Metabolic Engineering of Wine Strains of Saccharomyces cerevisiae
Abstract
1. Introduction
2. Directions of Metabolic Engineering of Wine Yeast Strains
2.1. Wine Fermentation and Processing Efficiency
2.2. “Biocontrol” Strains
2.3. “Low Alcohol” Yeasts
2.4. Aroma and Taste of Wine
2.4.1. Volatile Esters
2.4.2. Monoterpenoids
2.4.3. Diacetyl Removal
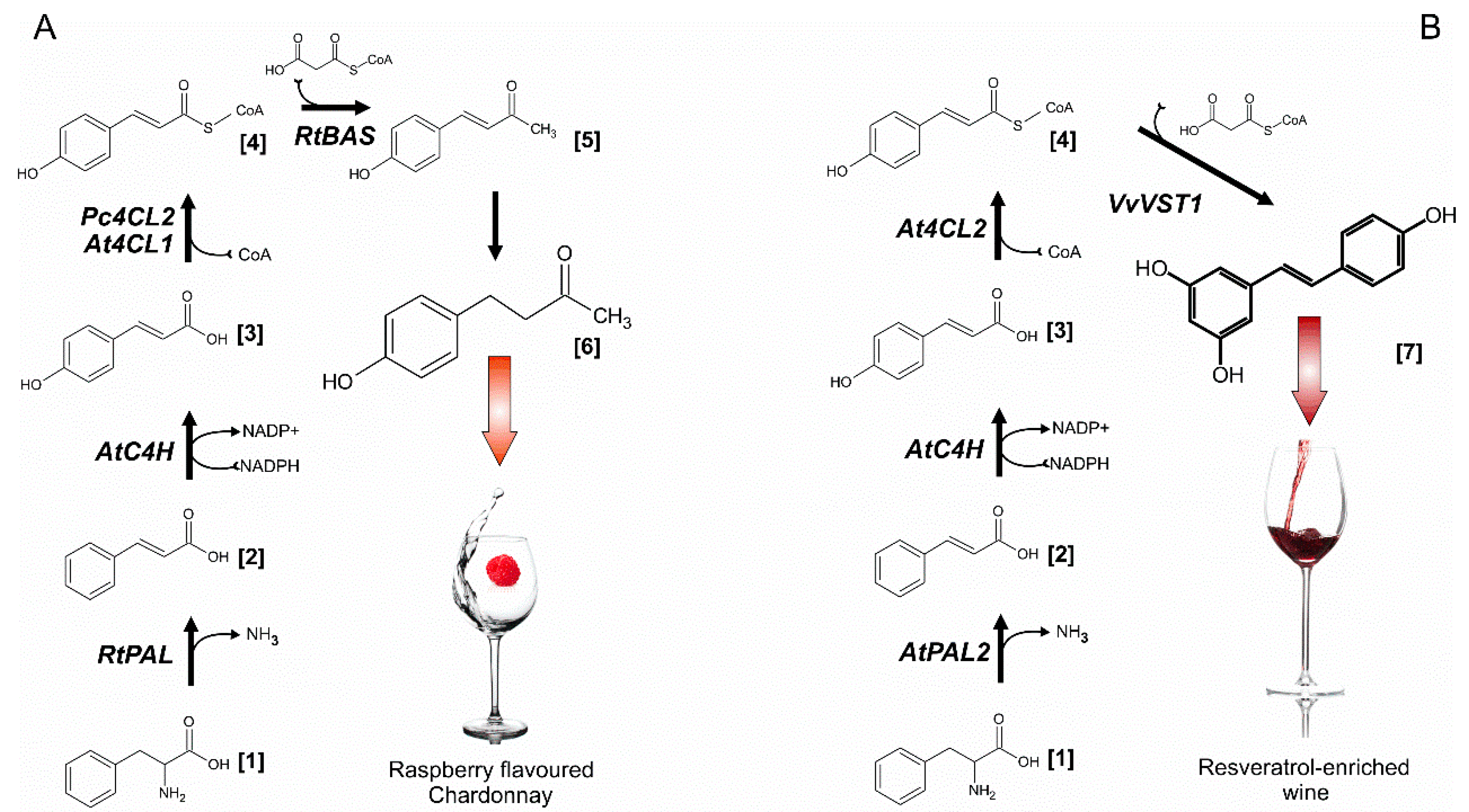
2.4.4. “Raspberry” Yeast
2.4.5. Resveratrol-Producing Yeast
2.5. Flor Yeast Strains
2.6. Commercial GM Wine Yeast Strains
3. CRISPR-Cas for Wine Yeast
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chambers, P.J.; Pretorius, I.S. Fermenting Knowledge: The History of Winemaking, Science and Yeast Research. EMBO Rep. 2010, 11, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Sicard, D.; Legras, J.L. Bread, Beer and Wine: Yeast Domestication in the Saccharomyces Sensu Stricto Complex. Comptes Rendus Biol. 2011, 334, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Fleet, G. Yeast Interactions and Wine Flavour. Int. J. Food Microbiol. 2003, 86, 11–22. [Google Scholar] [CrossRef]
- Fleet, G.H. Wine Microbiology and Biotechnology; CRC Press: Boca Raton, FL, USA, 1993; pp. 374–375. [Google Scholar]
- Divol, B.; Bauer, F.F. 2-Metabolic Engineering of Wine Yeast and Advances in Yeast Selection Methods for Improved Wine Quality. In Woodhead Publishing Series in Food Science, Technology and Nutrition; Reynolds, A.G.B.T.-M.W.Q., Ed.; Woodhead Publishing: Cambridge, UK, 2010; pp. 34–59. [Google Scholar] [CrossRef]
- Bisson, L.F. The Biotechnology of Wine Yeast. Food Biotechnol. 2004, 18, 63–96. [Google Scholar] [CrossRef]
- OIV—OIV 2019 Report on the World Vitivinicultural Situation. Available online: http://www.oiv.int/en/oiv-life/oiv-2019-report-on-the-world-vitivinicultural-situation (accessed on 31 March 2020).
- Steensels, J.; Snoek, T.; Meersman, E.; Nicolino, M.P.; Voordeckers, K.; Verstrepen, K.J. Improving Industrial Yeast Strains: Exploiting Natural and Artificial Diversity. FEMS Microbiol. Rev. 2014, 38, 947–995. [Google Scholar] [CrossRef]
- Marsit, S.; Dequin, S. Diversity and Adaptive Evolution of Saccharomyces Wine Yeast: A Review. FEMS Yeast Res. 2015, 15, fov067. [Google Scholar] [CrossRef]
- McBryde, C.; Gardner, J.M.; De Barros Lopes, M.; Jiranek, V. Generation of Novel Wine Yeast Strains by Adaptive Evolution. Am. J. Enol. Vitic. 2006, 57, 423–430. [Google Scholar]
- Kutyna, D.R.; Varela, C.; Stanley, G.A.; Borneman, A.R.; Henschke, P.A.; Chambers, P.J. Adaptive Evolution of Saccharomyces cerevisiae to Generate Strains with Enhanced Glycerol Production. Appl. Microbiol. Biotechnol. 2012, 93, 1175–1184. [Google Scholar] [CrossRef]
- Sandberg, T.E.; Salazar, M.J.; Weng, L.L.; Palsson, B.O.; Feist, A.M. The Emergence of Adaptive Laboratory Evolution as an Efficient Tool for Biological Discovery and Industrial Biotechnology. Metab. Eng. 2019, 56, 1–16. [Google Scholar] [CrossRef]
- Tilloy, V.; Ortiz-Julien, A.; Dequin, S. Reduction of Ethanol Yield and Improvement of Glycerol Formation by Adaptive Evolution of the Wine Yeast Saccharomyces cerevisiae under Hyperosmotic Conditions. Appl. Environ. Microbiol. 2014, 80, 2623–2632. [Google Scholar] [CrossRef]
- Betlej, G.; Bator, E.; Oklejewicz, B.; Potocki, L.; Górka, A.; Slowik-Borowiec, M.; Czarny, W.; Domka, W.; Kwiatkowska, A. Long-Term Adaption to High Osmotic Stress as a Tool for Improving Enological Characteristics in Industrial Wine Yeast. Genes 2020, 11, 576. [Google Scholar] [CrossRef] [PubMed]
- Borneman, A.R.; Schmidt, S.A.; Pretorius, I.S. At the Cutting-Edge of Grape and Wine Biotechnology. Trends Genet. 2013, 29, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Borneman, A.R.; Pretorius, I.S.; Chambers, P.J. Comparative Genomics: A Revolutionary Tool for Wine Yeast Strain Development. Curr. Opin. Biotechnol. 2013, 24, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, I.S. Synthetic Genome Engineering Forging New Frontiers for Wine Yeast. Crit. Rev. Biotechnol. 2017, 37, 112–136. [Google Scholar] [CrossRef] [PubMed]
- Bisson, L.F.; Karpel, J.E.; Ramakrishnan, V.; Joseph, L. Functional Genomics of Wine Yeast Saccharomyces cerevisiae. Adv. Food Nutr. Res. 2007, 53, 65–121. [Google Scholar] [CrossRef] [PubMed]
- Eldarov, M.A.; Kishkovskaia, S.A.; Tanaschuk, T.N.; Mardanov, A.V. Genomics and Biochemistry of Saccharomyces cerevisiae Wine Yeast Strains. Biochemistry 2016, 81, 1650–1668. [Google Scholar] [CrossRef]
- Husnik, J.I.; Volschenk, H.; Bauer, J.; Colavizza, D.; Luo, Z.; van Vuuren, H.J.J. Metabolic Engineering of Malolactic Wine Yeast. Metab. Eng. 2006, 8, 315–323. [Google Scholar] [CrossRef]
- Coulon, J.; Husnik, J.I.; Inglis, D.L.; van der Merwe, G.K.; Lonvaud, A.; Erasmus, D.J.; van Vuuren, H.J.J. Metabolic Engineering of Saccharomyces cerevisiae to Minimize the Production of Ethyl Carbamate in Wine. Am. J. Enol. Vitic. 2006, 57, 113–124. [Google Scholar]
- Liu, S.-Q. A Review: Malolactic Fermentation in Wine—Beyond Deacidification. J. Appl. Microbiol. 2002, 92, 589–601. [Google Scholar] [CrossRef]
- Pretorius, I.S. Tailoring Wine Yeast for the New Millennium: Novel Approaches to the Ancient Art of Winemaking. Yeast 2000, 16, 675–729. [Google Scholar] [CrossRef]
- Cebollero, E.; Gonzalez-Ramos, D.; Tabera, L.; Gonzalez, R. Transgenic Wine Yeast Technology Comes of Age: Is It Time for Transgenic Wine? Biotechnol. Lett. 2007, 29, 191–200. [Google Scholar] [CrossRef]
- Vigentini, I.; Gonzalez, R.; Tronchoni, J. Genetic Improvement of Wine Yeasts BT—Yeasts in the Production of Wine; Romano, P., Ciani, M., Fleet, G.H., Eds.; Springer: New York, NY, USA, 2019; pp. 315–342. [Google Scholar] [CrossRef]
- Donalies, U.E.B.; Nguyen, H.T.T.; Stahl, U.; Nevoigt, E. Improvement of Saccharomyces Yeast Strains Used in Brewing, Wine Making and Baking. Adv. Biochem. Eng. Biotechnol. 2008, 111, 67–98. [Google Scholar] [PubMed]
- Schuller, D.; Casal, M. The Use of Genetically Modified Saccharomyces cerevisiae Strains in the Wine Industry. Appl. Microbiol. Biotechnol. 2005, 68, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Bisson, L.F. Stuck and Sluggish Fermentations. Am. J. Enol. Vitic. 1999, 50, 107–119. [Google Scholar] [CrossRef]
- Mendes-Ferreira, A.; Barbosa, C.; Lage, P.; Mendes-Faia, A. The Impact of Nitrogen on Yeast Fermentation and Wine Quality. Cienc. E Tec. Vitivinic. 2011, 26, 17–32. [Google Scholar]
- Gobert, A.; Tourdot-Maréchal, R.; Sparrow, C.; Morge, C.; Alexandre, H. Influence of Nitrogen Status in Wine Alcoholic Fermentation. Food Microbiol. 2019, 83, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Tesnière, C.; Delobel, P.; Pradal, M.; Blondin, B. Impact of Nutrient Imbalance on Wine Alcoholic Fermentations: Nitrogen Excess Enhances Yeast Cell Death in Lipid-Limited Must. PLoS ONE 2013, 8, e61645. [Google Scholar] [CrossRef]
- Peter, J.J.; Watson, T.L.; Walker, M.E.; Gardner, J.M.; Lang, T.A.; Borneman, A.; Forgan, A.; Tran, T.; Jiranek, V. Use of a Wine Yeast Deletion Collection Reveals Genes That Influence Fermentation Performance under Low-Nitrogen Conditions. FEMS Yeast Res. 2018, 18, foy009. [Google Scholar] [CrossRef]
- Zhang, J.; Astorga, M.A.; Gardner, J.M.; Walker, M.E.; Grbin, P.R.; Jiranek, V. Disruption of the Cell Wall Integrity Gene ECM33 Results in Improved Fermentation by Wine Yeast. Metab. Eng. 2018, 45, 255–264. [Google Scholar] [CrossRef]
- Brown, S.L.; Stockdale, V.J.; Pettolino, F.; Pocock, K.F.; De Barros Lopes, M.; Williams, P.J.; Bacic, A.; Fincher, G.B.; Høj, P.B.; Waters, E.J. Reducing Haziness in White Wine by Overexpression of Saccharomyces cerevisiae Genes YOL155c and YDR055w. Appl. Microbiol. Biotechnol. 2007, 73, 1363–1376. [Google Scholar] [CrossRef]
- Gonzalez-Ramos, D.; Cebollero, E.; Gonzalez, R. A Recombinant Saccharomyces cerevisiae Strain Overproducing Mannoproteins Stabilizes Wine against Protein Haze. Appl. Environ. Microbiol. 2008, 74, 5533–5540. [Google Scholar] [CrossRef] [PubMed]
- van Rensburg, P.; Strauss, M.L.A.; Lambrechts, M.G.; Cordero Otero, R.R.; Pretorius, I.S. The Heterologous Expression of Polysaccharidase-Encoding Genes with Oenological Relevance in Saccharomyces cerevisiae. J. Appl. Microbiol. 2007, 103, 2248–2257. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Marti, E.; Zuzuarregui, A.; Ridaura, I.; Lozano, N.; del Olmo, M. Genetic Manipulation of HSP26 and YHR087W Stress Genes May Improve Fermentative Behaviour in Wine Yeasts under Vinification Conditions. Int. J. Food Microbiol. 2009, 130, 122–130. [Google Scholar] [CrossRef]
- Branco, P.; Sabir, F.; Diniz, M.; Carvalho, L.; Albergaria, H.; Prista, C. Biocontrol of Brettanomyces/Dekkera bruxellensis in Alcoholic Fermentations Using Saccharomycin-Overproducing Saccharomyces cerevisiae Strains. Appl. Microbiol. Biotechnol. 2019, 103, 3073–3083. [Google Scholar] [CrossRef] [PubMed]
- Malherbe, D.F.; du Toit, M.; Cordero Otero, R.R.; van Rensburg, P.; Pretorius, I.S. Expression of the Aspergillus niger Glucose Oxidase Gene in Saccharomyces cerevisiae and Its Potential Applications in Wine Production. Appl. Microbiol. Biotechnol. 2003, 61, 502–511. [Google Scholar] [CrossRef]
- Henricsson, C.; de Jesus Ferreira, M.C.; Hedfalk, K.; Elbing, K.; Larsson, C.; Bill, R.M.; Norbeck, J.; Hohmann, S.; Gustafsson, L. Engineering of a Novel Saccharomyces cerevisiae Wine Strain with a Respiratory Phenotype at High External Glucose Concentrations. Appl. Environ. Microbiol. 2005, 71, 6185–6192. [Google Scholar] [CrossRef]
- Drewke, C.; Thielen, J.; Ciriacy, M. Ethanol Formation in Adh0 Mutants Reveals the Existence of a Novel Acetaldehyde-Reducing Activity in Saccharomyces cerevisiae. J. Bacteriol. 1990, 172, 3909–3917. [Google Scholar] [CrossRef]
- Compagno, C.; Brambilla, L.; Capitanio, D.; Boschi, F.; Ranzi, B.M.; Porro, D. Alterations of the Glucose Metabolism in a Triose Phosphate Isomerase-Negative Saccharomyces cerevisiae Mutant. Yeast 2001, 18, 663–670. [Google Scholar] [CrossRef]
- Nevoigt, E.; Stahl, U. Reduced Pyruvate Decarboxylase and Increased Glycerol-3-Phosphate Dehydrogenase [NAD+] Levels Enhance Glycerol Production in Saccharomyces cerevisiae. Yeast 1996, 12, 1331–1337. [Google Scholar] [CrossRef]
- Varela, C.; Kutyna, D.R.; Solomon, M.R.; Black, C.A.; Borneman, A.; Henschke, P.A.; Pretorius, I.S.; Chambers, P.J. Evaluation of Gene Modification Strategies for the Development of Low-Alcohol-Wine Yeasts. Appl. Environ. Microbiol. 2012, 78, 6068–6077. [Google Scholar] [CrossRef]
- Rossouw, D.; Heyns, E.H.; Setati, M.E.; Bosch, S.; Bauer, F.F. Adjustment of Trehalose Metabolism in Wine Saccharomyces cerevisiae Strains to Modify Ethanol Yields. Appl. Environ. Microbiol. 2013, 79, 5197–5207. [Google Scholar] [CrossRef] [PubMed]
- Verstrepen, K.J.; Van Laere, S.D.M.; Vanderhaegen, B.M.P.; Derdelinckx, G.; Dufour, J.-P.; Pretorius, I.S.; Winderickx, J.; Thevelein, J.M.; Delvaux, F.R. Expression Levels of the Yeast Alcohol Acetyltransferase Genes ATF1, Lg-ATF1, and ATF2 Control the Formation of a Broad Range of Volatile Esters. Appl. Environ. Microbiol. 2003, 69, 5228–5237. [Google Scholar] [CrossRef] [PubMed]
- Pardo, E.; Rico, J.; Gil, J.V.; Orejas, M. De Novo Production of Six Key Grape Aroma Monoterpenes by a Geraniol Synthase-Engineered, S. cerevisiae Wine Strain. Microb. Cell Fact. 2015, 14, 136. [Google Scholar] [CrossRef] [PubMed]
- Zietsman, A.J.J.; de Klerk, D.; van Rensburg, P. Coexpression of Alpha-l-Arabinofuranosidase and Beta-Glucosidase in Saccharomyces cerevisiae. FEMS Yeast Res. 2011, 11, 88–103. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Guo, X.; Shi, T.; Hu, Z.; Chen, Y.; Du, L.; Xiao, D. Reducing Diacetyl Production of Wine by Overexpressing BDH1 and BDH2 in Saccharomyces uvarum. J. Ind. Microbiol. Biotechnol. 2017, 44, 1541–1550. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Lloyd, N.D.R.; Pretorius, I.S.; Borneman, A.R. Heterologous Production of Raspberry Ketone in the Wine Yeast Saccharomyces cerevisiae via Pathway Engineering and Synthetic Enzyme Fusion. Microb. Cell Fact. 2016, 15, 49. [Google Scholar] [CrossRef]
- Li, M.; Schneider, K.; Kristensen, M.; Borodina, I.; Nielsen, J. Engineering Yeast for High-Level Production of Stilbenoid Antioxidants. Sci. Rep. 2016, 6, 36827. [Google Scholar] [CrossRef]
- Fidalgo, M.; Barrales, R.R.; Ibeas, J.I.; Jimenez, J. Adaptive Evolution by Mutations in the FLO11 Gene. Proc. Natl. Acad. Sci. USA 2006, 103, 11228–11233. [Google Scholar] [CrossRef]
- Moreno-García, J.; Coi, A.L.; Zara, G.; García-Martínez, T.; Mauricio, J.C.; Budroni, M. Study of the Role of the Covalently Linked Cell Wall Protein (Ccw14p) and Yeast Glycoprotein (Ygp1p) within Biofilm Formation in a Flor Yeast Strain. FEMS Yeast Res. 2018, 18, foy005. [Google Scholar] [CrossRef]
- Fierro-Risco, J.; Rincón, A.M.; Benítez, T.; Codón, A.C. Overexpression of Stress-Related Genes Enhances Cell Viability and Velum Formation in Sherry Wine Yeasts. Appl. Microbiol. Biotechnol. 2013, 97, 6867–6881. [Google Scholar] [CrossRef]
- Vigentini, I.; Gebbia, M.; Belotti, A.; Foschino, R.; Roth, F.P. CRISPR/Cas9 System as a Valuable Genome Editing Tool for Wine Yeasts with Application to Decrease Urea Production. Front. Microbiol. 2017, 8, 2194. [Google Scholar] [CrossRef] [PubMed]
- Trindade de Carvalho, B.; Holt, S.; Souffriau, B.; Lopes Brandão, R.; Foulquié-Moreno, M.R.; Thevelein, J.M. Identification of Novel Alleles Conferring Superior Production of Rose Flavor Phenylethyl Acetate Using Polygenic Analysis in Yeast. MBio 2017, 8, e01173-17. [Google Scholar] [CrossRef] [PubMed]
- Mertens, S.; Gallone, B.; Steensels, J.; Herrera-Malaver, B.; Cortebeek, J.; Nolmans, R.; Saels, V.; Vyas, V.K.; Verstrepen, K.J. Reducing Phenolic Off-Flavors through CRISPR-Based Gene Editing of the FDC1 Gene in Saccharomyces cerevisiae x Saccharomyces eubayanus Hybrid Lager Beer Yeasts. PLoS ONE 2019, 14, e0209124. [Google Scholar] [CrossRef]
- Michaelis, S.; Herskowitz, I. The A-Factor Pheromone of Saccharomyces cerevisiae Is Essential for Mating. Mol. Cell. Biol. 1988, 8, 1309–1318. [Google Scholar] [CrossRef]
- Michaelis, S.; Barrowman, J. Biogenesis of the Saccharomyces cerevisiae Pheromone A-Factor, from Yeast Mating to Human Disease. Microbiol. Mol. Biol. Rev. 2012, 76, 626–651. [Google Scholar] [CrossRef]
- Kurjan, J.; Herskowitz, I. Structure of a Yeast Pheromone Gene (MF Alpha): A Putative Alpha-Factor Precursor Contains Four Tandem Copies of Mature Alpha-Factor. Cell 1982, 30, 933–943. [Google Scholar] [CrossRef]
- Ferreira, R.B.; Piçarra-Pereira, M.A.; Monteiro, S.; Loureiro, V.B.; Teixeira, A.R. The Wine Proteins. Trends Food Sci. Technol. 2001, 12, 230–239. [Google Scholar] [CrossRef]
- Dupin, I.V.S.; McKinnon, B.M.; Ryan, C.; Boulay, M.; Markides, A.J.; Jones, G.P.; Williams, P.J.; Waters, E.J. Saccharomyces cerevisiae Mannoproteins That Protect Wine from Protein Haze: Their Release during Fermentation and Lees Contact and a Proposal for Their Mechanism of Action. J. Agric. Food Chem. 2000, 48, 3098–3105. [Google Scholar] [CrossRef]
- Marks, V.D.; Ho Sui, S.J.; Erasmus, D.; Van Der Merwe, G.K.; Brumm, J.; Wasserman, W.W.; Bryan, J.; Van Vuuren, H.J.J. Dynamics of the Yeast Transcriptome during Wine Fermentation Reveals a Novel Fermentation Stress Response. Fems Yeast Res. 2008, 8, 35–52. [Google Scholar] [CrossRef]
- Bartowsky, E.J. Bacterial Spoilage of Wine and Approaches to Minimize It. Lett. Appl. Microbiol. 2009, 48, 149–156. [Google Scholar] [CrossRef]
- Du Toit, M.; Pretorius, I.S. Microbial Spoilage and Preservation of Wine: Using Weapons from Nature’s Own Arsenal—A Review. S. Afr. J. Enol. Vitic. 2000, 21. [Google Scholar] [CrossRef][Green Version]
- Malfeito-Ferreira, M.; Silva, A.C. Spoilage Yeasts in Wine Production BT—Yeasts in the Production of Wine; Romano, P., Ciani, M., Fleet, G.H., Eds.; Springer: New York, NY, USA, 2019; pp. 375–394. [Google Scholar] [CrossRef]
- Raybaudi-Massilia, R.M.; Mosqueda-Melgar, J.; Soliva-Fortuny, R.; Martín-Belloso, O. Control of Pathogenic and Spoilage Microorganisms in Fresh-Cut Fruits and Fruit Juices by Traditional and Alternative Natural Antimicrobials. Compr. Rev. Food Sci. Food Saf. 2009, 8, 157–180. [Google Scholar] [CrossRef]
- OUGH, C.; Crowell, E.A. Use of Sulfur Dioxide in Winemaking. J. Food Sci. 2006, 52, 386–388. [Google Scholar] [CrossRef]
- de Ullivarri, M.F.; Mendoza, L.M.; Raya, R.R. Killer Activity of Saccharomyces cerevisiae Strains: Partial Characterization and Strategies to Improve the Biocontrol Efficacy in Winemaking. Antonie Van Leeuwenhoek 2014, 106, 865–878. [Google Scholar] [CrossRef]
- Mehlomakulu, N.N.; Prior, K.J.; Setati, M.E.; Divol, B. Candida Pyralidae Killer Toxin Disrupts the Cell Wall of Brettanomyces bruxellensis in Red Grape Juice. J. Appl. Microbiol. 2017, 122, 747–758. [Google Scholar] [CrossRef]
- Schoeman, H.; Vivier, M.A.; Du Toit, M.; Dicks, L.M.; Pretorius, I.S. The Development of Bactericidal Yeast Strains by Expressing the Pediococcus acidilactici Pediocin Gene (PedA) in Saccharomyces cerevisiae. Yeast 1999, 15, 647–656. [Google Scholar] [CrossRef]
- Carstens, M.; Vivier, M.; RENSBURG, P.; Pretorius, I. Overexpression, Secretion and Antifungal Activity of the Saccharomyces cerevisiae Chitinase. Ann. Microbiol. 2003, 53, 15–28. [Google Scholar]
- Branco, P.; Francisco, D.; Monteiro, M.; Almeida, M.G.; Caldeira, J.; Arneborg, N.; Prista, C.; Albergaria, H. Antimicrobial Properties and Death-Inducing Mechanisms of Saccharomycin, a Biocide Secreted by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2017, 101, 159–171. [Google Scholar] [CrossRef]
- Albergaria, H.; Francisco, D.; Gori, K.; Arneborg, N.; Gírio, F. Saccharomyces cerevisiae CCMI 885 Secretes Peptides That Inhibit the Growth of Some Non-Saccharomyces Wine-Related Strains. Appl. Microbiol. Biotechnol. 2010, 86, 965–972. [Google Scholar] [CrossRef]
- Caballero, A.; Segura, A. The Quest for Lower Alcoholic Wines. Microb. Biotechnol. 2017, 10, 238–241. [Google Scholar] [CrossRef]
- Goold, H.D.; Kroukamp, H.; Williams, T.C.; Paulsen, I.T.; Varela, C.; Pretorius, I.S. Yeast’s Balancing Act between Ethanol and Glycerol Production in Low-Alcohol Wines. Microb. Biotechnol. 2017, 10, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Dequin, S.; Baptista, E.; Barre, P. Acidification of Grape Musts by Saccharomyces cerevisiae Wine Yeast Strains Genetically Engineered to Produce Lactic Acid. Am. J. Enol. Vitic. 1999, 50, 45–50. [Google Scholar]
- De Deken, R.H. The Crabtree Effect: A Regulatory System in Yeast. J. Gen. Microbiol. 1966, 44, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Cambon, B.; Monteil, V.; Remize, F.; Camarasa, C.; Dequin, S. Effects of GPD1 Overexpression in Saccharomyces cerevisiae Commercial Wine Yeast Strains Lacking ALD6 Genes. Appl. Environ. Microbiol. 2006, 72, 4688–4694. [Google Scholar] [CrossRef] [PubMed]
- Lambrechts, M.; Pretorius, I. Yeast and Its Importance to Wine Aroma. S. Afr. J. Enol. Vitic. 2000, 21, 97–129. [Google Scholar] [CrossRef]
- HG, S.; SD, K.; Paltauf, F. Molecular Cloning, Primary Structure and Disruption of the Structural Gene of Aldolase from Saccharomyces cerevisiae. Eur. J. Biochem. 1989, 180, 301–308. [Google Scholar]
- Compagno, C.; BM, R.; Martegani, E. The Promoter of Saccharomyces cerevisiae FBA1 Gene Contains a Single Positive Upstream Regulatory Element. FEBS Lett. 1991, 293, 97–100. [Google Scholar] [CrossRef]
- WU, C.-F.; Yang, J.; WANG, F.; WANG, X.-X. Resveratrol: Botanical Origin, Pharmacological Activity and Applications. Chin. J. Nat. Med. 2013, 11, 1–15. [Google Scholar] [CrossRef]
- Jeandet, P.; Bessis, R.; Sbaghi, M.; Meunier, P. Production of the Phytoalexin Resveratrol by Grapes as a Response to Botrytis Attack Under Natural Conditions. J. Phytopathol. 1995, 143, 135–139. [Google Scholar] [CrossRef]
- Goldberg, D.M.; Yan, J.; Ng, E.; Diamandis, E.P.; Karumanchiri, A.; Soleas, G.; Waterhouse, A.L. A Global Survey of Trans-Resveratrol Concentrations in Commercial Wines. Am. J. Enol. Vitic. 1995, 46, 159–165. [Google Scholar]
- Guerrero, R.; Garcia-Parrilla, M.; Puertas, B.; Cantos-Villar, E. Wine, Resveratrol and Health: A Review. Nat. Prod. Commun. 2009, 4, 635–658. [Google Scholar] [CrossRef] [PubMed]
- Jeandet, P.; Delaunois, B.; Conreux, A.; Donnez, D.; Nuzzo, V.; Cordelier, S.; Clément, C.; Courot, E. Biosynthesis, Metabolism, Molecular Engineering, and Biological Functions of Stilbene Phytoalexins in Plants. Biofactors 2010, 36, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.V.W.; Armstrong, G.O.; van der Merwe, M.J.; Lambrechts, M.G.; Vivier, M.A.; Pretorius, I.S. Metabolic Engineering of Saccharomyces cerevisiae for the Synthesis of the Wine-Related Antioxidant Resveratrol. FEMS Yeast Res. 2003, 4, 79–85. [Google Scholar] [CrossRef]
- Li, M.; Kildegaard, K.R.; Chen, Y.; Rodriguez, A.; Borodina, I.; Nielsen, J. De Novo Production of Resveratrol from Glucose or Ethanol by Engineered Saccharomyces cerevisiae. Metab. Eng. 2015, 32, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, H. Flor Yeasts of Saccharomyces cerevisiae-Their Ecology, Genetics and Metabolism. Int. J. Food Microbiol. 2013, 167, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Legras, J.-L.; Moreno-Garcia, J.; Zara, S.; Zara, G.; Garcia-Martinez, T.; Mauricio, J.C.; Mannazzu, I.; Coi, A.L.; Bou Zeidan, M.; Dequin, S.; et al. Flor Yeast: New Perspectives Beyond Wine Aging. Front. Microbiol. 2016, 7, 503. [Google Scholar] [CrossRef] [PubMed]
- Ángeles Pozo-Bayón, M.; Victoria Moreno-Arribas, M. Sherry Wines. Adv. Food Nutr. Res. 2011, 63, 17–40. [Google Scholar] [CrossRef]
- Ssaenko, N.F.; Kozub, G.I.; Averbuh, B.Y.; Shur, I.M. Vino Herez I Tehnologyia Proizvodstav; Karta Moldavanska: Kishinev, Russian, 1975. [Google Scholar]
- Kishkovskaia, S.A.; Eldarov, M.A.; Dumina, M.V.; Tanashchuk, T.N.; Ravin, N.V.; Mardanov, A.V. Flor Yeast Strains from Culture Collection: Genetic Diversity and Physiological and Biochemical Properties. Appl. Biochem. Microbiol. 2017, 53, 359–367. [Google Scholar] [CrossRef]
- Benítez, T.; Rincón, A.M.; Codón, A.C. Chapter 3—Yeasts Used in Biologically Aged Wines; Carrascosa, A.V., Muñoz, R., González, R.B.T.-M.W.M., Eds.; Academic Press: San Diego, UK, 2011; pp. 51–84. [Google Scholar] [CrossRef]
- Charpentier, C.; Colin, A.; Alais, A.; Legras, J.L. French Jura Flor Yeasts: Genotype and Technological Diversity. Antonie Van Leeuwenhoekint. J. Gen. Mol. Microbiol. 2009, 95, 263–273. [Google Scholar] [CrossRef]
- Legras, J.-L.; Galeote, V.; Bigey, F.; Camarasa, C.; Marsit, S.; Nidelet, T.; Sanchez, I.; Couloux, A.; Guy, J.; Franco-Duarte, R.; et al. Adaptation of S. cerevisiae to Fermented Food Environments Reveals Remarkable Genome Plasticity and the Footprints of Domestication. Mol. Biol. Evol. 2018, 35, 1712–1727. [Google Scholar] [CrossRef]
- Eldarov, M.A.; Beletsky, A.V.; Tanashchuk, T.N.; Kishkovskaya, S.A.; Ravin, N.V.; Mardanov, A.V. Whole-Genome Analysis of Three Yeast Strains Used for Production of Sherry-like Wines Revealed Genetic Traits Specific to Flor Yeasts. Front. Microbiol. 2018, 9, 965. [Google Scholar] [CrossRef] [PubMed]
- Coi, A.L.; Bigey, F.; Mallet, S.; Marsit, S.; Zara, G.; Gladieux, P.; Galeote, V.; Budroni, M.; Dequin, S.; Legras, J.L. Genomic Signatures of Adaptation to Wine Biological Ageing Conditions in Biofilm-Forming Flor Yeasts. Mol. Ecol. 2017, 26, 2150–2166. [Google Scholar] [CrossRef] [PubMed]
- Fidalgo, M.; Barrales, R.R.; Jimenez, J. Coding Repeat Instability in the FLO11 Gene of Saccharomyces Yeasts. Yeast 2008, 25, 879–889. [Google Scholar] [CrossRef] [PubMed]
- Espinazo-Romeu, M.; Cantoral, J.M.; Matallana, E.; Aranda, A. Btn2p Is Involved in Ethanol Tolerance and Biofilm Formation in Flor Yeast. FEMS Yeast Res. 2008, 8, 1127–1136. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zara, S.; Antonio Farris, G.; Budroni, M.; Bakalinsky, A.T. HSP12 Is Essential for Biofilm Formation by a Sardinian Wine Strain of S. cerevisiae. Yeast 2002, 19, 269–276. [Google Scholar] [CrossRef]
- Kovács, M.; Stuparevic, I.; Mrsa, V.; Maráz, A. Characterization of Ccw7p Cell Wall Proteins and the Encoding Genes of Saccharomyces cerevisiae Wine Yeast Strains: Relevance for Flor Formation. FEMS Yeast Res. 2008, 8, 1115–1126. [Google Scholar] [CrossRef][Green Version]
- Grossmann, M.; Kießling, F.; Singer, J.; Schoeman, H.; Schröder, M.-B.; von Wallbrunn, C. Genetically Modified Wine Yeasts and Risk Assessment Studies Covering Different Steps within the Wine Making Process. Ann. Microbiol. 2011, 61, 103–115. [Google Scholar] [CrossRef]
- Fraczek, M.G.; Naseeb, S.; Delneri, D. History of Genome Editing in Yeast. Yeast 2018, 35, 361–368. [Google Scholar] [CrossRef]
- Le Borgne, S. Genetic Engineering of Industrial Strains of Saccharomyces cerevisiae. Methods Mol. Biol. 2012, 824, 451–465. [Google Scholar] [CrossRef]
- DiCarlo, J.E.; Norville, J.E.; Mali, P.; Rios, X.; Aach, J.; Church, G.M. Genome Engineering in Saccharomyces cerevisiae Using CRISPR-Cas Systems. Nucleic Acids Res. 2013, 41, 4336–4343. [Google Scholar] [CrossRef]
- Giersch, R.M.; Finnigan, G.C. Yeast Still a Beast: Diverse Applications of CRISPR/Cas Editing Technology in S. Cerevisiae. Yale J. Biol. Med. 2017, 90, 643–651. [Google Scholar] [PubMed]
- Stovicek, V.; Holkenbrink, C.; Borodina, I. CRISPR/Cas System for Yeast Genome Engineering: Advances and Applications. FEMS Yeast Res. 2017, 17, fox030. [Google Scholar] [CrossRef]
- Lian, J.; HamediRad, M.; Zhao, H. Advancing Metabolic Engineering of Saccharomyces cerevisiae Using the CRISPR/Cas System. Biotechnol. J. 2018, 13, e1700601. [Google Scholar] [CrossRef] [PubMed]
- Raschmanová, H.; Weninger, A.; Glieder, A.; Kovar, K.; Vogl, T. Implementing CRISPR-Cas Technologies in Conventional and Non-Conventional Yeasts: Current State and Future Prospects. Biotechnol. Adv. 2018, 36, 641–665. [Google Scholar] [CrossRef] [PubMed]
- Varela, C.; Cárdenas, J.; Melo, F.; Agosin, E. Quantitative Analysis of Wine Yeast Gene Expression Profiles under Winemaking Conditions. Yeast 2005, 22, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Mardanov, A.V.; Eldarov, M.A.; Beletsky, A.V.; Tanashchuk, T.N.; Kishkovskaya, S.A.; Ravin, N.V. Transcriptome Profile of Yeast Strain Used for Biological Wine Aging Revealed Dynamic Changes of Gene Expression in Course of Flor Development. Front. Microbiol. 2020, 11, 538. [Google Scholar] [CrossRef]
- Wight, A.J. Strict EU Ruling on Gene-Edited Crops Squeezes Science. Nature 2018, 563, 15–16. [Google Scholar] [CrossRef]

| Alcohol Fermentation | |
| The efficiency of sugar assimilation and the fermentation process itself | Resistance to osmotic and ethanol stresses |
| Efficiency of nitrogen assimilation | Reduced foam formation |
| General “endurance” and stress resistance | Moderate biomass accumulation |
| The Nutritional Qualities of Wines | |
| Increased Resveratrol content | Reduced content of biogenic amines |
| Reduced Ethyl Carbamate | Reduced alcohol content for low alcohol wines |
| Pest Protection | |
| Optimum sulfur dioxide production | Optimal antimicrobial enzyme production |
| Resistance to antimicrobial agents | Optimal antimicrobial peptide production |
| Wine Processing Technologies | |
| Simplification of wine clarification | Film formation (for technologies of sherry wines) |
| Compact sediment (for champagne technology) | |
| Organoleptic Properties | |
| Ability to release aromatic terpenoids | Ability to release and convert aromatic thiols |
| Increased Glycerol Productiona | Optimized fusel oil production |
| Reduced Volatile Acid Production | Reduced hydrogen sulfide production |
| Strain | Genetic Modification | Oenology-Related Trait | Ref. |
|---|---|---|---|
| ML01 | Overexpression of S. pombe mae1 gene O. oeni mleA gene | Malolactic fermentation | [20] |
| ECMo01 | Overexpression of S. cerevisiae DUR1,2 gene | Reduced ethyl carbamate content | [21] |
| AWRI 1631 | Deletion of MFA2 gene | Improved fermentation efficiency under nitrogen limitation | [32] |
| C911D | Deletion of ECM33 gene | Improved fermentation efficiency under nitrogen limitation | [33] |
| S288C | Overexpression of S. cerevisiae YOL155c and YDR055w genes | reduced haziness during fermentation | [34] |
| EC1118 | Deletion of KNR4 gene | reduced haziness during fermentation, retaining good fermentation performance | [35] |
| VIN13 | Overexpression of Butyrivibrio fibrisolvens end1 gene, Aspergillus niger xynC gene | decrease in wine turbidity, increase in colour intensity, increase in phenolic compounds | [36] |
| VIN13 | Overexpression of Erwinia chrysanthemi pelE gene, Erwinia carotovora peh1 gene | decrease in phenolic compounds | [36] |
| ICV16, ICV27 | Overexpression of S. cerevisiae HSP26 and YHR087W genes | Improved Stress resistance and fermentation efficiency | [37] |
| PYCC 5484 | Overexpression of 925–963 segments of TDH1 and TDH2/3 ORFs | Secretion of AMPs, inhibiting D. bruxellensis growth | [38] |
| Sigma1278 | Overexression of A. niger GOX gene | Reduction of sugar content in juice | [39] |
| V5.TM6 *P. | Overexpression of chimeric HXT1-HXT7 gene in a hxt null strain | decreased ethanol production, increased biomass under high glucose conditions | [40] |
| MC42 | Deletion of ADH1, ADH3, ADH4 genes, ADSH2 gene mutations | 66% reduction of ethanol yield, increased glycerol production | [41] |
| CEN.PK 113-7D | Deletion of TPI1 gene | Unable to grow on glucose, growth on mixed substrates | [42] |
| YSH l.l.-6B | Deletion of PDC2 gene, overexpression of GPD1 gene | Reduction of glucose catabolism, 6-7-fold increase in glycerol formation | [43] |
| AWRI1631 | GPD1 overexpression, ALD6 deletion * | Decreased ethanol production | [44] |
| BY4742, VIN13 | Screening of EOROSCARF deletion collection, weak TPS overexpression | 10% reduction in ethanol yield, increased glycerol, trehalose production | [45] |
| CMBS33, BY4742 | Analysis of ATF1,2 knockouts in the lab strain, constitutive ATF1,2 overexpression in lager strains | Reduction in acetate esters production in ATF1,2 deletion strains, enhanced production of volatile esters in overexpression strains | [46] |
| T73-4 | Overexpression of Ocimum basilicum (sweet basil) geraniol synthase (GES) gene | Increased geraniol production during fermentation, 230-fold increased total monoterpene content | [47] |
| VIN13 | Overexpression of A. awamori arabinofuranosidase, A. kawachii β-glucosidase. | increased release of citronellol, linalool, nerol and α-terpineol. | [48] |
| WY1 | Overexpression of BDH1,2 genes | Decreased diacetyl, increased acetoin, butanediol contents | [49] |
| AWRI | Overexpression of RtPAL, AtC4H, At4CL, RtBAS genes for frambion biosynthesis | Frambion production at 0.68 mg/L simultaneously with chardonnay wine fermentation | [50] |
| CEN.PK 113-7D | Overexpression of AtPAL2, AtC4H, At4CL, VvVST1 gene for resveratrol biosynthesis, complex strain and cultivation optimization strategy | Yeast-based de novo resveratrol production from glucose at 800 mg/l level | [51] |
| 133d | Overexpression of FLO11 gene using different promoter variamts | Improved velum formation | [52] |
| P3-D5 | Deletion of CCW14, YGP1 genes in a flor strain | Impaired velum formation | [53] |
| FJF206, FJF414, B16 | Overexpression of SOD1, SOD2, HSP12 in flor strains | increased superoxide dismutase, catalase, gluthathione peroxidase activities, increased oxidative stress resistance, quicker velum formation, slight decrease in ethanol and increase in acetaldehyde content | [54] |
| EC1118, AWRI1796 | Crispr-cas9 mediated inactivation of CAN1 gene | Reduced ethyl-carbamate formation | [55] |
| BTC.1D | Crispr-cas9 mediated allele exchange for FAS2 and TOR1 genes in wine strain | Increased phenyl-ethyl acetate formation | [56] |
| W34/70 | Crispr-cas9 mediated allele exchange for FDC1 gene in lager strain | Decreased 4-vinyl guaiacol formation | [57] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eldarov, M.A.; Mardanov, A.V. Metabolic Engineering of Wine Strains of Saccharomyces cerevisiae. Genes 2020, 11, 964. https://doi.org/10.3390/genes11090964
Eldarov MA, Mardanov AV. Metabolic Engineering of Wine Strains of Saccharomyces cerevisiae. Genes. 2020; 11(9):964. https://doi.org/10.3390/genes11090964
Chicago/Turabian StyleEldarov, Mikhail A., and Andrey V. Mardanov. 2020. "Metabolic Engineering of Wine Strains of Saccharomyces cerevisiae" Genes 11, no. 9: 964. https://doi.org/10.3390/genes11090964
APA StyleEldarov, M. A., & Mardanov, A. V. (2020). Metabolic Engineering of Wine Strains of Saccharomyces cerevisiae. Genes, 11(9), 964. https://doi.org/10.3390/genes11090964





