Interaction between the Circadian Clock and Regulators of Heat Stress Responses in Plants
Abstract
:1. Introduction
2. Regulation of Clock Genes by Heat Stress in Plants
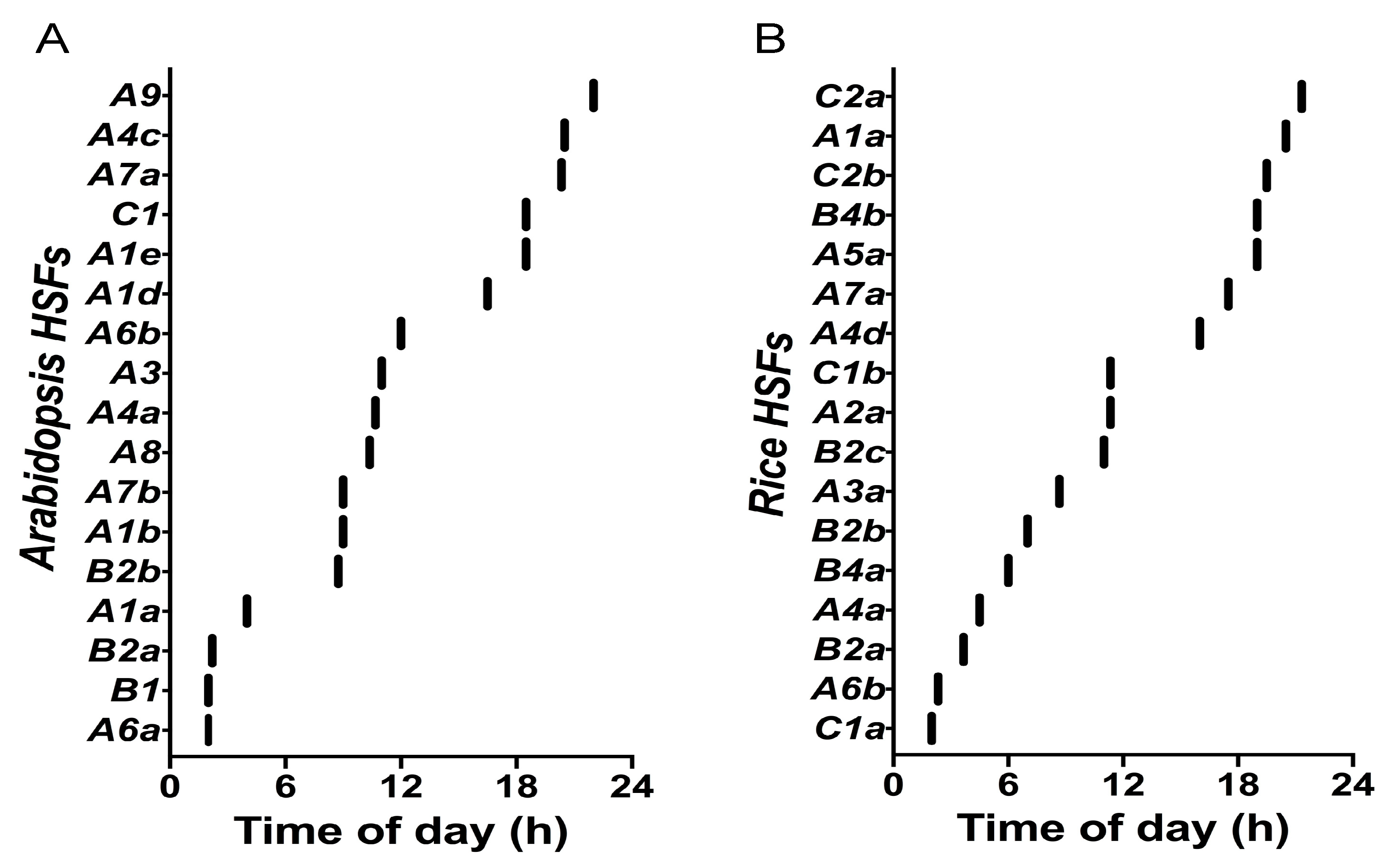
3. Time-of-Day Regulation of Heat Shock Transcription Factors in Plants
4. Time of Day and other Clock-Regulated Heat Responsive Transcription Factor Families
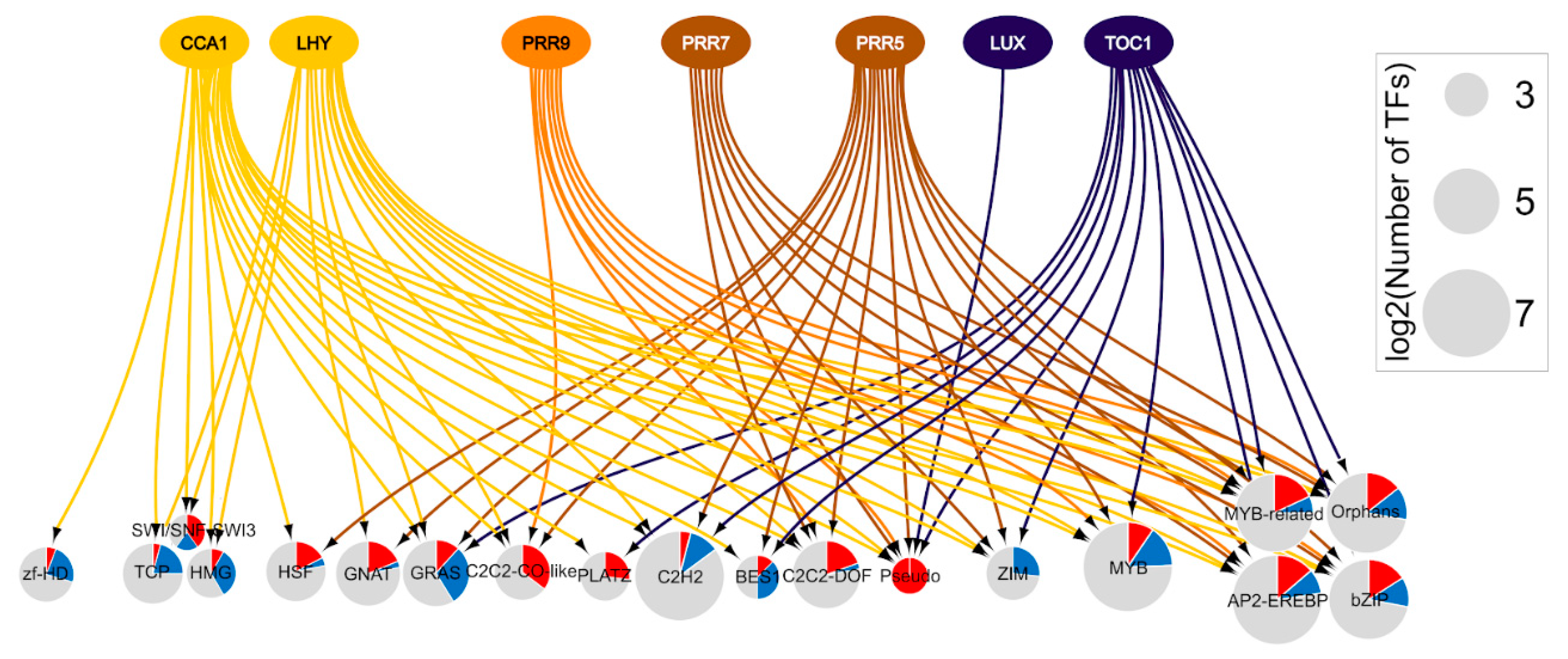
5. Integrated Time of day and Heat Stress Transcriptional Networks
6. Future Perspective
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef] [PubMed]
- Lamaoui, M.; Jemo, M.; Datla, R.; Bekkaoui, F. Heat and Drought Stresses in Crops and Approaches for Their Mitigation. Front. Chem. 2018, 6, 26. [Google Scholar] [CrossRef]
- Penfield, S. Temperature perception and signal transduction in plants. New Phytol. 2008, 179, 615–628. [Google Scholar] [CrossRef]
- Nohales, M.A.; Kay, S.A. Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct. Mol. Biol. 2016, 23, 1061–1069. [Google Scholar] [CrossRef]
- Harmer, S.L. The circadian system in higher plants. Annu. Rev. Plant Biol. 2009, 60, 357–377. [Google Scholar] [CrossRef] [Green Version]
- Greenham, K.; McClung, C.R. Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 2015, 16, 598–610. [Google Scholar] [CrossRef]
- Dodd, A.N.; Salathia, N.; Hall, A.; Kévei, E.; Tóth, R.; Nagy, F.; Hibberd, J.M.; Millar, A.J.; Webb, A.A.R. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005, 309, 630–633. [Google Scholar] [CrossRef] [Green Version]
- McClung, C.R.; Davis, S.J. Ambient thermometers in plants: from physiological outputs towards mechanisms of thermal sensing. Curr. Biol. CB 2010, 20, R1086–R1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil, K.-E.; Park, C.-M. Thermal adaptation and plasticity of the plant circadian clock. New Phytol. 2019, 221, 1215–1229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinoza, C.; Degenkolbe, T.; Caldana, C.; Zuther, E.; Leisse, A.; Willmitzer, L.; Hincha, D.K.; Hannah, M.A. Interaction with Diurnal and Circadian Regulation Results in Dynamic Metabolic and Transcriptional Changes during Cold Acclimation in Arabidopsis. PLoS ONE 2010, 5, e14101. [Google Scholar] [CrossRef] [PubMed]
- Rienth, M.; Torregrosa, L.; Luchaire, N.; Chatbanyong, R.; Lecourieux, D.; Kelly, M.T.; Romieu, C. Day and night heat stress trigger different transcriptomic responses in green and ripening grapevine (vitis vinifera) fruit. BMC Plant Biol. 2014, 14, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, W.; Yin, X.; Struik, P.C.; Solis, C.; Xie, F.; Schmidt, R.C.; Huang, M.; Zou, Y.; Ye, C.; Jagadish, S.V.K. High day- and night-time temperatures affect grain growth dynamics in contrasting rice genotypes. J. Exp. Bot. 2017, 68, 5233–5245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagel, D.H.; Kay, S.A. Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. CB 2012, 22, R648–R657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, P.Y.; Harmer, S.L. Wheels within wheels: the plant circadian system. Trends Plant Sci. 2014, 19, 240–249. [Google Scholar] [CrossRef] [Green Version]
- Alabadí, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Más, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef]
- Sanchez, S.E.; Kay, S.A. The Plant Circadian Clock: From a Simple Timekeeper to a Complex Developmental Manager. Cold Spring Harb. Perspect. Biol. 2016, 8. [Google Scholar] [CrossRef]
- Huang, W.; Pérez-García, P.; Pokhilko, A.; Millar, A.J.; Antoshechkin, I.; Riechmann, J.L.; Mas, P. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 2012, 336, 75–79. [Google Scholar] [CrossRef] [Green Version]
- Pokhilko, A.; Fernández, A.P.; Edwards, K.D.; Southern, M.M.; Halliday, K.J.; Millar, A.J. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 2012, 8, 574. [Google Scholar] [CrossRef] [PubMed]
- Alabadí, D.; Yanovsky, M.J.; Más, P.; Harmer, S.L.; Kay, S.A. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. CB 2002, 12, 757–761. [Google Scholar] [CrossRef] [Green Version]
- Pruneda-Paz, J.L.; Breton, G.; Para, A.; Kay, S.A. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 2009, 323, 1481–1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawat, R.; Takahashi, N.; Hsu, P.Y.; Jones, M.A.; Schwartz, J.; Salemi, M.R.; Phinney, B.S.; Harmer, S.L. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 2011, 7, e1001350. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kiba, T.; Henriques, R.; Mizuno, T.; Chua, N.-H.; Sakakibara, H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 Are Transcriptional Repressors in the Arabidopsis Circadian Clock. Plant Cell 2010, 22, 594–605. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Nusinow, D.A. Into the Evening: Complex Interactions in the Arabidopsis Circadian Clock. Trends Genet. 2016, 32, 674–686. [Google Scholar] [CrossRef]
- Adams, S.; Manfield, I.; Stockley, P.; Carré, I.A. Revised Morning Loops of the Arabidopsis Circadian Clock Based on Analyses of Direct Regulatory Interactions. PLoS ONE 2015, 10, e0143943. [Google Scholar] [CrossRef] [Green Version]
- Matsushika, A.; Makino, S.; Kojima, M.; Mizuno, T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol. 2000, 41, 1002–1012. [Google Scholar] [CrossRef] [Green Version]
- Mockler, T.C.; Michael, T.P.; Priest, H.D.; Shen, R.; Sullivan, C.M.; Givan, S.A.; McEntee, C.; Kay, S.A.; Chory, J. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Eckardt, N.A. Temperature Entrainment of the Arabidopsis Circadian Clock. Plant Cell 2005, 17, 645–647. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yuan, L.; Su, T.; Wang, Q.; Gao, Y.; Zhang, S.; Jia, Q.; Yu, G.; Fu, Y.; Cheng, Q.; et al. Light- and temperature-entrainable circadian clock in soybean development. Plant Cell Environ. 2019. [Google Scholar] [CrossRef]
- Panter, P.E.; Muranaka, T.; Cuitun-Coronado, D.; Graham, C.A.; Yochikawa, A.; Kudoh, H.; Dodd, A.N. Circadian Regulation of the Plant Transcriptome Under Natural Conditions. Front. Genet. 2019, 10, 1239. [Google Scholar] [CrossRef] [Green Version]
- Fankhauser, C.; Staiger, D. Photoreceptors in Arabidopsis thaliana: light perception, signal transduction and entrainment of the endogenous clock. Planta 2002, 216, 1–16. [Google Scholar] [CrossRef]
- Gould, P.D.; Locke, J.C.W.; Larue, C.; Southern, M.M.; Davis, S.J.; Hanano, S.; Moyle, R.; Milich, R.; Putterill, J.; Millar, A.J.; et al. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 2006, 18, 1177–1187. [Google Scholar] [CrossRef] [Green Version]
- Avello, P.A.; Davis, S.J.; Ronald, J.; Pitchford, J.W. Heat the Clock: Entrainment and Compensation in Arabidopsis Circadian Rhythms. J. Circadian Rhythms 2019, 17, 5. [Google Scholar] [CrossRef] [Green Version]
- Thines, B.; Harmon, F.G. Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 2010, 107, 3257–3262. [Google Scholar] [CrossRef] [Green Version]
- Salomé, P.A.; Weigel, D.; McClung, C.R. The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell 2010, 22, 3650–3661. [Google Scholar] [CrossRef] [Green Version]
- Legris, M.; Klose, C.; Burgie, E.S.; Rojas, C.C.R.; Neme, M.; Hiltbrunner, A.; Wigge, P.A.; Schäfer, E.; Vierstra, R.D.; Casal, J.J. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 2016, 354, 897–900. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.-H.; Domijan, M.; Klose, C.; Biswas, S.; Ezer, D.; Gao, M.; Khattak, A.K.; Box, M.S.; Charoensawan, V.; Cortijo, S.; et al. Phytochromes function as thermosensors in Arabidopsis. Science 2016, 354, 886–889. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.-Y.; Oh, E.; Wang, T.; Wang, Z.-Y. TOC1–PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat. Commun. 2016, 7, 13692. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Cumbie, J.S.; Dharmawadhana, J.P.; Jaiswal, P.; Chang, J.H.; Palusa, S.G.; Reddy, A.S.N.; Megraw, M.; Mockler, T.C. Environmental Stresses Modulate Abundance and Timing of Alternatively Spliced Circadian Transcripts in Arabidopsis. Mol. Plant 2014. [Google Scholar] [CrossRef] [PubMed]
- Kangisser, S.; Yakir, E.; Green, R.M. Proteasomal regulation of CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) stability is part of the complex control of CCA1. Plant Signal. Behav. 2013, 8, e23206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grinevich, D.O.; Desai, J.S.; Stroup, K.P.; Duan, J.; Slabaugh, E.; Doherty, C.J. Novel transcriptional responses to heat revealed by turning up the heat at night. Plant Mol. Biol. 2019, 101, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fowler, S.G.; Cook, D.; Thomashow, M.F. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005, 137, 961–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinoza, C.; Bieniawska, Z.; Hincha, D.K.; Hannah, M.A. Interactions between the circadian clock and cold-response in Arabidopsis. Plant Signal. Behav. 2008, 3, 593–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bieniawska, Z.; Espinoza, C.; Schlereth, A.; Sulpice, R.; Hincha, D.K.; Hannah, M.A. Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 2008, 147, 263–279. [Google Scholar] [CrossRef] [Green Version]
- Grundy, J.; Stoker, C.; Carré, I.A. Circadian regulation of abiotic stress tolerance in plants. Front. Plant Sci. 2015, 6, 648. [Google Scholar] [CrossRef]
- Seo, P.J.; Mas, P. STRESSing the role of the plant circadian clock. Trends Plant Sci. 2015, 20, 230–237. [Google Scholar] [CrossRef]
- Davis, A.M.; Ronald, J.; Ma, Z.; Wilkinson, A.J.; Philippou, K.; Shindo, T.; Queitsch, C.; Davis, S.J. HSP90 Contributes to Entrainment of the Arabidopsis Circadian Clock via the Morning Loop. Genetics 2018, 210, 1383–1390. [Google Scholar] [CrossRef] [Green Version]
- Blair, E.J.; Bonnot, T.; Hummel, M.; Hay, E.; Marzolino, J.M.; Quijada, I.A.; Nagel, D.H. Contribution of time of day and the circadian clock to the heat stress responsive transcriptome in Arabidopsis. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Gil, K.-E.; Kim, W.-Y.; Lee, H.-J.; Faisal, M.; Saquib, Q.; Alatar, A.A.; Park, C.-M. ZEITLUPE Contributes to a Thermoresponsive Protein Quality Control System in Arabidopsis. Plant Cell 2017, 29, 2882–2894. [Google Scholar] [CrossRef] [PubMed]
- Filichkin, S.A.; Mockler, T.C. Unproductive alternative splicing and nonsense mRNAs: A widespread phenomenon among plant circadian clock genes. Biol. Direct 2012, 7, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Covington, M.F.; Maloof, J.N.; Straume, M.; Kay, S.A.; Harmer, S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008, 9, R130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamichi, N.; Kusano, M.; Fukushima, A.; Kita, M.; Ito, S.; Yamashino, T.; Saito, K.; Sakakibara, H.; Mizuno, T. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009, 50, 447–462. [Google Scholar] [CrossRef] [Green Version]
- Keily, J.; MacGregor, D.R.; Smith, R.W.; Millar, A.J.; Halliday, K.J.; Penfield, S. Model selection reveals control of cold signalling by evening-phased components of the plant circadian clock. Plant J. 2013, 76, 247–257. [Google Scholar] [CrossRef] [Green Version]
- Cao, S.; Ye, M.; Jiang, S. Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 2005, 24, 683–690. [Google Scholar] [CrossRef]
- Chow, B.Y.; Sanchez, S.E.; Breton, G.; Pruneda-Paz, J.L.; Krogan, N.T.; Kay, S.A. Transcriptional regulation of LUX by CBF1 mediates cold input to the circadian clock in Arabidopsis. Curr. Biol. CB 2014, 24, 1518–1524. [Google Scholar] [CrossRef] [Green Version]
- Dong, M.A.; Farré, E.M.; Thomashow, M.F. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 7241–7246. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Gao, Z.; Liu, X.; Sun, D.; Tang, W. Transcriptional Profiling Reveals a Time-of-Day-Specific Role of REVEILLE 4/8 in Regulating the First Wave of Heat Shock-Induced Gene Expression in Arabidopsis. Plant Cell 2019, 31, 2353–2369. [Google Scholar] [CrossRef]
- McClung, C.R. Beyond Arabidopsis: The circadian clock in non-model plant species. Semin. Cell Dev. Biol. 2013, 24, 430–436. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Breton, G.; Priest, H.D.; Dharmawardhana, P.; Jaiswal, P.; Fox, S.E.; Michael, T.P.; Chory, J.; Kay, S.A.; Mockler, T.C. Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS ONE 2011, 6, e16907. [Google Scholar] [CrossRef] [Green Version]
- Hudson, K.A. The Circadian Clock-controlled Transcriptome of Developing Soybean Seeds. Plant Genome J. 2010, 3, 3. [Google Scholar] [CrossRef]
- Khan, S.; Rowe, S.C.; Harmon, F.G. Coordination of the maize transcriptome by a conserved circadian clock. BMC Plant Biol. 2010, 10, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calixto, C.P.G.; Waugh, R.; Brown, J.W.S. Evolutionary Relationships Among Barley and Arabidopsis Core Circadian Clock and Clock-Associated Genes. J. Mol. Evol. 2015, 80, 108–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, M.; Tago, Y.; Yamashino, T.; Mizuno, T. Comparative Overviews of Clock-Associated Genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007, 48, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, L.; Zhang, S.; Wu, L.; Ku, L.; Wei, X.; Xie, L.; Chen, Y. Robust expression and association of ZmCCA1 with circadian rhythms in maize. Plant Cell Rep. 2011, 30, 1261–1272. [Google Scholar] [CrossRef] [PubMed]
- Ko, D.K.; Rohozinski, D.; Song, Q.; Taylor, S.H.; Juenger, T.E.; Harmon, F.G.; Chen, Z.J. Temporal Shift of Circadian-Mediated Gene Expression and Carbon Fixation Contributes to Biomass Heterosis in Maize Hybrids. PLoS Genet. 2016, 12, e1006197. [Google Scholar] [CrossRef]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008, 2008, 420747. [Google Scholar] [CrossRef]
- Li, M.; Cao, L.; Mwimba, M.; Zhou, Y.; Li, L.; Zhou, M.; Schnable, P.S.; O’Rourke, J.A.; Dong, X.; Wang, W. Comprehensive mapping of abiotic stress inputs into the soybean circadian clock. Proc. Natl. Acad. Sci. USA 2019, 116, 23840–23849. [Google Scholar] [CrossRef] [Green Version]
- Marcolino-Gomes, J.; Rodrigues, F.A.; Fuganti-Pagliarini, R.; Bendix, C.; Nakayama, T.J.; Celaya, B.; Molinari, H.B.C.; de Oliveira, M.C.N.; Harmon, F.G.; Nepomuceno, A. Diurnal Oscillations of Soybean Circadian Clock and Drought Responsive Genes. PLoS ONE 2014, 9, e86402. [Google Scholar] [CrossRef] [Green Version]
- Habte, E.; Müller, L.M.; Shtaya, M.; Davis, S.J.; Von Korff, M. Osmotic stress at the barley root affects expression of circadian clock genes in the shoot: Osmotic stress changes the barley circadian clock. Plant Cell Environ. 2014, 37, 1321–1337. [Google Scholar] [CrossRef] [PubMed]
- Borland, A.M.; Griffiths, H.; Hartwell, J.; Smith, J.A.C. Exploiting the potential of plants with crassulacean acid metabolism for bioenergy production on marginal lands. J. Exp. Bot. 2009, 60, 2879–2896. [Google Scholar] [CrossRef] [PubMed]
- Boxall, S.F.; Foster, J.M.; Bohnert, H.J.; Cushman, J.C.; Nimmo, H.G.; Hartwell, J. Conservation and Divergence of Circadian Clock Operation in a Stress-Inducible Crassulacean Acid Metabolism Species Reveals Clock Compensation against Stress. Plant Physiol. 2005, 137, 969–982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endogenous Plant Rhythms; Hall, A.J.W.; McWatterss, H. (Eds.) Blackwell Publishing Ltd: Oxford, UK, 2006; ISBN 978-0-470-98852-7. [Google Scholar]
- Hartwell, J. The co-ordination of central plant metabolism by the circadian clock. Biochem. Soc. Trans. 2005, 33, 945. [Google Scholar] [CrossRef] [Green Version]
- Ming, R.; VanBuren, R.; Wai, C.M.; Tang, H.; Schatz, M.C.; Bowers, J.E.; Lyons, E.; Wang, M.-L.; Chen, J.; Biggers, E.; et al. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015, 47, 1435–1442. [Google Scholar] [CrossRef] [Green Version]
- Mallona, I.; Egea-Cortines, M.; Weiss, J. Conserved and Divergent Rhythms of Crassulacean Acid Metabolism-Related and Core Clock Gene Expression in the Cactus Opuntia ficus-indica. Plant Physiol. 2011, 156, 1978–1989. [Google Scholar] [CrossRef] [Green Version]
- Lindquist, S. The heat-shock response. Annu. Rev. Biochem. 1986, 55, 1151–1191. [Google Scholar] [CrossRef]
- Lindquist, S.; Craig, E.A. The heat-shock proteins. Annu. Rev. Genet. 1988, 22, 631–677. [Google Scholar] [CrossRef]
- Morimoto, R.I. Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998, 12, 3788–3796. [Google Scholar] [CrossRef] [Green Version]
- Scharf, K.-D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim. Biophys. Acta 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Albihlal, W.S.; Obomighie, I.; Blein, T.; Persad, R.; Chernukhin, I.; Crespi, M.; Bechtold, U.; Mullineaux, P.M. Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b regulates multiple developmental genes under benign and stress conditions. J. Exp. Bot. 2018, 69, 2847–2862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.-C.; Liao, H.-T.; Charng, Y.-Y. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011, 34, 738–751. [Google Scholar] [CrossRef] [PubMed]
- Nagel, D.H.; Doherty, C.J.; Pruneda-Paz, J.L.; Schmitz, R.J.; Ecker, J.R.; Kay, S.A. Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, E4802–E4810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamioka, M.; Takao, S.; Suzuki, T.; Taki, K.; Higashiyama, T.; Kinoshita, T.; Nakamichi, N. Direct Repression of Evening Genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis Circadian Clock. Plant Cell 2016, 28, 696–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamichi, N.; Kiba, T.; Kamioka, M.; Suzuki, T.; Yamashino, T.; Higashiyama, T.; Sakakibara, H.; Mizuno, T. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. USA 2012, 109, 17123–17128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.L.; Newton, L.; Liu, M.-J.; Shiu, S.-H.; Farré, E.M. A G-Box-Like Motif Is Necessary for Transcriptional Regulation by Circadian Pseudo-Response Regulators in Arabidopsis. Plant Physiol. 2016, 170, 528–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Carlsson, J.; Takeuchi, T.; Newton, L.; Farré, E.M. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. Cell Mol. Biol. 2013, 76, 101–114. [Google Scholar] [CrossRef]
- Kolmos, E.; Chow, B.Y.; Pruneda-Paz, J.L.; Kay, S.A. HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proc. Natl. Acad. Sci. USA 2014, 111, 16172–16177. [Google Scholar] [CrossRef] [Green Version]
- Swindell, W.R.; Huebner, M.; Weber, A.P. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 2007, 8, 125. [Google Scholar] [CrossRef] [Green Version]
- Mittal, D.; Chakrabarti, S.; Sarkar, A.; Singh, A.; Grover, A. Heat shock factor gene family in rice: Genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol. Biochem. 2009, 47, 785–795. [Google Scholar] [CrossRef]
- Sharma, A.; Wai, C.M.; Ming, R.; Yu, Q. Diurnal Cycling Transcription Factors of Pineapple Revealed by Genome-Wide Annotation and Global Transcriptomic Analysis. Genome Biol. Evol. 2017, 9, 2170–2190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruneda-Paz, J.L.; Breton, G.; Nagel, D.H.; Kang, S.E.; Bonaldi, K.; Doherty, C.J.; Ravelo, S.; Galli, M.; Ecker, J.R.; Kay, S.A. A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell Rep. 2014, 8, 622–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirsch, S.; Oldroyd, G.E.D. GRAS-domain transcription factors that regulate plant development. Plant Signal. Behav. 2009, 4, 698–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Han, G.; Sun, C.; Sui, N. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 2019, 14, 1613131. [Google Scholar] [CrossRef]
- Torres-Galea, P.; Huang, L.-F.; Chua, N.-H.; Bolle, C. The GRAS protein SCL13 is a positive regulator of phytochrome-dependent red light signaling, but can also modulate phytochrome A responses. Mol. Genet. Genomics MGG 2006, 276, 13–30. [Google Scholar] [CrossRef]
- Torres-Galea, P.; Hirtreiter, B.; Bolle, C. Two GRAS proteins, SCARECROW-LIKE21 and PHYTOCHROME A SIGNAL TRANSDUCTION1, function cooperatively in phytochrome A signal transduction. Plant Physiol. 2013, 161, 291–304. [Google Scholar] [CrossRef] [Green Version]
- El-Kereamy, A.; Bi, Y.-M.; Ranathunge, K.; Beatty, P.H.; Good, A.G.; Rothstein, S.J. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PloS ONE 2012, 7, e52030. [Google Scholar] [CrossRef] [Green Version]
- Casaretto, J.A.; El-Kereamy, A.; Zeng, B.; Stiegelmeyer, S.M.; Chen, X.; Bi, Y.-M.; Rothstein, S.J. Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance. BMC Genomics 2016, 17, 312. [Google Scholar] [CrossRef] [Green Version]
- Adams, S.; Grundy, J.; Veflingstad, S.R.; Dyer, N.P.; Hannah, M.A.; Ott, S.; Carré, I.A. Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol. 2018. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822–18827. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schramm, F.; Larkindale, J.; Kiehlmann, E.; Ganguli, A.; Englich, G.; Vierling, E.; von Koskull-Döring, P. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. Cell Mol. Biol. 2008, 53, 264–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, M.; Zhang, W.; Zhang, Q.; Xu, Z.; Zhu, Z.; Duan, F.; Wu, R. Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem. PPB 2011, 49, 1384–1391. [Google Scholar] [CrossRef] [PubMed]
- Matsukura, S.; Mizoi, J.; Yoshida, T.; Todaka, D.; Ito, Y.; Maruyama, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics MGG 2010, 283, 185–196. [Google Scholar] [CrossRef]
- Yang, S.; Xu, K.; Chen, S.; Li, T.; Xia, H.; Chen, L.; Liu, H.; Luo, L. A stress-responsive bZIP transcription factor OsbZIP62 improves drought and oxidative tolerance in rice. BMC Plant Biol. 2019, 19, 260. [Google Scholar] [CrossRef] [Green Version]
- Das, P.; Lakra, N.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. A unique bZIP transcription factor imparting multiple stress tolerance in Rice. Rice N. Y. N 2019, 12, 58. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Zou, H.-F.; Wei, W.; Hao, Y.-J.; Tian, A.-G.; Huang, J.; Liu, Y.-F.; Zhang, J.-S.; Chen, S.-Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef]
- Oh, S.-J.; Song, S.I.; Kim, Y.S.; Jang, H.-J.; Kim, S.Y.; Kim, M.; Kim, Y.-K.; Nahm, B.H.; Kim, J.-K. Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 2005, 138, 341–351. [Google Scholar] [CrossRef] [Green Version]
- Ezer, D.; Jung, J.-H.; Lan, H.; Biswas, S.; Gregoire, L.; Box, M.S.; Charoensawan, V.; Cortijo, S.; Lai, X.; Stöckle, D.; et al. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants 2017, 3, 17087. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, N.; Bajad, S.; Shuman, J.; Shulaev, V.; Mittler, R. The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J. Biol. Chem. 2008, 283, 9269–9275. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, N.; Sejima, H.; Tam, R.; Schlauch, K.; Mittler, R. Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2011, 66, 844–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, S.P.; Ort, D.R. More than taking the heat: crops and global change. Curr. Opin. Plant Biol. 2010, 13, 241–248. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mody, T.; Bonnot, T.; Nagel, D.H. Interaction between the Circadian Clock and Regulators of Heat Stress Responses in Plants. Genes 2020, 11, 156. https://doi.org/10.3390/genes11020156
Mody T, Bonnot T, Nagel DH. Interaction between the Circadian Clock and Regulators of Heat Stress Responses in Plants. Genes. 2020; 11(2):156. https://doi.org/10.3390/genes11020156
Chicago/Turabian StyleMody, Tejasvinee, Titouan Bonnot, and Dawn H. Nagel. 2020. "Interaction between the Circadian Clock and Regulators of Heat Stress Responses in Plants" Genes 11, no. 2: 156. https://doi.org/10.3390/genes11020156




