestication of the common bean (P haseolus vulgaris) in Mesoamerica and the Andes. New Phytol. 2013, 197, 300–313. [Google Scholar] [CrossRef]
- Bozoglu, H.; Sozen, O. A sample for biodiversity in Turkey: Common bean (Phaseolus vulgaris L.) landraces from Artvin. Afr. J. Biotechnol. 2011, 10, 13789–13796. [Google Scholar] [CrossRef]
- Nadeem, M.A.; Habyarimana, E.; Çiftçi, V.; Nawaz, M.A.; Karaköy, T.; Comertpay, G.; Shahid, M.Q.; Hatipoğlu, R.; Yeken, M.Z.; Ali, F.; et al. Characterization of genetic diversity in Turkish common bean gene pool using phenotypic and whole-genome DArTseq-generated silicoDArT marker information. PLoS ONE 2018, 13, 0205363. [Google Scholar] [CrossRef]
- Ozturk, I.; Kara, M.; Yildiz, C.; Ercisli, S. Physico-mechanical seed properties of the common Turkish bean (Phaseolus vulgaris) cultivars ‘Hinis’ and ‘Ispir’. N. Z. J. Crop Hort. Sci. 2009, 37, 41–50. [Google Scholar] [CrossRef]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotec. Eq. 2018, 32, 261–285. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef]
- Moghaddam, S.M.; Mamidi, S.; Osorno, J.M.; Lee, R.; Brick, M.; Kelly, J.; Miklas, P.; Urrea, C.; Song, Q.; Cregan, P.; et al. Genome-wide association study identifies candidate loci underlying agronomic traits in a Middle American diversity panel of common bean. Plant Genome 2016, 9, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Hofmann, N.; Li, S.; Ferreira, M.E.; Song, B.; Jiang, G.; Ren, S.; Quigley, C.; Fickus, E.; Cregan, P.; et al. Identification of QTL with large effect on seed weight in a selective population of soybean with genome-wide association and fixation index analyses. BMC Genom. 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed]
- Perseguini, J.M.; Oblessuc, P.R.; Rosa, J.R.; Gomes, K.A.; Chiorato, A.F.; Carbonell, S.A.; Garcia, A.A.; Vianello, R.P.; Benchimol-Reis, L.L. Genome-wide association studies of anthracnose and angular leaf spot resistance in common bean (Phaseolus vulgaris L.). PLoS ONE 2016, 11, e0150506. [Google Scholar] [CrossRef] [PubMed]
- Cichy, K.A.; Wiesinger, J.A.; Mendoza, F.A. Genetic diversity and genome-wide association analysis of cooking time in dry bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2015, 128, 1555–1567. [Google Scholar] [CrossRef] [PubMed]
- Fritsche-Neto, R.; Souza, T.L.; Pereira, H.S.; Faria, L.C.; Melo, L.C.; Novaes, E.; Brum, I.J.; Jannink, J.L. Association mapping in common bean revealed regions associated with Anthracnose and Angular Leaf Spot resistance. Sci. Agric. 2019, 76, 321–327. [Google Scholar] [CrossRef]
- Hoyos-Villegas, V.; Song, Q.; Kelly, J.D. Genome-wide association analysis for drought tolerance and associated traits in common bean. Plant Genome 2016, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Mamta, M.K.; Dhillon, G.S.; Brar, S.K.; Verma, M. Antioxidants. In Biotransformation of Waste Biomass into High Value Biochemicals; Brar, S., Dhillon, G., Soccol, C., Eds.; Springer: New York, NY, USA, 2014; pp. 117–138. [Google Scholar]
- Pooja, V.; Sunita, M. Antioxidants and disease prevention. Int. J. Adv. Sci. Tech. Res. 2014, 2, 903–911. [Google Scholar]
- Sudhir, S.; Budhiraja, R.D.; Miglani, G.P.; Arora, B.; Gupta, L.C.; Garg, K.N. Pharmacological studies on leaves of Withania somnifera. Planta Med. 1986, 52, 61–63. [Google Scholar] [CrossRef]
- Hajhashemi, V.; Vaseghi, G.; Pourfarzam, M.; Abdollahi, A. Are antioxidants helpful for disease prevention? Res. Pharm. Sci. 2010, 5, 1–8. [Google Scholar]
- Halliwell, B. Biochemistry of oxidative stress. Biochem. Soc. Trans. 2007, 35, 1147–1150. [Google Scholar] [CrossRef]
- Uttara, B.; Singh, A.V.; Zamboni, P.; Mahajan, R.T. Oxidative Stress and Neurodegenerative Diseases: A Review of Upstream and Downstream Antioxidant Therapeutic Options. Curr. Neuropharmacol. 2009, 7, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Sikora, E.; Cieślik, E.; Topolska, K. The sources of natural antioxidants. Acta Sci. Pol. Technol. Aliment. 2008, 7, 5–17. [Google Scholar]
- Carlsen, M.H.; Halvorsen, B.L.; Holte, K.; Bøhn, S.K.; Dragland, S.; Sampson, L.; Willey, C.; Senoo, H.; Umezono, Y.; Sanada, C.; et al. The total antioxidant content of more than 3100 foods, beverages, spices, herbs and supplements used worldwide. Nutr. J. 2010, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- La Vecchia, C.; Altieri, A.; Tavani, A. Vegetables, fruit, antioxidants and cancer: A review of Italian studies. Eur. J. Nutr. 2001, 40, 261–267. [Google Scholar] [CrossRef]
- Kusvuran, S.; Kiran, S.; Ellialtioglu, S.S. Antioxidant enzyme activities and abiotic stress tolerance relationship in vegetable crops. In Abiotic and Biotic Stress in Plants-Recent Advances and Future Perspectives, 1st ed.; InTech: London, UK, 2016; pp. 481–503. [Google Scholar] [CrossRef]
- Dolatabadian, A.; Jouneghani, R.S. Impact of exogenous ascorbic acid on antioxidant activity and some physiological traits of common bean subjected to salinity stress. Not. Bot. Horti Agrobot. Cluj Napoca. 2009, 37, 165–172. [Google Scholar] [CrossRef]
- Silvana, B.D.; Susanna, M.G.; María, P.B.; María, L.T. Behaviour of antioxidant defense system in the adaptive response to salt stress in Helianthus annuus L. cells. Plant Growth Regul. 2003, 40, 81–88. [Google Scholar] [CrossRef]
- Han, Z.; Zhang, J.; Cai, S.; Chen, X.; Quan, X.; Zhang, G. Association mapping for total polyphenol content, total flavonoid content and antioxidant activity in barley. BMC Genom. 2018, 19, 81. [Google Scholar] [CrossRef]
- Xu, F.; Bao, J.; Kim, T.S.; Park, Y.J. Genome-wide association mapping of polyphenol contents and antioxidant capacity in whole-grain rice. J. Agric. Food Chem. 2016, 64, 4695–4703. [Google Scholar] [CrossRef]
- Cardador-Martinez, A.; Castano-Tostado, E.; Loarca-Pina, G. Antimutagenic activity of natural phenolic compounds present in the common bean (Phaseolus vulgaris) against aflatoxin B 1. Food Addit. Contam. 2002, 19, 62–69. [Google Scholar] [CrossRef]
- Weidner, S.; Król, A.; Karamać, M.; Amarowicz, R. Phenolic compounds and the antioxidant properties in seeds of green-and yellow-podded bean (Phaseolus vulgaris L.) varieties. CyTA J. Food 2018, 16, 373–380. [Google Scholar] [CrossRef]
- Khaidizar, M.I.; Haliloglu, K.; Elkoca, E.; Aydin, M.; Kantar, F. Genetic diversity of common bean (Phaseolus vulgaris L.) landraces grown in northeast Anatolia of Turkey assessed with simple sequence repeat markers. Turk. J. Field Crops. 2012, 17, 145–150. [Google Scholar]
- Ceylan, A.; Öcal, N.; Akbulut, M. Genetic diversity among the Turkish common bean cultivars (Phaseolus vulgaris L.) as assessed by SRAP, POGP and cpSSR markers. Biochem. Syst. Ecol. 2014, 54, 219–229. [Google Scholar] [CrossRef]
- Federer, W.T.; Reynolds, M.; Crossa, J. Combining results from augmented designs over sites. Agron. J. 2001, 93, 389–395. [Google Scholar] [CrossRef]
- Boyle, C.R.; Montgomery, R.D. An application of the augmented randomized complete block design to poultry research. Poult. Sci. 1996, 75, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Baloch, F.S.; Alsaleh, A.; Andeden, E.E.; Hatipoğlu, R.; Nachit, M.; Özkan, H. High levels of segregation distortion in the molecular linkage map of bread wheat representing the West Asia and North Africa region. Turk. J. Agric. For. 2016, 40, 352–364. [Google Scholar] [CrossRef]
- Saracoglu, O.; Ozturk, B.; Yildiz, K.; Kucuker, E. Pre-harvest methyl jasmonate treatments delayed ripening and improved quality of sweet cherry fruits. Sci. Hort. 2017, 226, 19–23. [Google Scholar] [CrossRef]
- Rathore, A.; Parsad, R.; Gupta, V.K. Computer aided construction and analysis of augmented designs. J. Indian Soc. Agric. Stat. 2004, 57, 320–344. [Google Scholar]
- Gomez, K.A.; Gomez, A.A. Statistical Procedures for Agricultural Research, 2nd ed.; John Wiley & Sons Inc.: New York, NY, USA, 1984. [Google Scholar]
- Habyarimana, E. Genomic prediction for yield improvement and safeguarding of genetic diversity in CIMMYT spring wheat (Triticum aestivum L.). Aust. J. Crop. Sci. 2016, 10, 127–136. [Google Scholar]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; ISBN 3-900051-07-0. Available online: http://www.R-project.org/ (accessed on 15 April 2019).
- Payne, R.W.; Harding, S.A.; Murray, D.A.; Soutar, D.M.; Baird, D.B.; Glaser, A.I.; Welham, S.J.; Gilmour, A.R.; Thompson, R.; Webster, R. The Guide to Gen Stat Release 14, Part 2: Statistics; VSN International: Hemel Hempstead, UK, 2011. [Google Scholar]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Achenbach, U.; Paulo, J.; Ilarionova, E.; Lübeck, J.; Strahwald, J.; Tacke, E.; Hofferbert, H.R.; Gebhardt, C. Using SNP markers to dissect linkage disequilibrium at a major quantitative trait locus for resistance to the potato cyst nematode Globodera pallida on potato chromosome V. Theor. Appl. Genet. 2009, 118, 619–629. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hill, W.G.; Robertson, A. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 1968, 38, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Birlea, S.A.; Fain, P.R.; Gowan, K.; Riccardi, S.L.; Holland, P.J.; Bennett, D.C.; Herbstman, D.M.; Wallace, M.R.; McCormack, W.T.; et al. Genome-wide analysis identifies a quantitative trait locus in the MHC class II region associated with generalized vitiligo age of onset. J. Investig. Dermatol. 2011, 131, 1308–1312. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.D. qqman: An R package for visualizing GWAS results using QQ and manhattan plots. Biorxiv 2014, 005165. [Google Scholar] [CrossRef]
- Huber, K.; Brigide, P.; Bretas, E.B.; Canniatti-Brazaca, S.G. Phenolic Acid, Flavonoids and Antioxidant Activity of Common Beans (Phaseolus vulgaris L.) Before and After Cooking. J. Nutr. Food Sci. 2016, 6, 551. [Google Scholar] [CrossRef]
- Bojilov, D.G.; Manolov, S.P.; Bozadzhiev, B.V.; Stremski, J.I.; Ivanov, I.I. Antioxidant activity in two species common beans (Phaseolus vulgaris L.) from village of smilyan, Bulgaria. Agric. Food 2018, 6, 314–324. [Google Scholar]
- García-Díaz, Y.D.; Aquino-Bolaños, E.N.; Chávez-Servia, J.L.; Vera-Guzmán, A.M.; Carrillo-Rodríguez, J.C. Bioactive compounds and antioxidant activity in the common bean are influenced by cropping season and genotype. Chil. J. Agric. Res. 2018, 78, 255–265. [Google Scholar] [CrossRef]
- Ullah, F.; Alam, A.; Yamamoto, S.; Kahn, N.; Honna, T. Screening for agronomic performance of six indigenous cultivars of alfalfa (Medicago sativa) at Karina northern areas of Pakistan. Electron. J. Environ. Agric. Food Chem. 2009, 8, 950–968. [Google Scholar]
- Di Silvestro, R.; Di Loreto, A.; Bosi, S.; Bregola, V.; Marotti, I.; Benedettelli, S.; Segura-Carretero, A.; Dinelli, G. Environment and genotype effects on antioxidant properties of organically grown wheat varieties: A 3-year study. J. Sci. Food Agric. 2017, 97, 641–649. [Google Scholar] [CrossRef]
- Khamphasan, P.; Lomthaisong, K.; Harakotr, B.; Ketthaisong, D.; Scott, M.; Lertrat, K.; Suriharn, B. Genotypic variation in anthocyanins, phenolic compounds, and antioxidant activity in cob and husk of purple field corn. Agronomy 2018, 8, 271. [Google Scholar] [CrossRef]
- Boateng, J.; Verghese, M.; Walker, L.T.; Ogutu, S. Effect of processing on antioxidant contents in selected dry beans (Phaseolus spp. L.). LWT-Food Sci. Technol. 2008, 41, 1541–1547. [Google Scholar] [CrossRef]
- Ranilla, L.G.; Genovese, M.I.; Lajolo, F.M. Polyphenols and antioxidant capacity of seed coat and cotyledon from Brazilian and Peruvian bean cultivars (Phaseolus vulgaris L.). J. Agric. Food Chem. 2007, 55, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Capistrán-Carabarin, A.; Aquino-Bolaños, E.N.; García-Díaz, Y.D.; Chávez-Servia, J.L.; Vera-Guzmán, A.M.; Carrillo-Rodríguez, J.C. Complementarity in Phenolic Compounds and the Antioxidant Activities of Phaseolus coccineus L. and P. vulgaris L. Landraces. Foods 2019, 8, 295. [Google Scholar] [CrossRef] [PubMed]
- Briñez, B.; Blair, M.W.; Kilian, A.; Carbonell, S.A.; Chiorato, A.F.; Rubiano, L.B. A whole genome DArT assay to assess germplasm collection diversity in common beans. Mol. Breed. 2012, 30, 181–193. [Google Scholar] [CrossRef]
- Mogga, M.; Sibiya, J.; Shimelis, H.; Lamo, J.; Yao, N. Diversity analysis and genome-wide association studies of grain shape and eating quality traits in rice (Oryza sativa L.) using DArT markers. PloS ONE 2018, 13, e0198012. [Google Scholar] [CrossRef]
- Aydin, M.F.; Baloch, F.S. Exploring the genetic diversity and population structure of Turkish common bean germplasm by the iPBS-retrotransposons markers. Legum. Res. 2019, 42, 18–24. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Thornsberry, J.M.; Buckler, E.S., 4th. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef]
- Hamblin, M.T.; Mitchell, S.E.; White, G.M.; Gallego, J.; Kukatla, R.; Wing, R.A.; Paterson, A.H.; Kresovich, S. Comparative population genetics of the panicoid grasses: Sequence polymorphism, linkage disequilibrium and selection in a diverse sample of Sorghum bicolor. Genetics 2004, 167, 471–483. [Google Scholar] [CrossRef]
- Remington, D.L.; Thornsberry, J.M.; Matsuoka, Y.; Wilson, L.M.; Whitt, S.R.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 2001, 98, 11479–11484. [Google Scholar] [CrossRef]
- Campa, A.; Murube, E.; Ferreira, J. Genetic diversity, population structure, and linkage disequilibrium in a spanish common bean diversity panel revealed through genotyping-by-sequencing. Genes 2018, 9, 518. [Google Scholar] [CrossRef]
- Ates, D.; Asciogul, T.K.; Nemli, S.; Erdogmus, S.; Esiyok, D.; Tanyolac, M.B. Association mapping of days to flowering in common bean (Phaseolus vulgaris L.) revealed by DArT markers. Mol. Breed. 2018, 38, 113–127. [Google Scholar] [CrossRef]
- Wang, H.; Xu, X.; Vieira, F.G.; Xiao, Y.; Li, Z.; Wang, J.; Nielsen, R.; Chu, C. The power of inbreeding: NGS-based GWAS of rice reveals convergent evolution during rice domestication. Mol. Plant 2016, 9, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Sun, J.; Gong, D.; Kong, Y. The Roles of Arabidopsis C1-2i Subclass of C2H2-type Zinc-Finger Transcription Factors. Genes 2019, 10, 653. [Google Scholar] [CrossRef] [PubMed]
- Hichri, I.; Muhovski, Y.; Žižková, E.; Dobrev, P.I.; Franco-Zorrilla, J.M.; Solano, R.; Lopez-Vidriero, I.; Motyka, V.; Lutts, S. The Solanum lycopersicum Zinc Finger2 cysteine-2/histidine-2 repressor-like transcription factor regulates development and tolerance to salinity in tomato and Arabidopsis. Plant Physiol. 2014, 164, 1967–1990. [Google Scholar] [CrossRef]
- Cao, H.; Huang, P.; Zhang, L.; Shi, Y.; Sun, D.; Yan, Y.; Liu, X.; Dong, B.; Chen, G.; Snyder, J.H.; et al. Characterization of 47 Cys2-His2 zinc finger proteins required for the development and pathogenicity of the rice blast fungus Magnaporthe oryzae. New Phytol. 2016, 211, 1035–1051. [Google Scholar] [CrossRef]
- Cheuk, A.; Houde, M. Genome wide identification of C1-2i zinc finger proteins and their response to abiotic stress in hexaploid wheat. Mol. Genet. Genom. 2016, 291, 873–890. [Google Scholar] [CrossRef]
- Zang, D.; Li, H.; Xu, H.; Zhang, W.; Zhang, Y.; Shi, X.; Wang, Y. An Arabidopsis zinc finger protein increases abiotic stress tolerance by regulating sodium and potassium homeostasis, reactive oxygen species scavenging and osmotic potential. Front. Plant Sci. 2016, 7, 1272. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Y.; Wen, F.; Yao, D.; Wang, L.; Guo, J.; Ni, L.; Zhang, A.; Tan, M.; Jiang, M. A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. J. Exp. Bot. 2014, 65, 5795–5809. [Google Scholar] [CrossRef]
- Bowles, D.; Lim, E.K.; Poppenberger, B.; Vaistij, F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006, 57, 567–597. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef]
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Ludwig-Müller, J.; Chung, G.; Ahmad, M.Q.; Yang, S.H.; Lee, S.I. Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci. Rep. 2018, 8, 1875. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.G.; Wang, B.; Jin, S.H.; Qu, X.X.; Li, Y.J.; Hou, B.K. Ectopic expression of Arabidopsis glycosyltransferase UGT85A5 enhances salt stress tolerance in tobacco. PLoS ONE 2013, 8, 59924. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.R.; Lima, R.P.; Lisboa, J.V.; Chaves, T.R.; Luna, R.C.; do Nascimento, R.A.; de Oliveira, Y.; Persuhn, D.C.; Sérgio da Silva, A.; Gonçalves, M.D.; et al. Influence of the C677T polymorphism of the MTHFR gene on oxidative stress in women with overweight or obesity: Response to a dietary folate intervention. J. Am. Coll. Nutr. 2018, 37, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Raeisi, J.; Pakkish, Z.; Saffari, V.R. Efficiency of Folic Acid in Improving Yield and Fruit Quality of Strawberry. J. Plant Physiol. Breed. 2017, 7, 15–25. [Google Scholar]
- Small, I.D.; Peeters, N. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000, 25, 45–47. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, H.; Jia, Z.; Fang, Q.; Luo, K. Combined expression of antimicrobial genes (Bbchit1 and LJAMP2) in transgenic poplar enhances resistance to fungal pathogens. Tree Physiol. 2012, 32, 1313–1320. [Google Scholar] [CrossRef]
- Zsigmond, L.; Rigó, G.; Szarka, A.; Székely, G.; Ötvös, K.; Darula, Z.; Medzihradszky, K.F.; Koncz, C.; Koncz, Z.; Szabados, L. Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport. Plant Physiol. 2008, 146, 1721–1737. [Google Scholar] [CrossRef]
- Chen, G.; Zou, Y.; Hu, J.; Ding, Y. Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genom. 2018, 19, 720. [Google Scholar] [CrossRef]
- Yaldiz, G.; Camlica, M.; Nadeem, M.A.; Nawaz, M.A.; Baloch, F.S. Genetic diversity assessment in Nicotiana tabacum L. with iPBS-retrotransposons. Turk. J. Agric. For. 2018, 42, 154–164. [Google Scholar] [CrossRef]
- Guliyev, N.; Sharifova, S.; Ojaghi, J.; Abbasov, M.; Akparov, Z. Genetic diversity among melon (Cucumis melo L.) accessions revealed by morphological traits and ISSR markers. Turk. J. Agric. For. 2018, 42, 393–401. [Google Scholar] [CrossRef]
- Gramazio, P.; Plesa, I.M.; Truta, A.M.; Sestras, A.F.; Vilanova, S.; Plazas, M.; Vicente, O.; Boscaiu, M.; Prohens, J.; Sestras, R.E. Highly informative SSR genotyping reveals large genetic diversity and limited differentiation in European larch (Larix decidua) populations from Romania. Turk. J. Agric. For. 2018, 42, 165–175. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).