OneStopRNAseq: A Web Application for Comprehensive and Efficient Analyses of RNA-Seq Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Implementation
2.2. RNA-Seq Data
3. Results
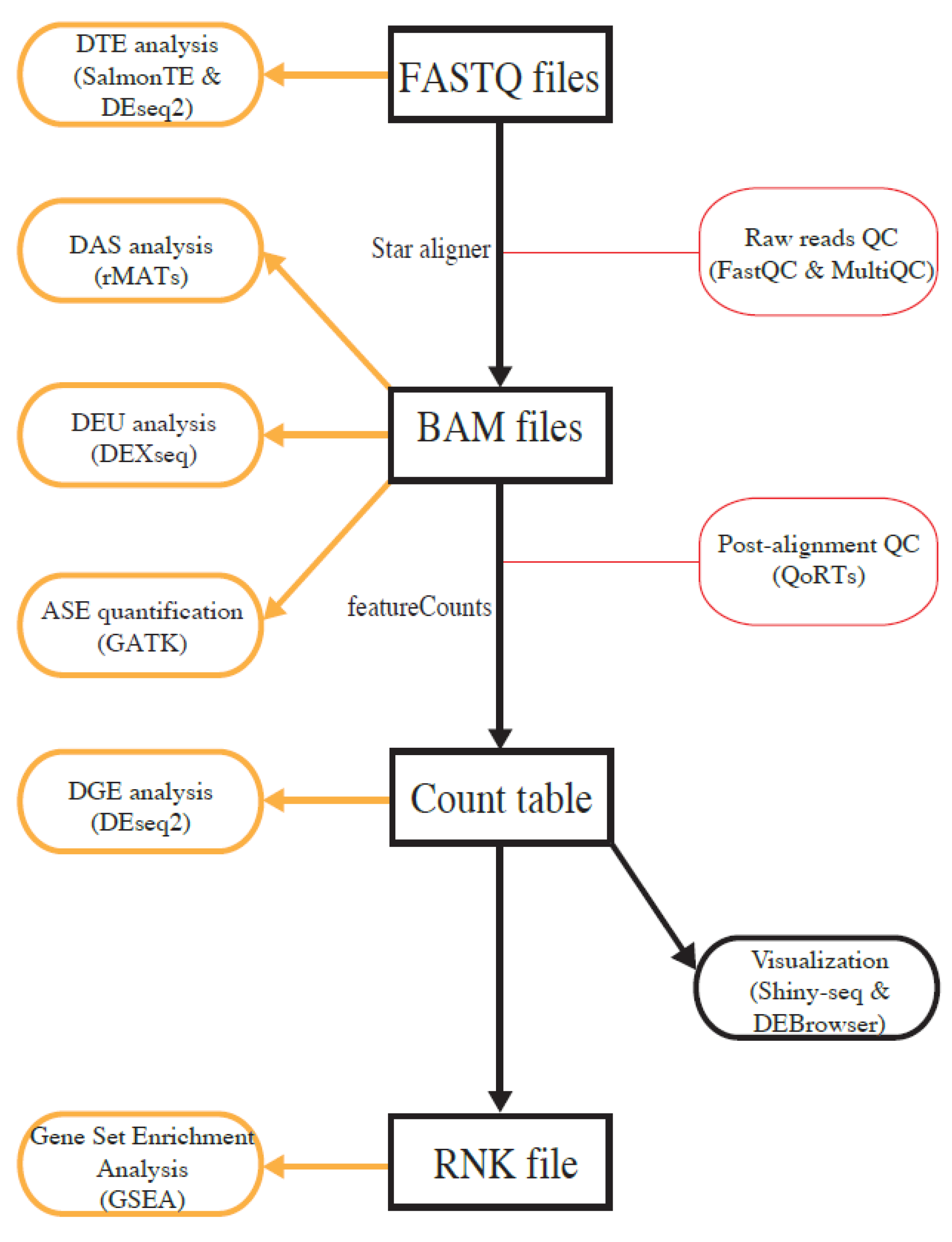
3.1. Functionality Summary of the OneStopRNAseq Application
3.2. Case Study Validating OneStopRNAseq Application Functionalities
3.3. Runtime of OneStopRNAseq Application
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Lowe, R.G.T.; Shirley, N.J.; Bleackley, M.R.; Dolan, S.K.; Shafee, T.M.A. Transcriptomics technologies. PLoS Comput. Boil. 2017, 13, e1005457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Boil. 2013, 14, 699–712. [Google Scholar] [CrossRef] [Green Version]
- Yao, R.-W.; Wang, Y.; Chen, L.-L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Sagan, S.M.; Macrae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Boil. 2018, 20, 21–37. [Google Scholar] [CrossRef]
- Weber, A.P.M. Discovering New Biology through Sequencing of RNA1. Plant Physiol. 2015, 169, 1524–1531. [Google Scholar] [CrossRef]
- Madsen, J.G.S.; Schmidt, S.F.; Larsen, B.D.; Loft, A.; Nielsen, R.; Mandrup, S. iRNA-seq: Computational method for genome-wide assessment of acute transcriptional regulation from total RNA-seq data. Nucleic Acids Res. 2015, 43, e40. [Google Scholar] [CrossRef] [Green Version]
- Abreu, R.D.S.; Penalva, L.O.; Marcotte, E.; Vogel, C. Global signatures of protein and mRNA expression levels. Mol. BioSyst. 2009, 5, 1512–1526. [Google Scholar] [CrossRef] [Green Version]
- Vogel, C.; Marcotte, E. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [Green Version]
- Borràs, D.M.; Janssen, B. The Use of Transcriptomics in Clinical Applications. In Integration of Omics Approaches and Systems Biology for Clinical Applications; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; pp. 49–66. ISBN 9781119183952. [Google Scholar]
- Consortium, S.M.-I. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat. Biotechnol. 2014, 32, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, E.L.; Jaszczyszyn, Y.; Thermes, C. Library preparation methods for next-generation sequencing: Tone down the bias. Exp. Cell Res. 2014, 322, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Dard-Dascot, C.; Naquin, D.; D’Aubenton-Carafa, Y.; Alix, K.; Thermes, C.; Van Dijk, E.L. Systematic comparison of small RNA library preparation protocols for next-generation sequencing. BMC Genom. 2018, 19, 118. [Google Scholar] [CrossRef] [Green Version]
- Wright, C.; Rajpurohit, A.; Burke, E.E.; Williams, C.; Collado-Torres, L.; Kimos, M.; Brandon, N.J.; Cross, A.J.; Jaffe, A.E.; Weinberger, D.R.; et al. Comprehensive assessment of multiple biases in small RNA sequencing reveals significant differences in the performance of widely used methods. BMC Genom. 2019, 20, 513. [Google Scholar] [CrossRef] [Green Version]
- Chao, H.-P.; Chen, Y.; Takata, Y.; Tomida, M.W.; Lin, K.; Kirk, J.; Simper, M.S.; Mikulec, C.D.; Rundhaug, J.E.; Fischer, S.M.; et al. Systematic evaluation of RNA-Seq preparation protocol performance. BMC Genom. 2019, 20, 571. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Boil. 2016, 17, 13. [Google Scholar] [CrossRef] [Green Version]
- Koen, V.D.B.; Katharina, M.H.; Charlotte, S.; Simone, T.; Lieven, C.; Michael, I.L.; Rob, P.; Mark, D.R. RNA Sequencing Data: Hitchhiker’s Guide to Expression Analysis. Annu. Rev. Biomed. Data Sci. 2019, 2, 139–173. [Google Scholar]
- Han, Y.; Gao, S.; Muegge, K.; Zhang, W.; Zhou, B. Advanced Applications of RNA Sequencing and Challenges. Bioinform. Boil. Insights 2015, 9, BBI–S28991. [Google Scholar] [CrossRef] [Green Version]
- Byron, S.A.; Van Keuren-Jensen, K.R.; Engelthaler, D.M.; Carpten, J.D.; Craig, D.W. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat. Rev. Genet. 2016, 17, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Rose, C.M.; Cass, A.A.; Williams, A.; Darwish, M.; Lianoglou, S.; Haverty, P.M.; Tong, A.-J.; Blanchette, C.; Albert, M.L.; et al. Transposable element expression in tumors is associated with immune infiltration and increased antigenicity. Nat. Commun. 2019, 10, 5228. [Google Scholar] [CrossRef] [PubMed]
- Hancks, D.C.; Kazazian, H.H. Active human retrotransposons: Variation and disease. Curr. Opin. Genet. Dev. 2012, 22, 191–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffith, M.; Walker, J.R.; Spies, N.C.; Ainscough, B.J.; Griffith, O.L. Informatics for RNA Sequencing: A Web Resource for Analysis on the Cloud. PLoS Comput. Boil. 2015, 11, e1004393. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Mortazavi, A. Integrating ChIP-seq with other functional genomics data. Briefings Funct. Genom. 2018, 17, 104–115. [Google Scholar] [CrossRef]
- Yan, F.; Powell, D.R.; Curtis, D.J.; Wong, N.C. From reads to insight: A hitchhiker’s guide to ATAC-seq data analysis. Genome Boil. 2020, 21, 1–16. [Google Scholar] [CrossRef]
- Nica, A.C.; Dermitzakis, E.T. Expression quantitative trait loci: Present and future. Philos. Trans. R. Soc. B Boil. Sci. 2013, 368, 20120362. [Google Scholar] [CrossRef]
- Knight, J.C. Allele-specific gene expression uncovered. Trends Genet. 2004, 20, 113–116. [Google Scholar] [CrossRef]
- Haider, S.; Pal, R. Integrated Analysis of Transcriptomic and Proteomic Data. Curr. Genom. 2013, 14, 91–110. [Google Scholar] [CrossRef]
- Cavill, R.; Jennen, D.; Kleinjans, J.; Briedé, J.J. Transcriptomic and metabolomic data integration. Briefings Bioinform. 2015, 17, 891–901. [Google Scholar] [CrossRef] [Green Version]
- Lightbody, G.; Haberland, V.; Browne, F.; Taggart, L.; Zheng, H.; Parkes, E.; Blayney, J.K. Review of applications of high-throughput sequencing in personalized medicine: Barriers and facilitators of future progress in research and clinical application. Brief. Bioinform. 2019, 20, 1795–1811. [Google Scholar] [CrossRef] [PubMed]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. Methods Mol. Biol. 2016, 1418, 93–110. [Google Scholar] [PubMed] [Green Version]
- Kodama, Y.; Shumway, M.; Leinonen, R.; on behalf of the International Nucleotide Sequence Database Collaboration. The sequence read archive: Explosive growth of sequencing data. Nucleic Acids Res. 2011, 40, D54–D56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, I.S.; Kim, S. Analysis of Whole Transcriptome Sequencing Data: Workflow and Software. Genom. Inform. 2015, 13, 119–125. [Google Scholar] [CrossRef]
- Köster, J.; Rahmann, S. Snakemake—A scalable bioinformatics workflow engine. Bioinformatics 2012, 28, 2520–2522. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 3 April 2020).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2012, 29, 15–21. [Google Scholar] [CrossRef]
- Hartley, S.W.; Mullikin, J.C. QoRTs: A comprehensive toolset for quality control and data processing of RNA-Seq experiments. BMC Bioinform. 2015, 16, 224. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Shen, S.; Park, J.W.; Lu, Z.-X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Reyes, A.; Huber, W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012, 22, 2008–2017. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.-H.; Yalamanchili, H.K.; Guo, C.; Shulman, J.M.; Liu, Z. An ultra-fast and scalable quantification pipeline for transposable elements from next generation sequencing data. Pac. Symp. Biocomput. Pac. Symp. Biocomput. 2018, 23, 168–179. [Google Scholar] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 002832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Der Auwera, G.A.; O Carneiro, M.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Ohol, Y.M.; Sun, M.T.; Cutler, G.; Leger, P.R.; Hu, D.X.; Biannic, B.; Rana, P.; Cho, C.; Jacobson, S.; Wong, S.T.; et al. Novel, Selective Inhibitors of USP7 Uncover Multiple Mechanisms of Antitumor Activity in Vitro and in Vivo. Mol. Cancer Ther. 2020. [Google Scholar] [CrossRef]
- Kucukural, A.; Yukselen, O.; Ozata, D.M.; Moore, M.J.; Garber, M. DEBrowser: Interactive differential expression analysis and visualization tool for count data. BMC Genom. 2019, 20, 6. [Google Scholar] [CrossRef] [Green Version]
- Sundararajan, Z.; Knoll, R.; Hombach, P.; Becker, M.; Schultze, J.L.; Ulas, T. Shiny-Seq: Advanced guided transcriptome analysis. BMC Res. Notes 2019, 12, 432. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Geisler, S.J.; Paro, R. Trithorax and Polycomb group-dependent regulation: A tale of opposing activities. Development 2015, 142, 2876–2887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Kang, W.; You, Y.; Pang, J.; Ren, H.; Suo, Z.; Liu, H.; Zheng, Y. USP7: Novel Drug Target in Cancer Therapy. Front. Pharmacol. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Boil. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Li, Y.; Rao, X.; Mattox, W.; Amos, C.I.; Liu, B. RNA-Seq Analysis of Differential Splice Junction Usage and Intron Retentions by DEXSeq. PLoS ONE 2015, 10, e0136653. [Google Scholar] [CrossRef]
- Pirinen, M.; Lappalainen, T.; Zaitlen, N.A.; Dermitzakis, E.T.; Donnelly, P.; McCarthy, M.I.; Rivas, M.A. Assessing allele-specific expression across multiple tissues from RNA-seq read data. Bioinformatics 2015, 31, 2497–2504. [Google Scholar] [CrossRef] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Tam, O.H.; Paniagua, E.; Hammell, M. TEtranscripts: A package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 2015, 31, 3593–3599. [Google Scholar] [CrossRef]
- Alhamdoosh, M.; Ng, M.; Wilson, N.; Sheridan, J.; Huynh, H.; Wilson, M.; Ritchie, M. Combining multiple tools outperforms individual methods in gene set enrichment analyses. Bioinformatics 2017, 33, 414–424. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [PubMed] [Green Version]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Hu, K.; Liu, H.; Green, M.R.; Zhu, L.J. OneStopRNAseq: A Web Application for Comprehensive and Efficient Analyses of RNA-Seq Data. Genes 2020, 11, 1165. https://doi.org/10.3390/genes11101165
Li R, Hu K, Liu H, Green MR, Zhu LJ. OneStopRNAseq: A Web Application for Comprehensive and Efficient Analyses of RNA-Seq Data. Genes. 2020; 11(10):1165. https://doi.org/10.3390/genes11101165
Chicago/Turabian StyleLi, Rui, Kai Hu, Haibo Liu, Michael R. Green, and Lihua Julie Zhu. 2020. "OneStopRNAseq: A Web Application for Comprehensive and Efficient Analyses of RNA-Seq Data" Genes 11, no. 10: 1165. https://doi.org/10.3390/genes11101165
APA StyleLi, R., Hu, K., Liu, H., Green, M. R., & Zhu, L. J. (2020). OneStopRNAseq: A Web Application for Comprehensive and Efficient Analyses of RNA-Seq Data. Genes, 11(10), 1165. https://doi.org/10.3390/genes11101165




