Chromosomal Differentiation in Genetically Isolated Populations of the Marsh-Specialist Crocidura suaveolens (Mammalia: Soricidae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Cell Culture and Chromosomal Harvest
2.3. Chromosomal Characterization
2.4. Karyotype Distribution across Lineages and Populations
3. Results
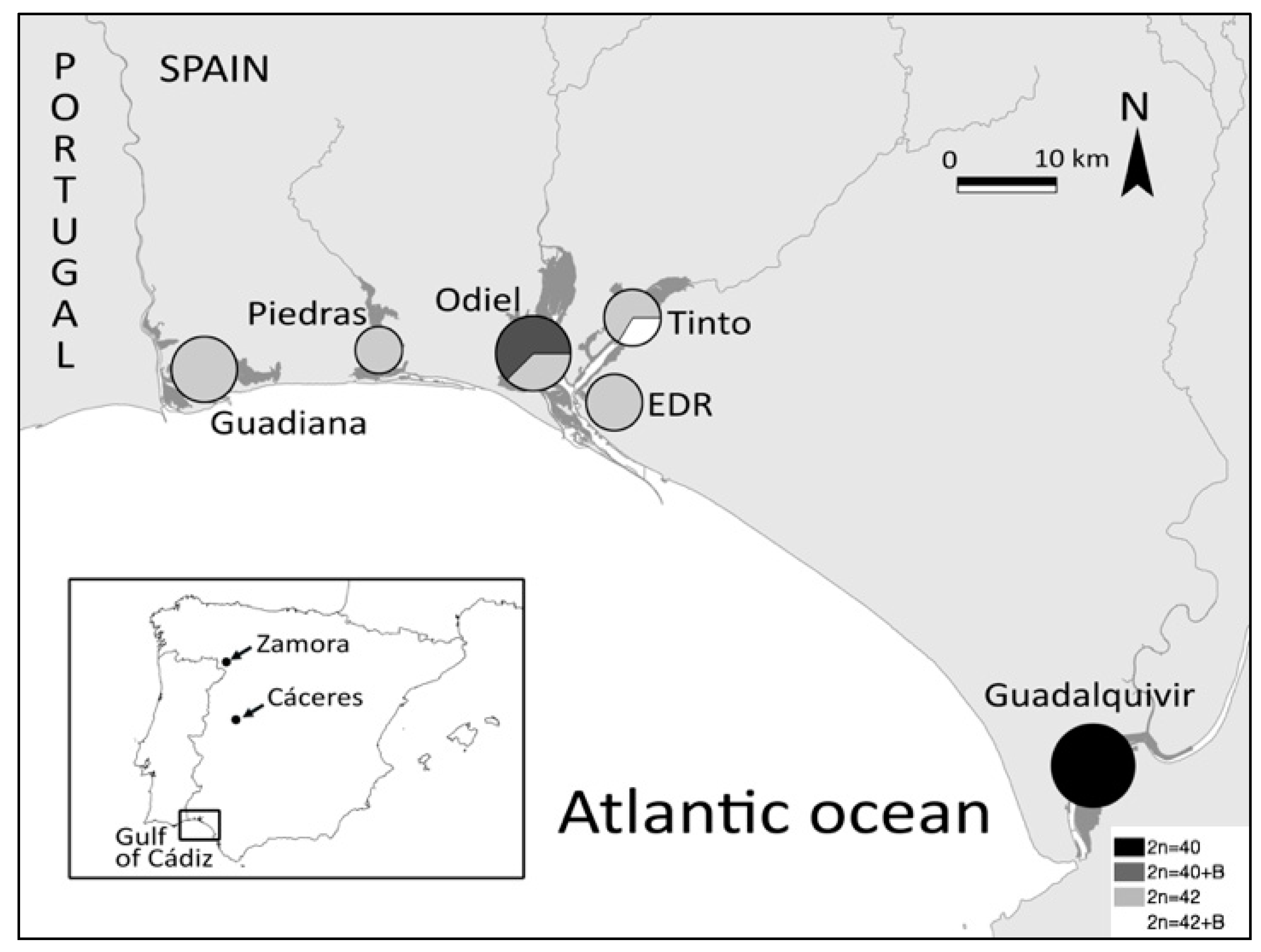
3.1. Chromosomal Diversity in C. suaveolens in the Gulf of Cádiz
3.2. Karyotype Distribution and Diversity
4. Discussion
4.1. Overview of Chromosomal Evolution in Crocidura
4.2. Karyotypic Diversity and Differentiation of C. suaveolens Populations in the Gulf of Cádiz
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Faria, R.; Johannesson, K.; Butlin, R.K.; Westram, A.M. Evolving Inversions. Trends Ecol. Evol. 2019, 34, 239–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, A.A.; Rieseberg, L.H. Revisiting the impact of inversions in evolution: From population genetic markers to drivers of adaptive shifts and speciation? Annu. Rev. Ecol. Evol. Syst. 2008, 39, 21–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapun, M.; Flatt, T. The adaptive significance of chromosomal inversion polymorphisms in Drosophila melanogaster. Mol. Ecol. 2019, 28, 1263–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayala, D.; Zhang, S.; Chateau, M.; Fouet, C.; Morlais, I.; Costantini, C.; Hahn, M.W.; Besansky, N.J. Association mapping desiccation resistance within chromosomal inversions in the African malaria vector Anopheles gambiae. Mol. Ecol. 2019, 28, 1333–1342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christmas, M.J.; Wallberg, A.; Bunikis, I.; Olsson, A.; Wallerman, O.; Webster, M.T. Chromosomal inversions associated with environmental adaptation in honeybees. Mol. Ecol. 2019, 28, 1358–1374. [Google Scholar] [CrossRef] [PubMed]
- Gündüz, I.; Auffray, J.C.; Britton-Davidian, J.; Catalan, J.; Ganem, G.; Ramalhinho, M.G.; Mathias, M.L.; Searle, J.B. Molecular studies on the colonization of the Madeiran archipelago by house mice. Mol. Ecol. 2001, 10, 2023–2029. [Google Scholar] [CrossRef]
- Dumas, D.; Britton-Davidian, J. Chromosomal rearrangements and evolution of recombination: Comparison of chiasma distribution patterns in standard and Robertsonian populations of the house mouse. Genetics. 2002, 162, 1355–1366. [Google Scholar]
- Giménez, M.D.; Mirol, P.M.; Bidau, C.J.; Searle, J.B. Molecular analysis of populations of Ctenomys. (Caviomorpha, Rodentia) with high karyotypic variability. Cytogenet. Genome Res. 2002, 96, 130–136. [Google Scholar] [CrossRef]
- Borodin, P.M.; Karamysheva, T.V.; Belonogova, N.M.; Torgasheva, A.A.; Rubtsov, N.B.; Searle, J.B. Recombination map of the common shrew, Sorex araneus. (Eulipotyphla, Mammalia). Genetics 2008, 178, 621–632. [Google Scholar] [CrossRef] [Green Version]
- Franchini, P.; Colangelo, P.; Solano, E.; Capanna, E.; Verheyen, E.; Castiglia, R. Reduced gene flow at pericentromeric loci in a hybrid zone involving chromosomal races of the house mouse Mus musculus domesticus. Evolution 2010, 64, 2020–2032. [Google Scholar]
- Förster, D.W.; Mathias, M.L.; Britton-Davidian, J.; Searle, J.B. Origin of the chromosomal radiation of Madeiran house mice: A microsatellite analysis of metacentric chromosomes. Heredity 2013, 110, 380–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capilla, L.; Medarde, N.; Alemany-Schmidt, A.; Oliver-Bonet, M.; Ventura, J.; Ruiz-Herrera, A. Genetic recombination variation in wild Robertsonian mice: On the role of chromosomal fusions and Prdm9 allelic background. Proc. R. Soc. B Biol. Sci. 2014, 281, 20140297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vara, C.; Capilla, L.; Ferretti, L.; Ledda, A.; Sánchez-Guillén, R.A.; Gabriel, S.I.; Albert-Lizandra, G.; Florit-Sabater, B.; Bello-Rodríguez, J.; Ventura, J.; et al. PRDM9 Diversity at fine geographical scale reveals contrasting evolutionary patterns and functional constraints in natural populations of house mice. Mol. Biol. Evol. 2019, 36, 1686–1700. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.J.; Menninger, J.C.; Nash, W.G. An Atlas of Mammalian Chromosomes; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2006. [Google Scholar]
- Dubey, S.; Salamin, N.; Ohdachi, S.D.; Barrière, P.; Vogel, P. Molecular phylogenetics of shrews (Mammalia: Soricidae) reveal timing of transcontinental colonizations. Mol. Phylogenet. Evol. 2007, 44, 126–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubey, S.; Salamin, N.; Ruedi, M.; Barrière, P.; Colyn, M.; Vogel, P. Biogeographic origin and radiation of the Old World crocidurine shrews (Mammalia: Soricidae) inferred from mitochondrial and nuclear genes. Mol. Phylogenet. Evol. 2008, 48, 953–963. [Google Scholar] [CrossRef] [Green Version]
- Maddalena, T.; Ruedi, M. Chromosomal Evolution in the Genus Crocidura. (Soricidae, Insectivora). In Advances in the Biology of Shrews; Merrit, J.F., Kirkland, G.L., Jr., Rose, R.K., Eds.; Carnegie Museum of Natural History: Pittsburgh, PA, USA, 1994; Volume 18, pp. 335–344. [Google Scholar]
- Castiglia, R.; Annesi, F.; Sichilima, A.M.; Hutterer, R. A molecular and chromosomal study of the moonshine shrew, Crocidura luna Dollman, 1910 from Zambia with a description of a new remarkable karyotype. Mammalia 2009, 73, 56–59. [Google Scholar] [CrossRef]
- Biltueva, L.; Vorobieva, N.; Perelman, P.; Trifonov, V.; Volobouev, V.; Panov, V.; Ilyashenko, V.; Onischenko, S.; O’Brien, P.; Yang, F.; et al. Karyotype evolution of eulipotyphla (insectivora): The genome homology of seven sorex species revealed by comparative chromosome painting and banding data. Cytogenet. Genome Res. 2001, 135, 51–64. [Google Scholar] [CrossRef]
- Meylan, A.; Hausser, J. Cytotaxonomic position of several shrews of the genus Crocidura in Tessin (Mammalia, Insectivora). Rev. Suisse Zool. 1974, 8, 701–710. [Google Scholar] [CrossRef]
- Zima, J.; Lukacova, L.; Machola, M. Chromosomal evolution in shrews. In Evolution of Shrews; Wojcik, J.M., Wolsan, M., Eds.; Mammal Research Institute, Polish Academy of Sciences: Bialowieza, Poland, 1998. [Google Scholar]
- Palomo, L.; Kryštufek, B.; Amori, G.; Hutterer, R. Crocidura suaveolens. The IUCN Red List of Threatened Species 2016: E.T29656A22296429. Available online: http://www.iucnredlist.org/details/29656/ (accessed on 16 November 2016).
- Libois, R.; Ramalhinho, M.G.; Fons, R. Crocidura. suaveolens. (Pallas, 1811), the lesser-white toothed shrew. In The Atlas of European Mammals; Mitchell-Jones, A.J., Amori, G., Bogdanowicz, W., Kryštufek, B., Reijnders, P.J.H., Spitzenberger, F., Stubbe, M., Thissen, J.B.M., Vohralik, V., Zima, J., Eds.; Poyser Natural History: London, UK, 1999; pp. 72–73. [Google Scholar]
- Biedma, L.; Calzada, J.; Román, J.; Godoy, J.A. Rare and rear: Population genetics of marsh-specialist Crocidura. suaveolens. populations in the Gulf of Cádiz. J. Mammal. 2019, 100, 92–102. [Google Scholar] [CrossRef]
- Biedma, L.; Román, J.; Calzada, J.; Friis, G.; Godoy, J.A. Phylogeography of Crocidura suaveolens (Mammalia: Soricidae) in Iberia has been shaped by competitive exclusion by C. russula. Biol. J. Linn. Soc. 2018, 123, 81–95. [Google Scholar] [CrossRef]
- Biedma, L.; Calzada, J.; Godoy, J.A.; Román, J. Local habitat specialization as an evolutionary response to interspecific competition between two sympatric shrews. J. Mammal. 2020, 101, 80–91. [Google Scholar] [CrossRef]
- Dobigny, G.; Britton-Davidian, J.; Robinson, T.J. Chromosomal polymorphism in mammals: An evolutionary perspective. Biol. Rev. 2004, 92, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Herrera, A.; Ponsà, M.; García, F.; Egozcue, J.; García, M. Fragile sites in human and Macaca fascicularis chromosomes are breakpoints in chromosome evolution. Chromosome Res. 2002, 10, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Raymond, M.; Rousset, F. GENEPOP (Version 1.2): Population genetics software for exact tests and ecumenicism. Heredity 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Ruiz-Herrera, A.; Farré, M.; Robinson, T.J. Molecular cytogenetic and genomic insights into chromosomal evolution. Heredity 2012, 108, 28–36. [Google Scholar] [CrossRef] [Green Version]
- Hutterer, R. Order Soricomorpha. In Mammal Species of the World: A Taxonomic and Geo-Graphic Reference; Wilson, D.E., Reeder, D.M., Eds.; Johns Hopkins University Press: Baltimore, MD, USA, 2005; pp. 220–311. [Google Scholar]
- Ruedi, M.; Vogel, P. Chromosomal evolution and zoogeographical origin of southeast Asian shrews (genus Crocidura). Experientia 1995, 51, 174–178. [Google Scholar] [CrossRef]
- Maddalena, T. Systematics and biogeography of Afrotropical and Paleartic hrews of the genus Crocidura (Insectivora: Soricidae): An electrophoretic approach. In Vertebrates in the Tropics; Peters, G., Hutterer, R., Eds.; Museum Alexander Koenig Publication: Bonn, Germany, 1990; pp. 297–308. [Google Scholar]
- Vujosevic, M.; Rajičić, M.; Blagojević, J.B. Chromosomes in Populations of Mammals Revisited. Genes 2018, 9, 487. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, S.; M’barek, S.B.; Dhillon, B.; Wittenberg, A.H.; Crane, C.F.; Hane, J.K.; Foster, A.J.; Van der Lee, T.A.; Grimwood, J.; Aerts, A.; et al. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 2011, 7, e1002070. [Google Scholar] [CrossRef] [Green Version]
- Patton, J.L. A complex system of chromosomal variation in the pocket mouse, Perognathus baileyi Merriam. Chromosoma 1972, 36, 241–255. [Google Scholar] [CrossRef]
- Dvorak, J.; Deal, K.R.; Luo, M.C. Discovery and mapping of wheat Ph1 suppressors. Genetics 2006, 174, 17–21. [Google Scholar] [CrossRef] [Green Version]
- Thomson, R.L. B chromosomes in Rattus fuscipes II. The transmission of B chromosomes to offspring and population studies: Support for the “parasitic” model. Heredity 1984, 52, 363–372. [Google Scholar] [CrossRef] [Green Version]
- Baker, R.J.; Bickham, J.W. Speciation by monobrachial centric fusions. Proc. Natl. Acad. Sci. USA 1979, 83, 8245–8248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, M.J.D. Animal Cytology and Evolution; Cambridge University Press: New York, NY, USA, 1973. [Google Scholar]
- King, M. Species Evolution: The Role of Chromosome Change; Cambridge University Press: New York, NY, USA, 1993. [Google Scholar]
- Hamilton, A.E.; Beuttner-Janusch, J.; Chu, E.H. Chromosomes of lemuriformes. II. Chromosome polymorphism in Lemur fulvus collaris (E. Geoffroy 1812). Am. J. Phys. Anthropol. 1977, 46, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Farré, M.; Micheletti, D.; Ruiz-Herrera, A. Recombination rates and genomic shuffling in human and chimpanzee: A new twist in the chromosomal speciation theory. Mol. Biol. Evol. 2013, 30, 853–864. [Google Scholar] [CrossRef] [Green Version]
- Ullastres, A.; Farré, M.; Capilla, L.; Ruiz-Herrera, A. Unraveling the effect of genomic structural changes in the rhesus macaque-implications for the adaptive role of inversions. BMC Genom. 2014, 15, 530. [Google Scholar] [CrossRef] [Green Version]
- Rubes, J.; Pagacova, E.; Kopecna, O.; Kubickova, S.; Cernohorska, H.; Vahala, J.; Di Berardino, D. Karyotype, centric fusion polymorphism and chromosomal aberrations in captive-born mountain reedbuck (Redunca fulvorufula). Cytogenet. Genome Res. 2007, 116, 263–268. [Google Scholar] [CrossRef]
- Medarde, N.; López-Fuster, M.J.; Muñoz-Muñoz, F.; Ventura, J. Spatio-temporal variation in the structure of a chromosomal polymorphism zone in the house mouse. Heredity 2012, 109, 78–89. [Google Scholar] [CrossRef] [Green Version]
- Qumsiyeh, M.B.; Coate, J.L.; Peppers, J.A.; Kennedy, P.K.; Kennedy, M.L. Roberstonian chromosomal rearrangements in the short-tailed shrew Blarina carolinensis, in Western Tennessee. Cytogenet. Genome Res. 1997, 76, 153–158. [Google Scholar] [CrossRef]



| Population | Region | Locality | N | 2n | FNa | B-chr |
|---|---|---|---|---|---|---|
| 1 | Guadalquivir | Eastern bank | 4 | 2n = 40 | 46 | 0 |
| 1 | Guadalquivir | Western bank | 3 | 2n = 40 | 46 | 0 |
| 1 | Guadalquivir | Faginao—Western bank | 2 | 2n = 40 | 46 | 0 |
| 2 | Guadiana | Isla de San Bruno | 3 | 2n = 42 | 50 | 0 |
| 2 | Guadiana | Salinas del Duque | 4 | 2n = 42 | 50 | 0 |
| 3 | Odiel | Cascajera | 4 | 2n = 41/42 | 48/50 | 1 |
| 3 | Odiel | Puntales | 4 | 2n = 41/42 | 48/50 | 1 |
| 4 | Piedras | Salinas | 2 | 2n = 42 | 50 | 0 |
| 4 | Piedras | El Terrón | 3 | 2n = 42 | 50 | 0 |
| 5 | Tinto | Eastern bank (Fosfoyesos) | 6 | 2n = 42/43 | 48/52 | 1 |
| 6 | EDR | Carabelas dock | 6 | 2n = 42 | 50 | 0 |
| 7 | Zamora | Flechas | 1 | 2n = 40 | 46 | 0 |
| 8 | Cáceres | Garganta de la Olla | 1 | 2n = 40 | 46 | 0 |
| Karyotype Frequency | A-Chromosome Number | B- Chromosome | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Population | N | mtDNA Clade | 40 | 40+B | 42 | 42+B | 42 freq | HE | B freq | H |
| Guadalquivir | 9 | C4 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Odiel | 8 | C3 | 0.00 | 0.63 | 0.38 | 0.00 | 0.38 | 0.47 | 0.63 | 0.47 |
| Tinto | 6 | C3 | 0.00 | 0.00 | 0.67 | 0.33 | 1.00 | 0.00 | 0.33 | 0.44 |
| EDR | 6 | C3 | 0.00 | 0.00 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| Piedras | 5 | C3 | 0.00 | 0.00 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| Guadiana | 7 | C3 | 0.00 | 0.00 | 1.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| Clade C4 | 9 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Clade C3 | 32 | 0.00 | 0.16 | 0.78 | 0.06 | 0.84 | 0.00 | 0.22 | 0.00 | |
| Overall | 41 | 0.11 | 0.06 | 0.30 | 0.02 | 0.66 | 0.45 | 0.17 | 0.28 | |
| mtDNA Clade | ||||||
|---|---|---|---|---|---|---|
| C4 | C3 | |||||
| Guadalquivir | Odiel | Tinto | EDR | Piedras | Guadiana | |
| Guadalquivir | 0.308 * | 1000 ** | 1000 ** | 1000 ** | 1000 ** | |
| Odiel | 0.591 * | 0.525 ** | 0.525 ** | 0.496 * | 0.550 ** | |
| Tinto | 0.287 | 0.013 | - | - | - | |
| EDR | - | 0.525 | 0.200 | - | - | |
| Piedras | - | 0.496 | 0.161 | - | - | |
| Guadiana | - | 0.550 | 0.233 | - | - | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia, F.; Biedma, L.; Calzada, J.; Román, J.; Lozano, A.; Cortés, F.; Godoy, J.A.; Ruiz-Herrera, A. Chromosomal Differentiation in Genetically Isolated Populations of the Marsh-Specialist Crocidura suaveolens (Mammalia: Soricidae). Genes 2020, 11, 270. https://doi.org/10.3390/genes11030270
Garcia F, Biedma L, Calzada J, Román J, Lozano A, Cortés F, Godoy JA, Ruiz-Herrera A. Chromosomal Differentiation in Genetically Isolated Populations of the Marsh-Specialist Crocidura suaveolens (Mammalia: Soricidae). Genes. 2020; 11(3):270. https://doi.org/10.3390/genes11030270
Chicago/Turabian StyleGarcia, Francisca, Luis Biedma, Javier Calzada, Jacinto Román, Alberto Lozano, Francisco Cortés, José A. Godoy, and Aurora Ruiz-Herrera. 2020. "Chromosomal Differentiation in Genetically Isolated Populations of the Marsh-Specialist Crocidura suaveolens (Mammalia: Soricidae)" Genes 11, no. 3: 270. https://doi.org/10.3390/genes11030270
APA StyleGarcia, F., Biedma, L., Calzada, J., Román, J., Lozano, A., Cortés, F., Godoy, J. A., & Ruiz-Herrera, A. (2020). Chromosomal Differentiation in Genetically Isolated Populations of the Marsh-Specialist Crocidura suaveolens (Mammalia: Soricidae). Genes, 11(3), 270. https://doi.org/10.3390/genes11030270






