Dissecting the Genetic Regulation of Yeast Growth Plasticity in Response to Environmental Changes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data
2.2. Quantifying Yeast Growth Plasticity
2.3. Heritability Estimation
2.4. Mapping Alleles Underlying Variation in Phenotypic Plasticity
2.5. Estimating the Significance Threshold for the Association Analyses
2.6. Connecting Plasticity QTL to Previously Detected Epistatic Networks
3. Results
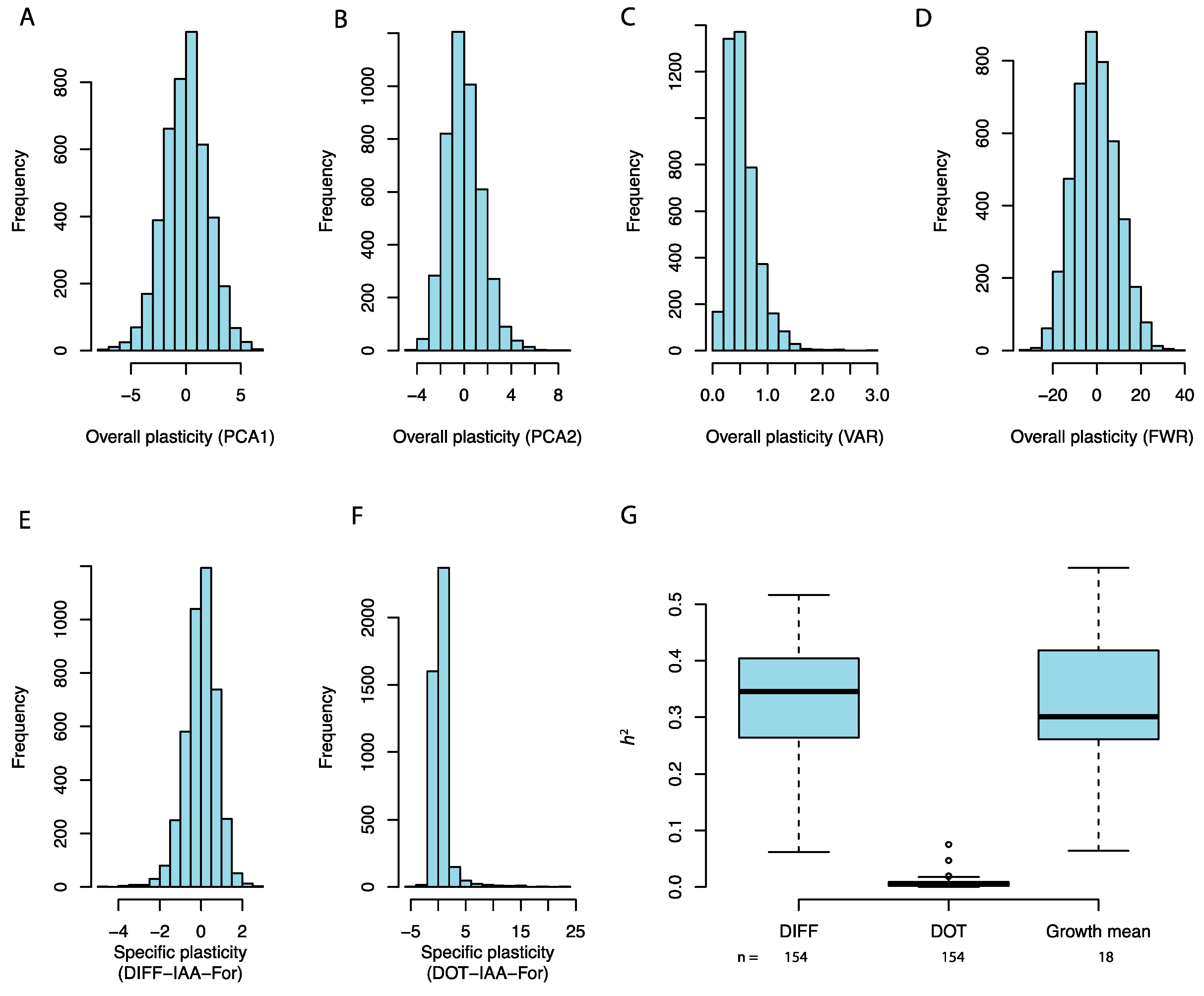
3.1. Heritable Variation in Yeast Growth Plasticity in Response to Chemical Stress
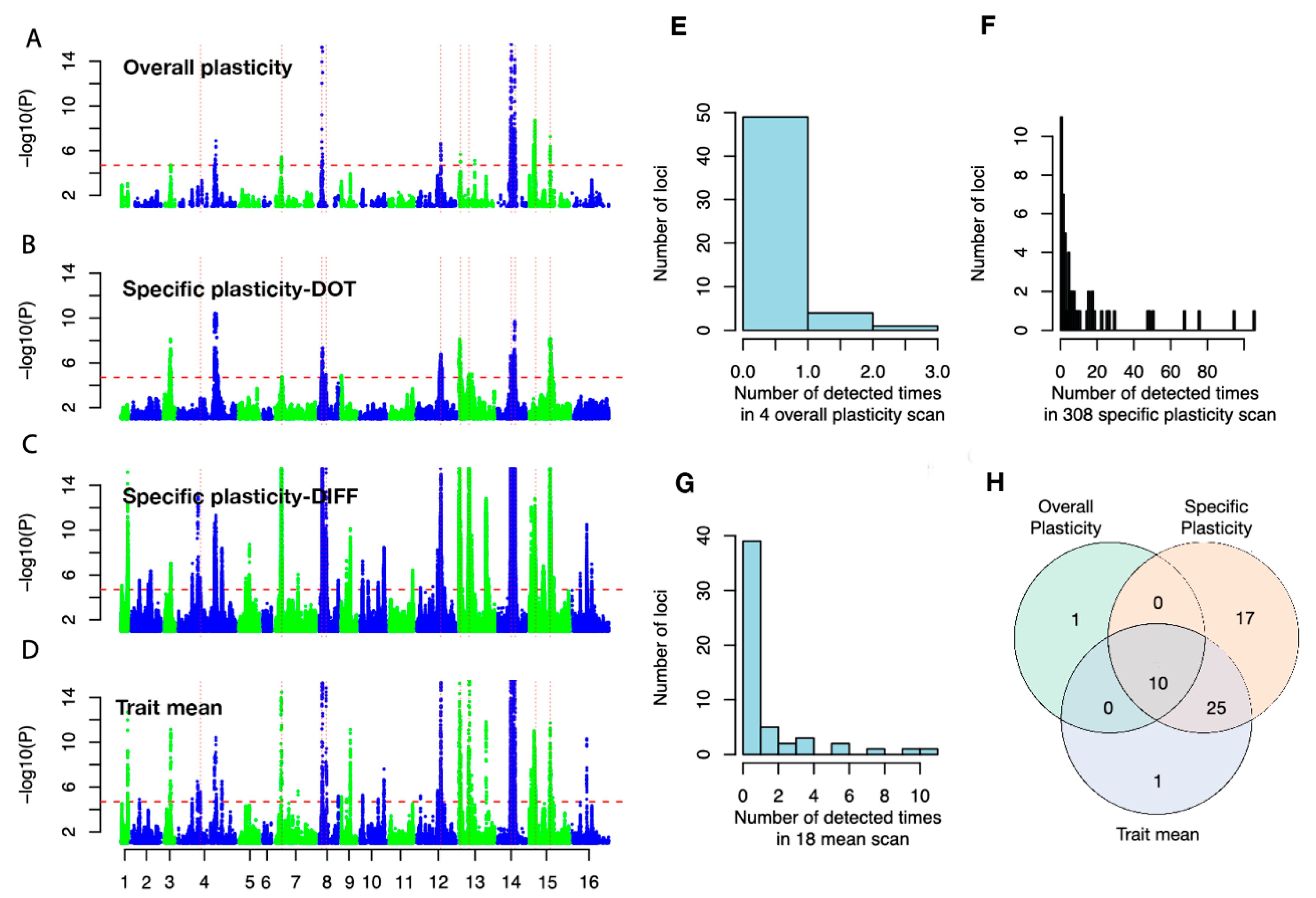
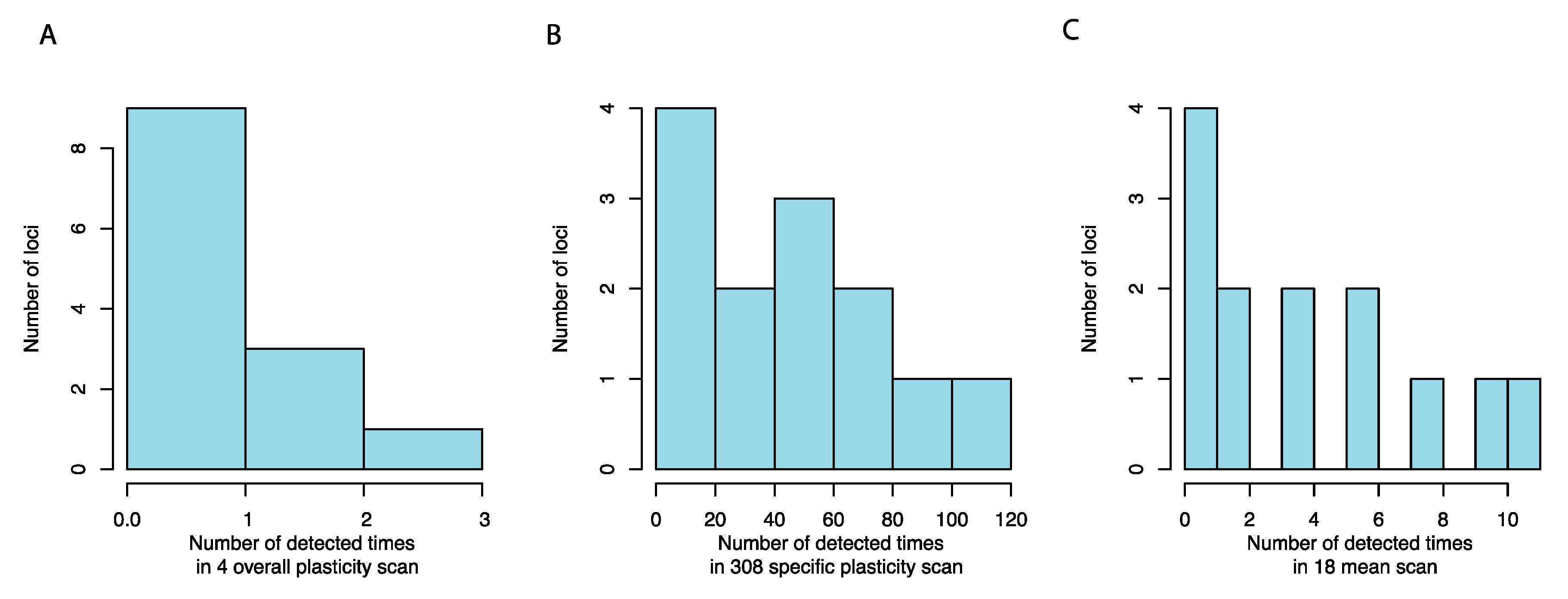
3.2. Mapping Alleles Contributing to the Variation in Growth Plasticity
3.3. Many Plasticity QTLs are Hub Loci in Previously Detected Epistatic Networks
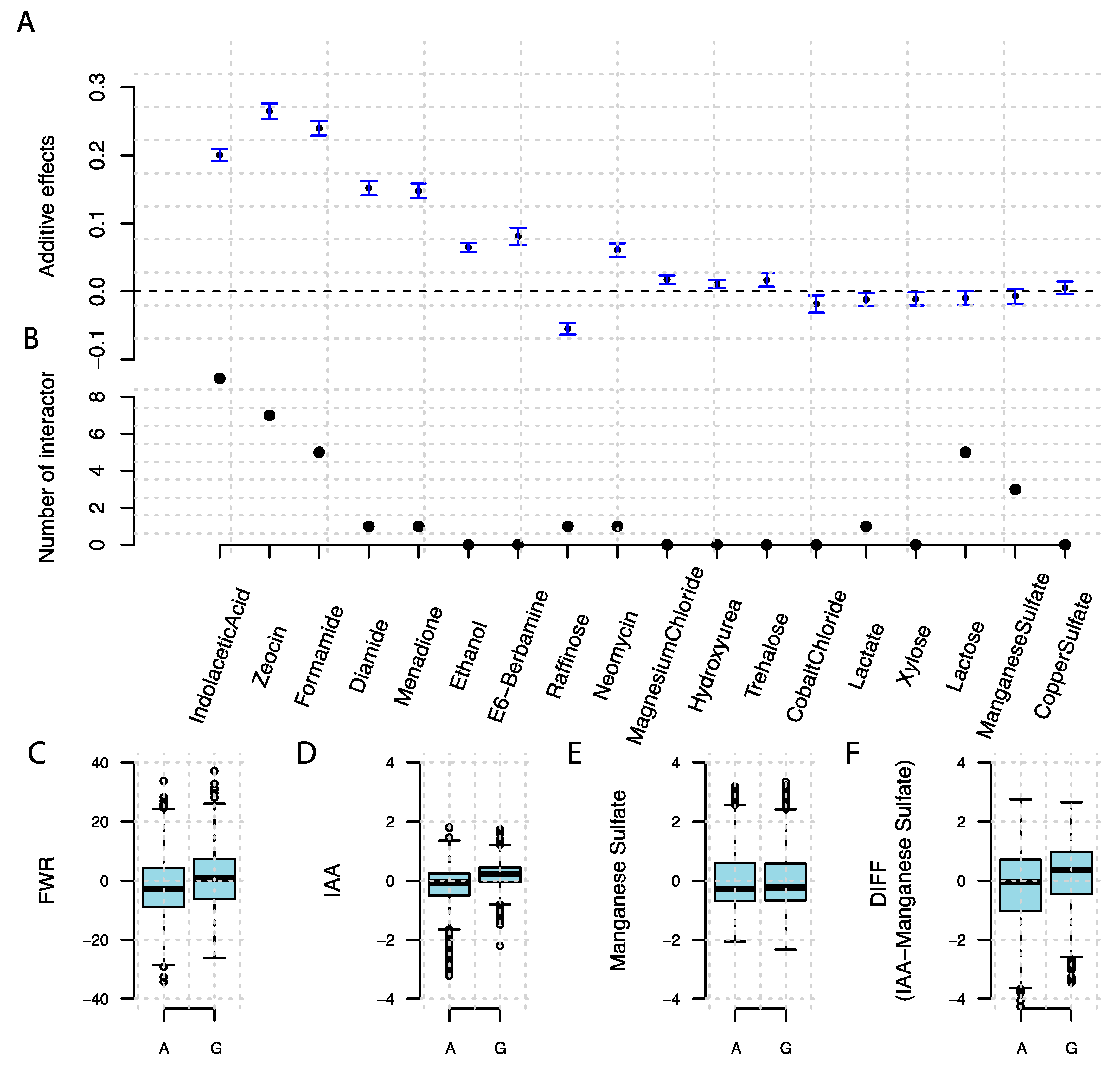
3.4. A Known Epistatic Growth Hub QTL Affecting Specific and Overall Plasticity
4. Discussion
4.1. Quantification of the Phenotypic Plasticity
4.2. The Role of QTL–QTL and QTL–Environment Interactions in Phenotype Plasticity
4.3. Relationship to Earlier Studies Evaluating the Role of G-by-G-by-E Interactions on Growth Variation in Individual Environments
4.4. Candidate Genes Contributing to Variation in Phenotype Plasticity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pelletier, F.; Réale, D.; Garant, D.; Coltman, D.W.; Festa-Bianchet, M. Selection on heritable seasonal phenotypic plasticity of body mass. Evolution 2007, 61, 1969–1979. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.R.; Chadee, D.D.; Mori, A.; Romero-Severson, J.; Severson, D.W. Heritability and adaptive phenotypic plasticity of adult body size in the mosquito Aedes aegypti with implications for dengue vector competence. Infect. Genet. Evol. 2011, 11, 11–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanous, A.; Gardner, C.; Blanco, M.; Martin-Schwarze, A.; Wang, J.; Li, X.; Lipka, A.E.; Flint-Garcia, S.; Bohn, M.; Edwards, J.; et al. Stability Analysis of Kernel Quality Traits in Exotic-Derived Doubled Haploid Maize Lines. Plant Genome 2019, 12, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangin, B.; Casadebaig, P.; Cadic, E.; Blanchet, N.; Boniface, M.-C.; Carrère, S.; Gouzy, J.; Legrand, L.; Mayjonade, B.; Pouilly, N.; et al. Genetic control of plasticity of oil yield for combined abiotic stresses using a joint approach of crop modeling and genome-wide association. Plant Cell Environ. 2017, 40, 2276–2291. [Google Scholar] [CrossRef] [PubMed]
- Diouf, I.; Derivot, L.; Koussevitzky, S.; Carretero, Y.; Bitton, F.; Moreau, L.; Causse, M. Genetic basis of phenotypic plasticity and genotype × environment interactions in a multi-parental tomato population. J. Exp. Bot. 2020, 71, 5365–5376. [Google Scholar] [CrossRef]
- Rong, M.; Zheng, X.; Ye, M.; Bai, J.; Xie, X.; Jin, Y.; He, X. Phenotypic Plasticity of Staphylococcus aureus in Liquid Medium Containing Vancomycin. Front. Microbiol. 2019, 10, 809. [Google Scholar] [CrossRef]
- Zheng, X.; Bai, J.; Ye, M.; Liu, Y.; Jin, Y.; He, X. Bivariate genome-wide association study of the growth plasticity of Staphylococcus aureus in coculture with Escherichia coli. Appl. Microbiol. Biotechnol. 2020, 104, 5437–5447. [Google Scholar] [CrossRef]
- Lacaze, X.; Hayes, P.M.; Korol, A. Genetics of phenotypic plasticity: QTL analysis in barley, Hordeum vulgare. Heredity 2009, 102, 163–173. [Google Scholar] [CrossRef] [Green Version]
- Stratton, D.A. Reaction norm functions and QTL-environment interactions for flowering time in Arabidopsis thaliana. Heredity 1998, 81, 144–155. [Google Scholar] [CrossRef]
- Forsberg, S.K.G.; Carlborg, Ö. On the relationship between epistasis and genetic variance heterogeneity. J. Exp. Bot. 2017, 68, 5431–5438. [Google Scholar] [CrossRef]
- Cordell, H.J. Epistasis: What it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum. Mol. Genet. 2002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlborg, Ö.; Haley, C.S. Epistasis: Too often neglected in complex trait studies? Nat. Rev. Genet. 2004, 5, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Carlborg, O.; Jacobsson, L.; Ahgren, P.; Siegel, P.; Andersson, L. Epistasis and the release of genetic variation during long-term selection. Nat. Genet. 2006, 38, 418–420. [Google Scholar] [CrossRef] [PubMed]
- Josephs, E.B. Determining the evolutionary forces shaping G × E. New Phytol. 2018, 219, 31–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, A.; Sinha, H. Gene-gene and gene-environment interactions in complex traits in yeast. Yeast 2018, 35, 403–416. [Google Scholar] [CrossRef] [Green Version]
- Bhatia, A.; Yadav, A.; Zhu, C.; Gagneur, J.; Radhakrishnan, A.; Steinmetz, L.M.; Bhanot, G.; Sinha, H. Yeast Growth Plasticity Is Regulated by Environment-Specific Multi-QTL Interactions. G3 Genes Genomes Genet. 2014, 4, 769–777. [Google Scholar] [CrossRef] [Green Version]
- Yadav, A.; Dhole, K.; Sinha, H. Differential regulation of cryptic genetic variation shapes the genetic interactome underlying complex traits. Genome Biol. Evol. 2016, 8, 3559–3573. [Google Scholar] [CrossRef] [Green Version]
- Zan, Y.; Carlborg, Ö. Dynamic genetic architecture of yeast response to environmental perturbation shed light on origin of cryptic genetic variation. PLoS Genet. 2020, 16, e1008801. [Google Scholar] [CrossRef]
- Forsberg, S.K.G.; Bloom, J.S.; Sadhu, M.J.; Kruglyak, L.; Carlborg, Ö. Accounting for genetic interactions improves modeling of individual quantitative trait phenotypes in yeast. Nat. Genet. 2017, 49, 497–503. [Google Scholar] [CrossRef] [Green Version]
- Bloom, J.S.; Kotenko, I.; Sadhu, M.J.; Treusch, S.; Albert, F.W.; Kruglyak, L. Genetic interactions contribute less than additive effects to quantitative trait variation in yeast. Nat. Commun. 2015, 6, 8712. [Google Scholar] [CrossRef] [Green Version]
- Yano, K.; Morinaka, Y.; Wang, F.; Huang, P.; Takehara, S.; Hirai, T.; Ito, A.; Koketsu, E.; Kawamura, M.; Kotake, K.; et al. GWAS with principal component analysis identifies a gene comprehensively controlling rice architecture. Proc. Natl. Acad. Sci. USA 2019, 116, 21262–21267. [Google Scholar] [CrossRef] [PubMed]
- Becker, H.C.; Leon, J. Stability Analysis in Plant Breeding. Plant Breed. 1988, 101, 1–23. [Google Scholar] [CrossRef]
- Lian, L.; De Los Campos, G. FW: An R package for Finlay-Wilkinson regression that incorporates genomic/pedigree information and covariance structures between environments. G3 Genes Genomes Genet. 2016, 6, 589–597. [Google Scholar] [CrossRef] [Green Version]
- Finlay, K.; Wilkinson, G. The analysis of adaptation in a plant-breeding programme. Aust. J. Agric. Res. 1963, 14, 742. [Google Scholar] [CrossRef] [Green Version]
- Weber, S.L.; Scheiner, S.M. The genetics of phenotypic plasticity. IV. Chromosomal localization. J. Evol. Biol. 1992, 5, 109–120. [Google Scholar] [CrossRef]
- Ungerer, M.C.; Halldorsdottir, S.S.; Purugganan, M.D.; Mackay, T.F.C. Genotype-environment interactions at quantitative trait loci affecting inflorescence development in Arabidopsis thaliana. Genetics 2003, 165, 353–365. [Google Scholar] [PubMed]
- Aulchenko, Y.S.; Ripke, S.; Isaacs, A.; van Duijn, C.M. GenABEL: An R library for genome-wide association analysis. Bioinformatics 2007, 23, 1294–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Ferreira, T.; Morris, A.P.; Medland, S.E.; Madden, P.A.F.; Heath, A.C.; Martin, N.G.; Montgomery, G.W.; Weedon, M.N.; Loos, R.J.; et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012, 44, 369–375. [Google Scholar] [CrossRef]
- Li, M.X.; Yeung, J.M.Y.; Cherny, S.S.; Sham, P.C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 2012, 131, 747–756. [Google Scholar] [CrossRef] [Green Version]
- Miyajima, I.; Nakafuku, M.; Nakayama, N.; Brenner, C.; Miyajima, A.; Kaibuchi, K.; Arai, K.; Kaziro, Y.; Matsumoto, K. GPA1, a haploid-specific essential gene, encodes a yeast homolog of mammalian G protein which may be involved in mating factor signal transduction. Cell 1987, 50, 1011–1019. [Google Scholar] [CrossRef]
- Bourgarel, D.; Nguyen, C.-C.; Bolotin-Fukuhara, M. HAP4, the glucose-repressed regulated subunit of the HAP transcriptional complex involved in the fermentation-respiration shift, has a functional homologue in the respiratory yeast Kluyveromyces lactis. Mol. Microbiol. 1999, 31, 1205–1215. [Google Scholar] [CrossRef]
- Zhang, L.; Hach, A. Molecular mechanism of heme signaling in yeast: The transcriptional activator Hap1 serves as the key mediator. Cell. Mol. Life Sci. 1999, 56, 415–426. [Google Scholar] [CrossRef]
- Creusot, F.; Verdière, J.; Gaisne, M.; Slonimski, P.P. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. J. Mol. Biol. 1988, 204, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Langhendries, J.-L.; Watzinger, P.; Kötter, P.; Entian, K.-D.; Lafontaine, D.L.J. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015, 43, 2242–2258. [Google Scholar] [CrossRef] [Green Version]
- Wickner, R.B. MKT1, a nonessential Saccharomyces cerevisiae gene with a temperature-dependent effect on replication of M2 double-stranded RNA. J. Bacteriol. 1987, 169, 4941–4945. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Lin, B.K.; Wood, D.R.; Tamanoi, F. IRA2, an upstream negative regulator of RAS in yeast, is a RAS GTPase-activating protein. Proc. Natl. Acad. Sci. USA 1991, 88, 468–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.T.; Coradini, A.L.V.; Shen, A.; Ehrenreich, I.M. Layers of Cryptic Genetic Variation Underlie a Yeast Complex Trait. Genetics 2019, 211, 1469–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albert, F.W.; Bloom, J.S.; Siegel, J.; Day, L.; Kruglyak, L. Genetics of trans-regulatory variation in gene expression. eLife 2018, 7, e35471. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zan, Y.; Carlborg, Ö. Dissecting the Genetic Regulation of Yeast Growth Plasticity in Response to Environmental Changes. Genes 2020, 11, 1279. https://doi.org/10.3390/genes11111279
Zan Y, Carlborg Ö. Dissecting the Genetic Regulation of Yeast Growth Plasticity in Response to Environmental Changes. Genes. 2020; 11(11):1279. https://doi.org/10.3390/genes11111279
Chicago/Turabian StyleZan, Yanjun, and Örjan Carlborg. 2020. "Dissecting the Genetic Regulation of Yeast Growth Plasticity in Response to Environmental Changes" Genes 11, no. 11: 1279. https://doi.org/10.3390/genes11111279




