Multi-Tissue Transcriptome Analysis Identifies Key Sexual Development-Related Genes of the Ornate Spiny Lobster (Panulirus ornatus)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation and Sequencing
2.2. Transcriptome Assembly and Quantification
2.3. Identification of Sex-Determination Pathway Orthologues
2.4. Phylogenetic Analyses of Dmrt Orthologues
2.5. Mapping Analyses
2.6. Differential Expression Analyses (DEA)
2.7. Annotation
2.8. PCR Using Genomic DNA
2.9. Sagmariasus Verreauxi Sample Collection and Quantitative RT-PCR Validation
3. Results and Discussion
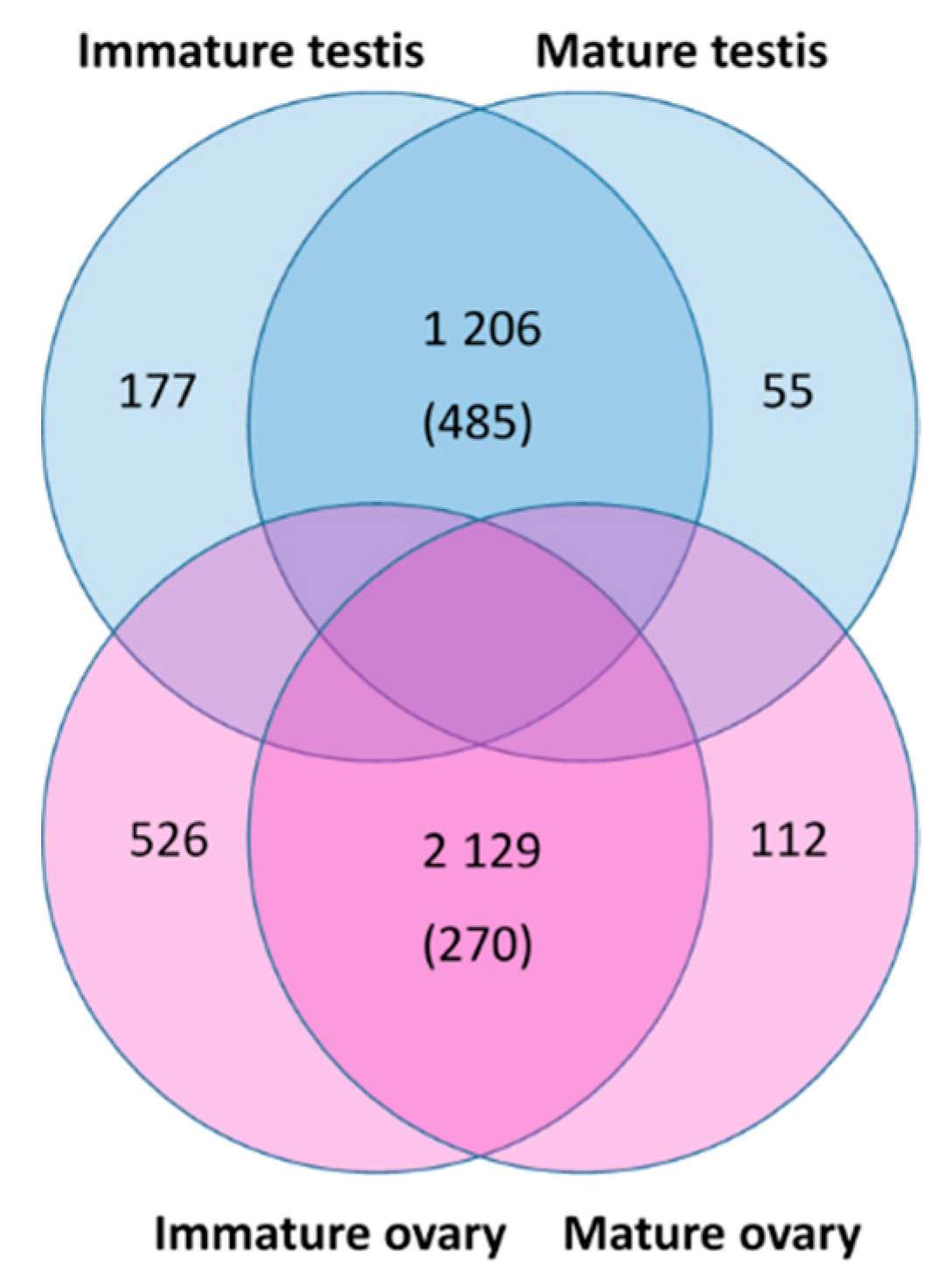
3.1. Transcriptome Sequencing and Assembly
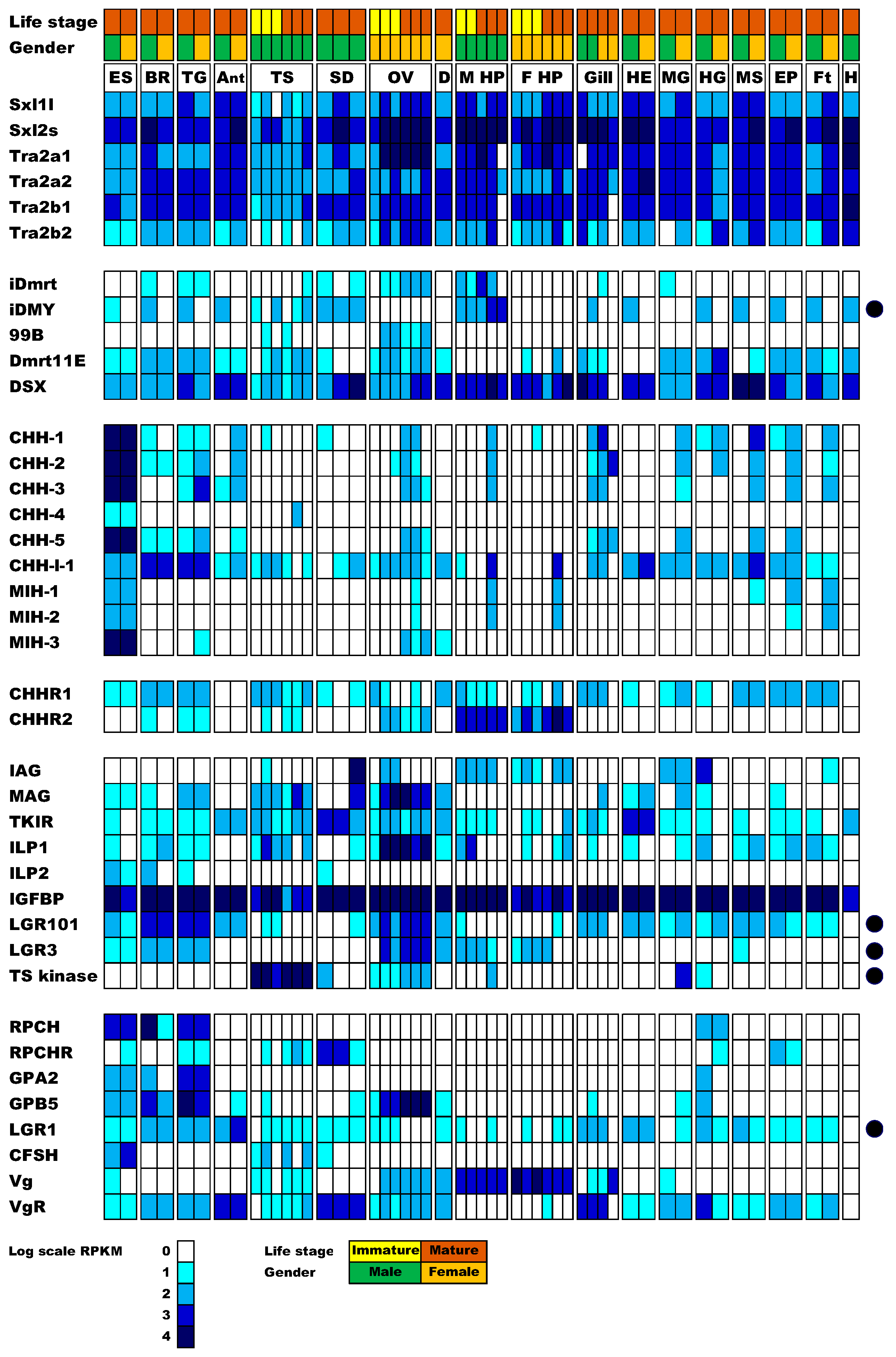
3.2. Identification of Sexual Development Factors
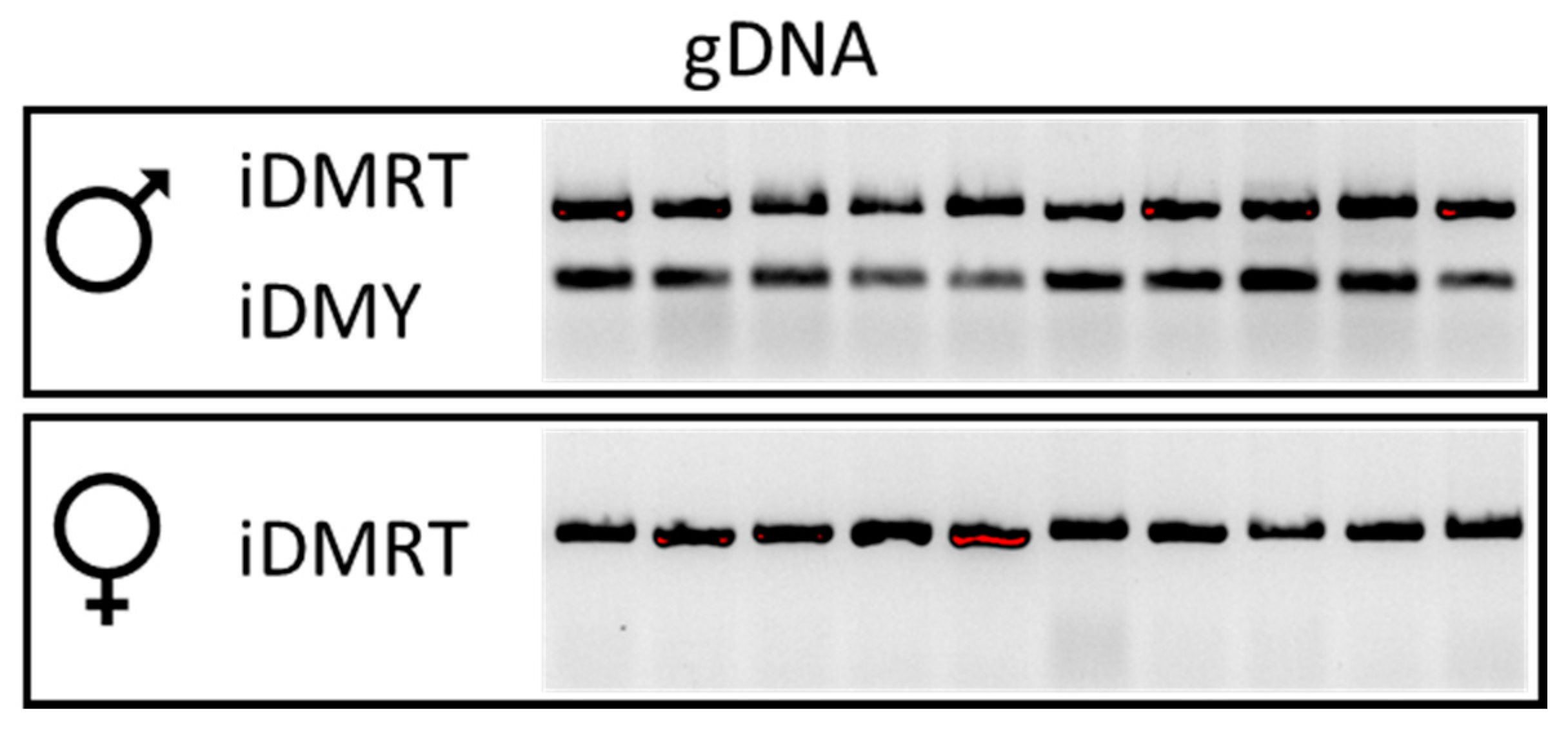
3.3. A Potential Sex-Determining Factor—The iDMY Gene Is Present Only in the Male Genome
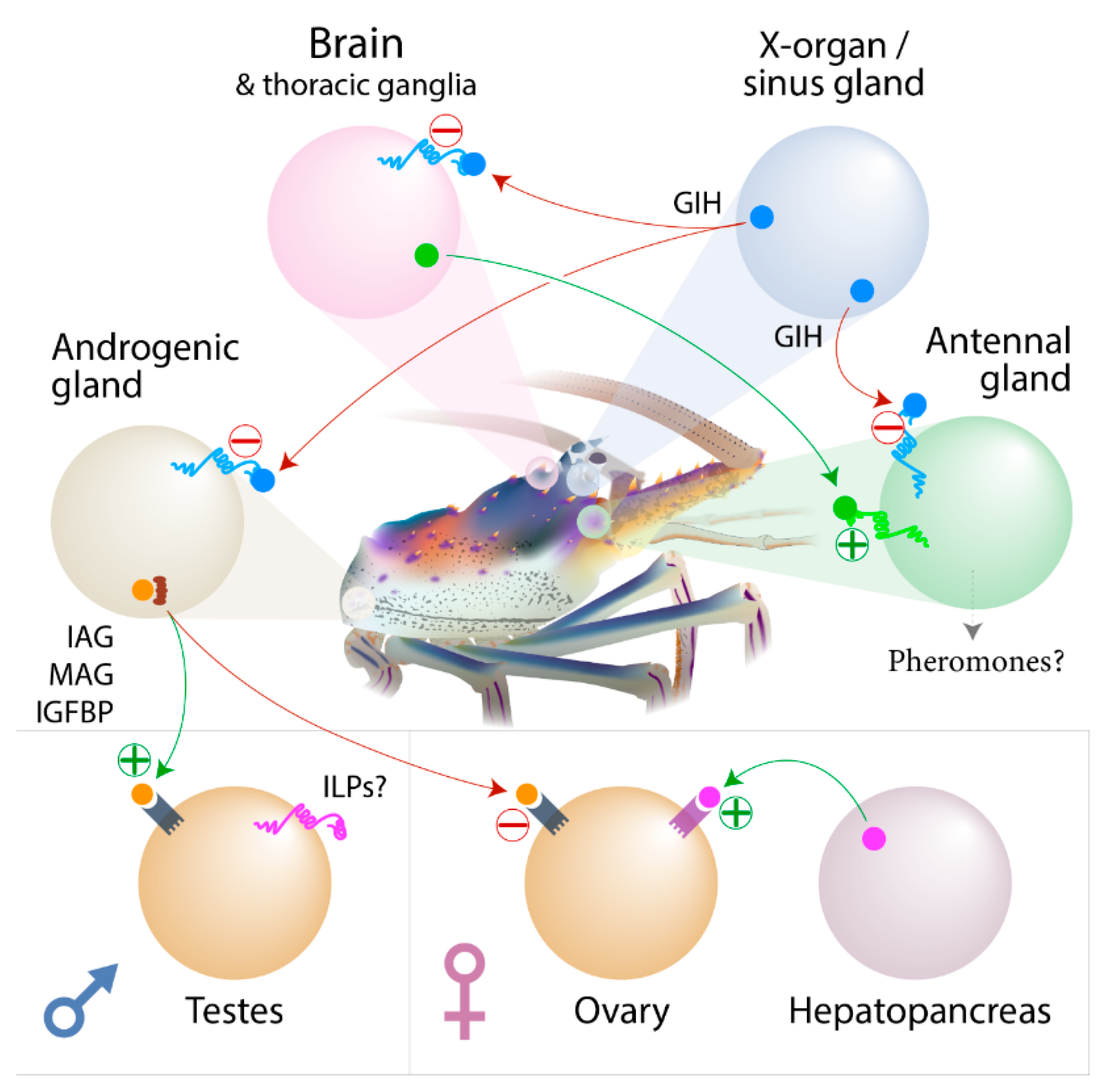
3.4. Expression of the CHH Family of Neuropeptides
3.5. Expression of the Insulin Endocrine Pathway
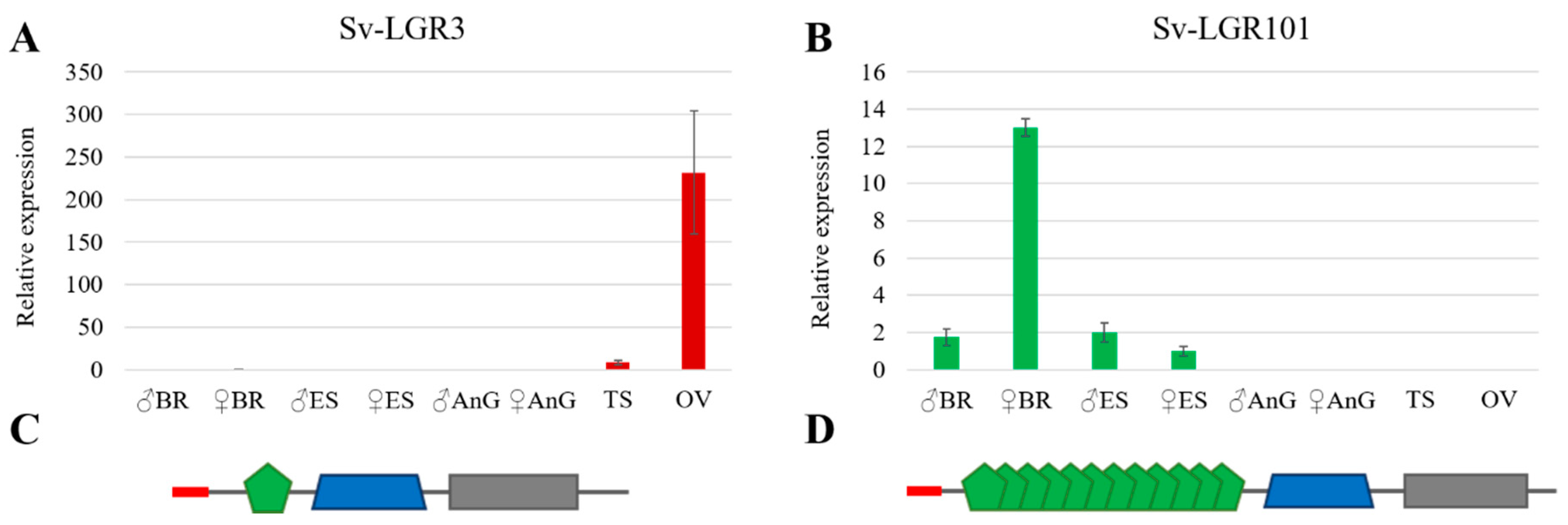
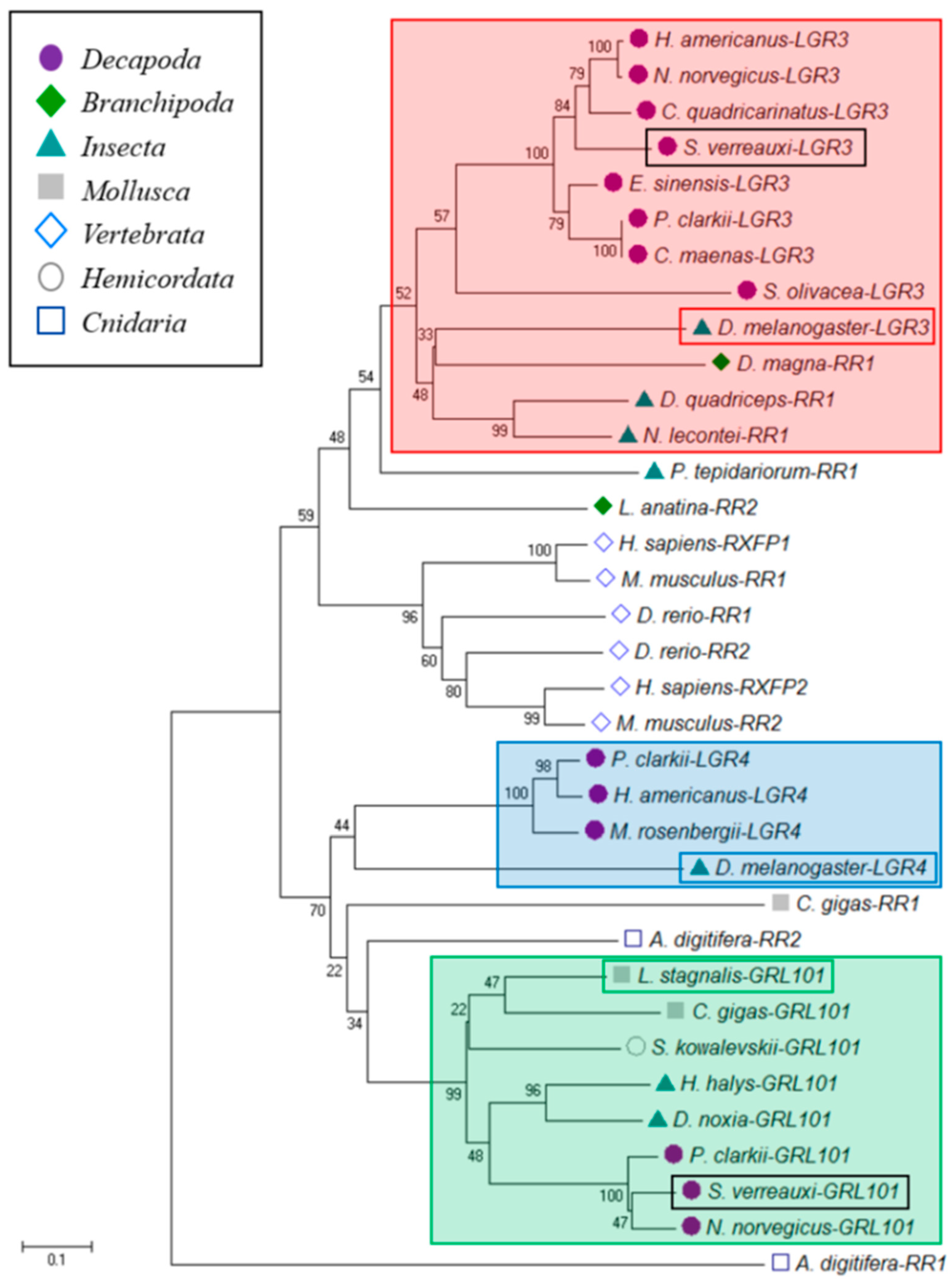
3.6. Insulin-Like Peptide Receptors Analysis
3.7. Ovarian Maturation Factors
3.8. Identification of Gonad-Specific Expressions
3.9. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ventura, T. Monosex in Aquaculture. In Marine Organisms as Model Systems; Kloc, M., Kubiak, J.Z., Eds.; Springer Nature: London, UK, 2018; pp. 91–101. [Google Scholar]
- Uller, T.; Helantera, H. From the origin of sex-determining factors to the evolution of sex-determining systems. Q. Rev. Biol. 2011, 86, 163–180. [Google Scholar] [CrossRef]
- Chandler, J.C.; Elizur, A.; Ventura, T. The decapod researcher’s guide to the galaxy of sex determination. Hydrobiologia 2018, 825, 61–80. [Google Scholar] [CrossRef]
- Schartl, M. A comparative view on sex determination in medaka. Mech. Dev. 2004, 121, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.V.; Rotllant, G.E.; Cummins, S.F.; Elizur, A.; Ventura, T. Insights into sexual maturation and reproduction in the Norway lobster (Nephrops norvegicus) via in silico prediction and characterization of neuropeptides and G protein-coupled receptors. Front. Endocrinol. 2018, 9, 430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotllant, G.; Nguyen, V.T.; Aizen, J.; Saowaros, S.; Ventura, T. Towards the identification of Female Gonad Stimulating Factor in crustaceans. Hydrobiologia 2018, 825, 91–119. [Google Scholar] [CrossRef]
- Chandler, J.C.; Fitzgibbon, Q.P.; Smith, G.; Elizur, A.; Ventura, T. Y-linked iDmrt1 paralogue (iDMY) in the Eastern spiny lobster, Sagmariasus verreauxi: The first invertebrate sex-linked Dmrt. Dev. Biol. 2017, 432, 337–345. [Google Scholar] [CrossRef]
- Chandler, C.J.; Gandhi, S.N.; Mancera, L.R.; Smith, G.; Elizur, A.; Ventura, T. Understanding insulin endocrinology in decapod crustacea: Molecular modelling characterization of an insulin-binding protein and insulin-like peptides in the Eastern spiny lobster, Sagmariasus verreauxi. Int. J. Mol. Sci. 2017, 18, 1832. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.V.; Cummins, S.F.; Elizur, A.; Ventura, T. Transcriptomic characterization and curation of candidate neuropeptides regulating reproduction in the eyestalk ganglia of the Australian crayfish, Cherax quadricarinatus. Sci. Rep. 2016, 6, 38658. Available online: http://www.nature.com/articles/srep38658 (accessed on 29 September 2020). [CrossRef] [Green Version]
- Chandler, J.C.; Aizen, J.; Fitzgibbon, Q.P.; Elizur, A.; Ventura, T. Applying the power of transcriptomics: Understanding male sexual development in decapod crustacea. Integr. Comp. Biol. 2016. [Google Scholar] [CrossRef]
- Chandler, J.C.; Aizen, J.; Bttaglene, S.C.; Elizur, A.; Ventura, T. Male sexual development and the androgenic gland: Novel insights through the de novo assembled transcriptome of the Eastern spiny lobster, Sagmariasus verreauxi. Sex. Dev. 2015, 9, 338–354. [Google Scholar] [CrossRef]
- Aizen, J.; Chandler, J.C.; Fitzgibbon, Q.P.; Sagi, A.; Battaglene, S.C.; Elizur, A.; Ventura, T. Production of recombinant insulin-like androgenic gland hormones from three decapod species: In vitro testicular phosphorylation and activation of a newly identified tyrosine kinase receptor from the Eastern spiny lobster, Sagmariasus verreauxi. Gen. Comp. Endocrinol. 2016, 229, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.C.; Aizen, J.; Elizur, A.; Hollander-Cohen, L.; Battaglene, S.; Ventura, T. Discovery of a novel insulin-like peptide and insulin binding proteins in the Eastern rock lobster Sagmariasus verreauxi. Gen. Comp. Endocrinol. 2015, 215, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Ventura, T.; Fitzgibbon, Q.; Battaglene, S.; Sagi, A.; Elizur, A. Identification and characterization of androgenic gland specific insulin-like peptide-encoding transcripts in two spiny lobster species: Sagmariasus verreauxi and Jasus edwardsii. Gen. Comp. Endocrinol. 2014, 214, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Ventura, T.; Cummins, S.F.; Fitzgibbon, Q.; Battaglene, S.; Elizur, A. Analysis of the central nervous system transcriptome of the eastern rock lobster Sagmariasus verreauxi reveals its putative neuropeptidome. PLoS ONE 2014, 9, e97323. [Google Scholar] [CrossRef]
- Kopp, A. Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 2012, 28, 175–184. [Google Scholar] [CrossRef] [Green Version]
- Zarkower, D. Establishing sexual dimorphism: Conservation amidst diversity? Nat. Rev. Genet. 2001, 2, 175–185. [Google Scholar] [CrossRef]
- Bopp, D.; Saccone, G.; Beye, M. Sex determination in insects: Variations on a common theme. Sex. Dev. 2014, 8, 20–28. [Google Scholar] [CrossRef] [Green Version]
- Sosnowski, B.A.; Belote, J.M.; McKeown, M. Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell 1989, 58, 449–459. [Google Scholar] [CrossRef]
- Matson, C.K.; Zarkower, D. Sex and the singular DM domain: Insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 2012, 13, 163–174. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.-Q.; Ma, W.-M.; Zeng, Q.-G.; Qian, Y.-Q.; Yang, J.-S.; Yang, W.-J. Molecular cloning and sexually dimorphic expression of two Dmrt genes in the giant freshwater prawn, Macrobrachium rosenbergii. Agric. Res. 2014, 3, 181–191. [Google Scholar] [CrossRef]
- Li, S.; Li, F.; Yu, K.; Xiang, J. Identification and characterization of a doublesex gene which regulates the expression of insulin-like androgenic gland hormone in Fenneropenaeus chinensis. Gene 2018, 649, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ventura, T.; Rosen, O.; Sagi, A. From the discovery of the crustacean androgenic gland to the insulin-like hormone in six decades. Gen. Comp. Endocrinol. 2011, 173, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Sroyraya, M.; Chotwiwatthanakun, C.; Stewart, M.J.; Soonklang, N.; Kornthong, N.; Phoungpetchara, I.; Hanna, P.J.; Sobhon, P. Bilateral eyestalk ablation of the blue swimmer crab, Portunus pelagicus, produces hypertrophy of the androgenic gland and an increase of cells producing insulin-like androgenic gland hormone. Tissue Cell 2010, 42, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Jo, Q.; Kim, B.K.; Han, C.H. Eyestalk ablation-induced androgenic gland activity and gonad development in the freshwater prawns Macrobrachium nipponense (De Haan, 1849). Invertebr. Reprod. Dev. 2002, 42, 35–42. [Google Scholar] [CrossRef]
- Khalaila, I.; Manor, R.; Weil, S.; Granot, Y.; Keller, R.; Sagi, A. The eyestalk-androgenic gland-testis endocrine axis in the crayfish Cherax quadricarinatus. Gen. Comp. Endocrinol. 2002, 127, 147–156. [Google Scholar] [CrossRef]
- Gross, P.S.; Knowlton, R.E. Effects of timed eyestalk ablation on molting in larvae of the snapping shrimp, Alpheus heterochaelis Say. Invertebr. Reprod. Dev. 1997, 32, 119–126. [Google Scholar] [CrossRef]
- Wilder, M.N.; Okumura, T.; Suzuki, Y.; Fusetani, N.; Aida, K. Vitellogenin production induced by eyestalk ablation in juvenile giant freshwater prawn Macrobrachium rosenbergii and trial methyl farnesoate administration. Zool. Sci. 1994, 11, 45–53. [Google Scholar]
- Tan-Fermin, J.D. Effects of unilateral eyestalk ablation on ovarian histology and oocyte size frequency of wild and pond-reared Penaeus monodon (Fabricius) broodstock. Aquaculture 1991, 93, 77–86. [Google Scholar] [CrossRef]
- Webster, S.G.; Keller, R.; Dircksen, H. The CHH-superfamily of multifunctional peptide hormones controlling crustacean metabolism, osmoregulation, moulting, and reproduction. Gen. Comp. Endocrinol. 2012, 175, 217–233. [Google Scholar] [CrossRef]
- Keller, R. Crustacean neuropeptides: Structures, functions and comparative aspects. Experientia 1992, 48, 439–448. [Google Scholar] [CrossRef]
- Kleinholz, L.H.; Keller, R. Endocrine regulation in Crustacea. In Hormones and Evolution; Barrington, E.J.W., Ed.; Academic Press: New York, NY, USA, 1979; pp. 160–213. [Google Scholar]
- Veenstra, J.A. Similarities between decapod and insect neuropeptidomes. PeerJ 2016, 4, e2043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christie, A.E.; Roncalli, V.; Cieslak, M.C.; Pascual, M.G.; Yu, A.; Lameyer, T.J.; Stanhope, M.E.; Dickinson, P.S. Prediction of a neuropeptidome for the eyestalk ganglia of the lobster Homarus americanus using a tissue-specific de novo assembled transcriptome. Gen. Comp. Endocrinol. 2017, 243, 96–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckley, S.J.; Fitzgibbon, Q.P.; Smith, G.G.; Ventura, T. In silico prediction of the G-protein coupled receptors expressed during the metamorphic molt of Sagmariasus verreauxi (Crustacea: Decapoda) by mining transcriptomic data: RNA-seq to repertoire. Gen. Comp. Endocrinol. 2016, 228, 111–127. [Google Scholar] [CrossRef] [PubMed]
- Nagai, C.; Mabashi-Asazuma, H.; Nagasawa, H.; Nagata, S. Identification and characterization of receptors for ion transport peptide (ITP) and ITP-like (ITPL) in the silkworm Bombyx mori. J. Biol. Chem. 2014, 289, 32166–32177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharabi, O.; Manor, R.; Weil, S.; Aflalo, E.D.; Lezer, Y.; Levy, T.; Aizen, J.; Ventura, T.; Mather, P.B.; Khalaila, I.; et al. Identification and characterization of an insulin-like receptor involved in crustacean reproduction. Endocrinology 2015, 157, 928–941. [Google Scholar] [CrossRef] [Green Version]
- Rosen, O.; Weil, S.; Manor, R.; Roth, Z.; Khalaila, I.; Sagi, A. A crayfish insulin-like-binding protein: Another piece in the androgenic gland insulin-like hormone puzzle is revealed. J. Biol. Chem. 2013, 288, 22289–22298. [Google Scholar] [CrossRef] [Green Version]
- Rosen, O.; Manor, R.; Weil, S.; Aflalo, E.D.; Bakhrat, A.; Abdu, U.; Sagi, A. An androgenic gland membrane-anchored gene associated with the crustacean insulin-like androgenic gland hormone. J. Exp. Biol. 2013, 216, 2122. [Google Scholar] [CrossRef] [Green Version]
- Claeys, I.; Simonet, G.; Poels, J.; Van Loy, T.; Vercammen, L.; De Loof, A.; Vanden Broeck, J. Insulin-related peptides and their conserved signal transduction pathway. Peptides 2002, 23, 807–816. [Google Scholar] [CrossRef]
- Rosen, O.; Manor, R.; Weil, S.; Gafni, O.; Linial, A.; Aflalo, E.D.; Ventura, T.; Sagi, A. A sexual shift induced by silencing of a single insulin-like gene in crayfish: Ovarian upregulation and testicular degeneration. PLoS ONE 2010, 5, e15281. [Google Scholar] [CrossRef] [Green Version]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Raviv, S.; Glazer, L.; Sagi, A. Temporal silencing of an androgenic gland-specific insulin-like gene affecting phenotypical gender differences and spermatogenesis. Endocrinology 2009, 150, 1278–1286. [Google Scholar] [CrossRef] [Green Version]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Rosen, O.; Sagi, A. Timing sexual differentiation: Full functional sex reversal achieved through silencing of a single insulin-like gene in the prawn, Macrobrachium rosenbergii. Biol. Reprod. 2012, 86, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Levy, T.; Rosen, O.; Eilam, B.; Azulay, D.; Aflalo, E.D.; Manor, R.; Shechter, A.; Sagi, A. A single injection of hypertrophied androgenic gland cells produces all-female aquaculture. Mar. Biotechnol. 2016, 18, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Ventura, T.; Aflalo, E.D.; Weil, S.; Kashkush, K.; Sagi, A. Isolation and characterization of a female-specific DNA marker in the giant freshwater prawn Macrobrachium rosenbergii. Heredity 2011, 107, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Ventura, T.; Sagi, A. The insulin-like androgenic gland hormone in crustaceans: From a single gene silencing to a wide array of sexual manipulation-based biotechnologies. Biotechnol. Adv. 2012, 30, 1543–1550. [Google Scholar] [CrossRef]
- Levy, T.; Rosen, O.; Eilam, B.; Azulay, D.; Zohar, I.; Aflalo, E.D.; Benet, A.; Naor, A.; Shechter, A.; Sagi, A. All-female monosex culture in the freshwater prawn Macrobrachium rosenbergii—A comparative large-scale field study. Aquaculture 2017, 479, 857–862. [Google Scholar] [CrossRef]
- Savaya Alkalay, A.; Rosen, O.; Sokolow, S.H.; Faye, Y.P.; Faye, D.S.; Aflalo, E.D.; Jouanard, N.; Zilberg, D.; Huttinger, E.; Sagi, A. The prawn Macrobrachium vollenhovenii in the Senegal River basin: Towards sustainable restocking of all-male populations for biological control of schistosomiasis. PLoS Negl. Trop. Dis. 2014, 8, e3060. [Google Scholar] [CrossRef]
- Raviv, S.; Parnes, S.; Sagi, A. Coordination of Reproduction and Molt in Decapods. In Reproductive Biology of Crustaceans Case Studies of Decapod Crustaceans; Mente, E., Ed.; Science Publishers: New York, NY, USA, 2008; pp. 365–390. [Google Scholar]
- Fingerman, M. Roles of neurotransmitters in regulating reproductive hormone release and gonadal maturation in decapod crustaceans. Invertebr. Reprod. Dev. 1997, 31, 47–54. [Google Scholar] [CrossRef]
- Alexander, J.L.; Oliphant, A.; Wilcockson, D.C.; Audsley, N.; Down, R.E.; Lafont, R.; Webster, S.G. Functional characterization and signaling systems of corazonin and red pigment concentrating hormone in the green shore crab, Carcinus maenas. Front. Neurosci. 2018, 11, 752. [Google Scholar] [CrossRef] [Green Version]
- Zmora, N.; Chung, J.S. A novel hormone is required for the development of reproductive phenotypes in adult female crabs. Endocrinology 2014, 155, 230–239. [Google Scholar] [CrossRef]
- Fitzgibbon, Q.P.; Battaglene, S.C. Effect of photoperiod on the early-stage phyllosoma and metamorphosis of spiny lobster, Sagmariasus verreauxi. Aquaculture 2012, 368–369, 48–54. [Google Scholar] [CrossRef]
- Ventura, T.; Fitzgibbon, Q.P.; Battaglene, S.C.; Elizur, A. Redefining metamorphosis in spiny lobsters: Molecular analysis of the phyllosoma to puerulus transition in Sagmariasus verreauxi. Sci. Rep. 2015, 5, 13537. Available online: http://www.nature.com/articles/srep13537 (accessed on 29 September 2020). [CrossRef] [PubMed] [Green Version]
- Hyde, C.J.; Fitzgibbon, Q.P.; Elizur, A.; Smith, G.G.; Ventura, T. Transcriptional profiling of spiny lobster metamorphosis reveals three new additions to the nuclear receptor superfamily. BMC Genom. 2019, 20, 531. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Ye, H.; Chung, J.S. The presence of an insulin-like androgenic gland factor (IAG) and insulin-like peptide binding protein (ILPBP) in the ovary of the blue crab, Callinectes sapidus and their roles in ovarian development. Gen. Comp. Endocrinol. 2017, 249, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.S.; Manor, R.; Sagi, A. Cloning of an insulin-like androgenic gland factor (IAG) from the blue crab, Callinectes sapidus: Implications for eyestalk regulation of IAG expression. Gen. Comp. Endocrinol. 2011, 173, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Bathgate, R.A.D.; Halls, M.L.; van der Westhuizen, E.T.; Callander, G.E.; Kocan, M.; Summers, R.J. Relaxin family peptides and their receptors. Physiol. Rev. 2013, 93, 405–480. [Google Scholar] [CrossRef] [PubMed]
- Van Hiel, M.B.; Vandersmissen, H.P.; Van Loy, T.; Vanden Broeck, J. An evolutionary comparison of leucine-rich repeat containing G protein-coupled receptors reveals a novel LGR subtype. Peptides 2012, 34, 193–200. [Google Scholar] [CrossRef]
- Tensen, C.P.; Van Kesteren, E.R.; Planta, R.J.; Cox, K.J.; Burke, J.F.; van Heerikhuizen, H.; Vreugdenhil, E. A G protein-coupled receptor with low density lipoprotein-binding motifs suggests a role for lipoproteins in G-linked signal transduction. Proc. Natl. Acad. Sci. USA 1994, 91, 4816–4820. [Google Scholar] [CrossRef] [Green Version]
- Garelli, A.; Gontijo, A.M.; Miguela, V.; Caparros, E.; Dominguez, M. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science 2012, 336, 579–582. [Google Scholar] [CrossRef]
- Garelli, A.; Heredia, F.; Casimiro, A.P.; Macedo, A.; Nunes, C.; Garcez, M.; Dias, A.R.M.; Volonte, Y.A.; Uhlmann, T.; Caparros, E.; et al. Dilp8 requires the neuronal relaxin receptor Lgr3 to couple growth to developmental timing. Nat. Commun. 2015, 6, 8732. [Google Scholar] [CrossRef] [Green Version]
- Jaszczak, J.S.; Wolpe, J.B.; Bhandari, R.; Jaszczak, R.G.; Halme, A. Growth coordination during Drosophila melanogaster imaginal disc regeneration Is mediated by signaling through the relaxin receptor lgr3 in the prothoracic gland. Genetics 2016, 204, 703. [Google Scholar] [CrossRef] [Green Version]
- Denley, A.; Cosgrove, L.J.; Booker, G.W.; Wallace, J.C.; Forbes, B.E. Molecular interactions of the IGF system. Cytokine Growth Factor Rev. 2005, 16, 421–439. [Google Scholar] [CrossRef] [PubMed]
- Subramoniam, T. Crustacean ecdysteriods in reproduction and embryogenesis. Comp. Biochem. Physiol. Part C Pharmacol. Toxicol. Endocrinol. 2000, 125, 135–156. [Google Scholar] [CrossRef]
- Veenstra, J.A. The power of next-generation sequencing as illustrated by the neuropeptidome of the crayfish Procambarus clarkii. Gen. Comp. Endocrinol. 2015, 224, 84–95. [Google Scholar] [CrossRef] [PubMed]
| Primer Name | Gene/Amplicon Size (nt) | Primer Sequence 5′–3′; Accession Number |
|---|---|---|
| Po-iDMY F | Po-iDMY/322nt | ACACACTTAAGCCGTCTCCA |
| Po-iDMY R | TTTCATAACGCCGTGGTTCC | |
| Po-iDmrt F | Po-iDmrt/528nt | GCAGCCTGAATATGAGGGGT |
| Po-iDmrt R | AGTAAGGCAAGTTGACGGGA | |
| qSv-LGR3 F | Sv-LGR3/73nt | GACGGAGTGTCATCGTTCG |
| qSv-LGR3 R | CCACAATCACCAGCCACA Accession number: KY427011 (Sv-RR1) | |
| qSv-LGR101 F | Sv-LGR101/61nt | GACATCGTGGCTGTGTCG |
| qSv-LGR101 R | GCTGGACATTCCAACGACTT Accession number: KY427010 (Sv-GRL101) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ventura, T.; Chandler, J.C.; Nguyen, T.V.; Hyde, C.J.; Elizur, A.; Fitzgibbon, Q.P.; Smith, G.G. Multi-Tissue Transcriptome Analysis Identifies Key Sexual Development-Related Genes of the Ornate Spiny Lobster (Panulirus ornatus). Genes 2020, 11, 1150. https://doi.org/10.3390/genes11101150
Ventura T, Chandler JC, Nguyen TV, Hyde CJ, Elizur A, Fitzgibbon QP, Smith GG. Multi-Tissue Transcriptome Analysis Identifies Key Sexual Development-Related Genes of the Ornate Spiny Lobster (Panulirus ornatus). Genes. 2020; 11(10):1150. https://doi.org/10.3390/genes11101150
Chicago/Turabian StyleVentura, Tomer, Jennifer C. Chandler, Tuan V. Nguyen, Cameron J. Hyde, Abigail Elizur, Quinn P. Fitzgibbon, and Gregory G. Smith. 2020. "Multi-Tissue Transcriptome Analysis Identifies Key Sexual Development-Related Genes of the Ornate Spiny Lobster (Panulirus ornatus)" Genes 11, no. 10: 1150. https://doi.org/10.3390/genes11101150







