Natural Variation Uncovers Candidate Genes for Barley Spikelet Number and Grain Yield under Drought Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Setup and Phenotyping
2.2. Data Analysis
2.3. Genome-Wide Marker–Trait Associations
2.4. SNP-Gene-Based Association and Haplotype Analysis
3. Results
3.1. Phenotypic Characteristics and Natural Variation
3.2. Correlations Analysis
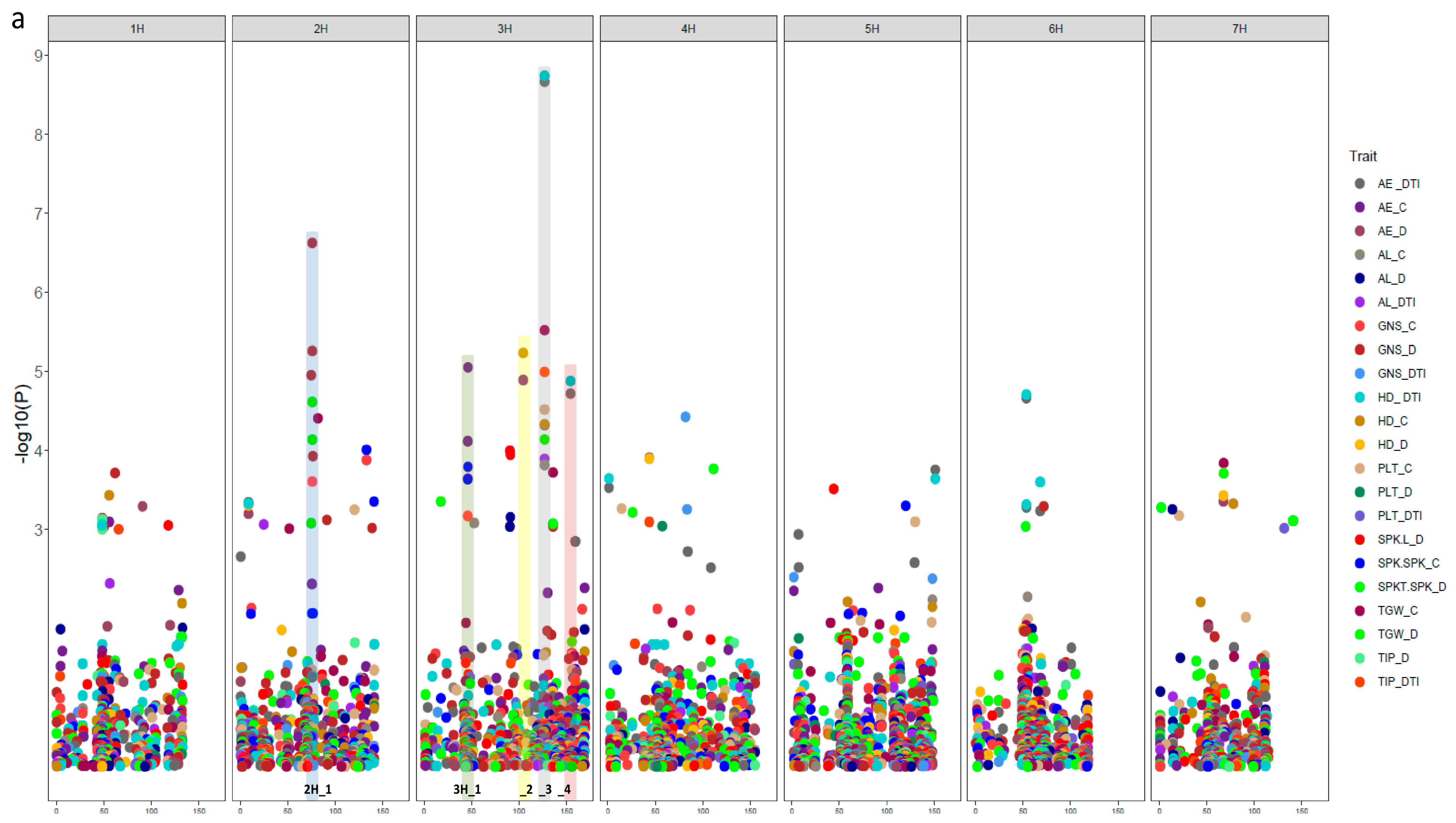
3.3. Natural Genetic Variation and Candidate Genes Potentially Underlying Drought Tolerance
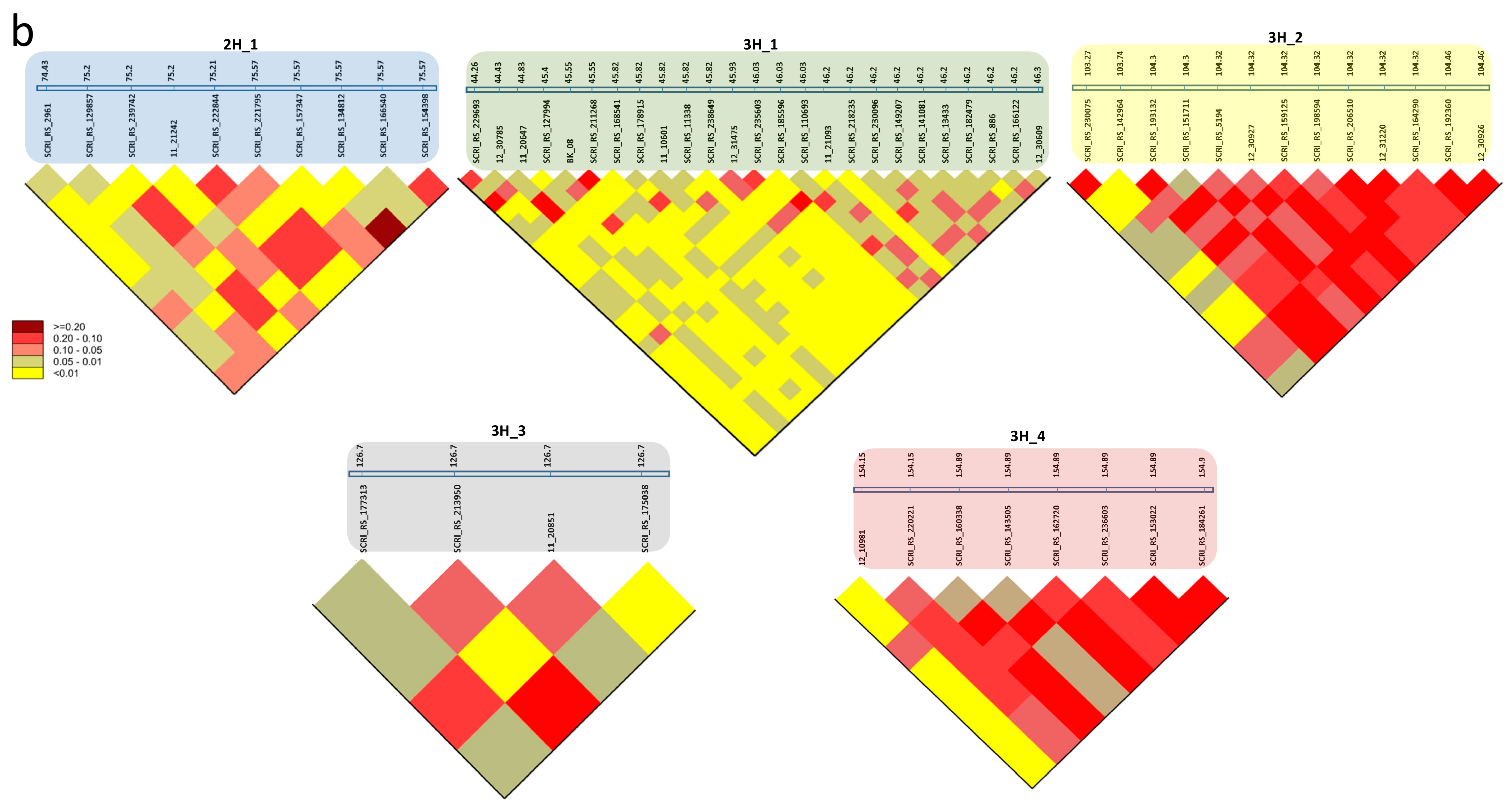
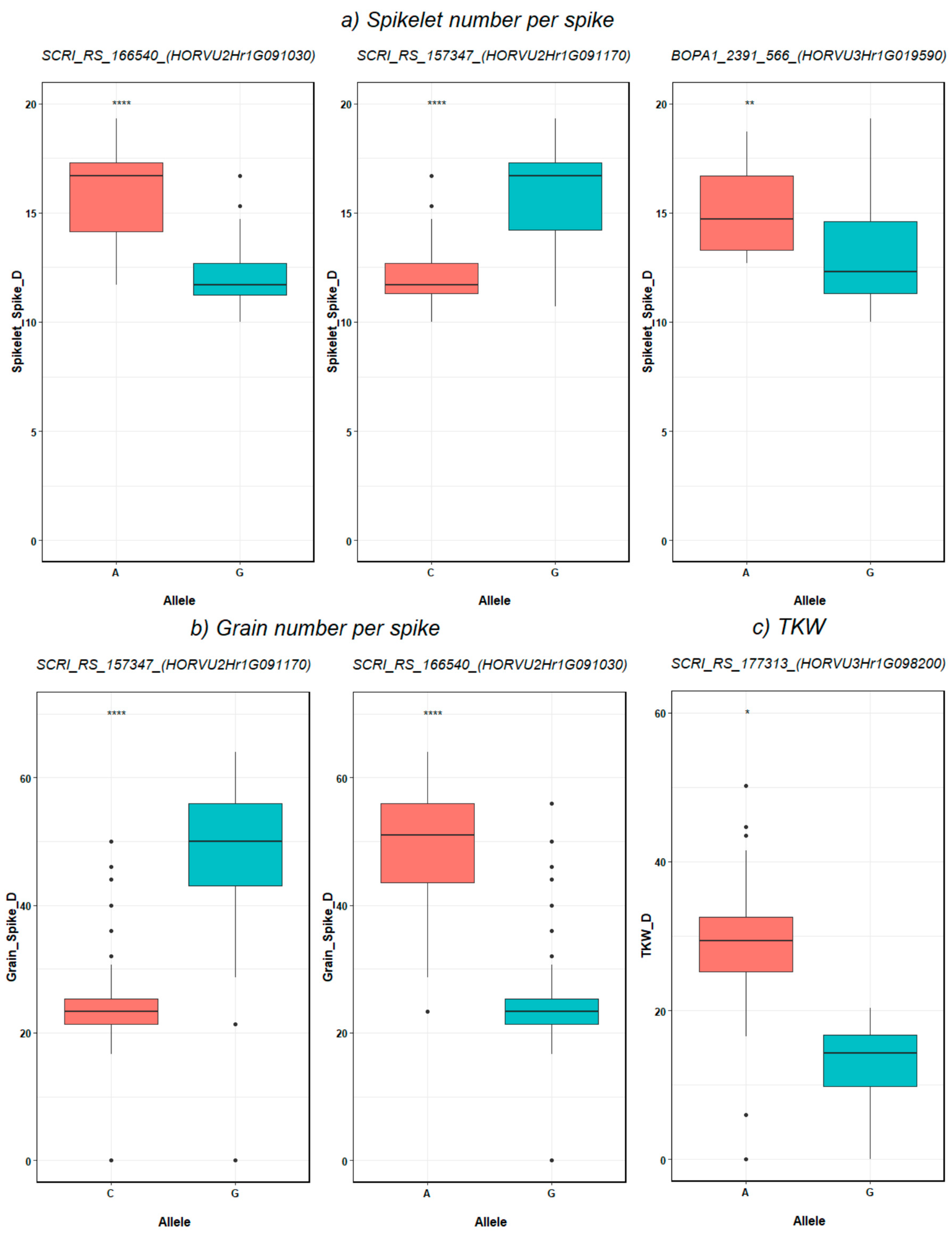
3.4. SNP-Gene-Based Analysis
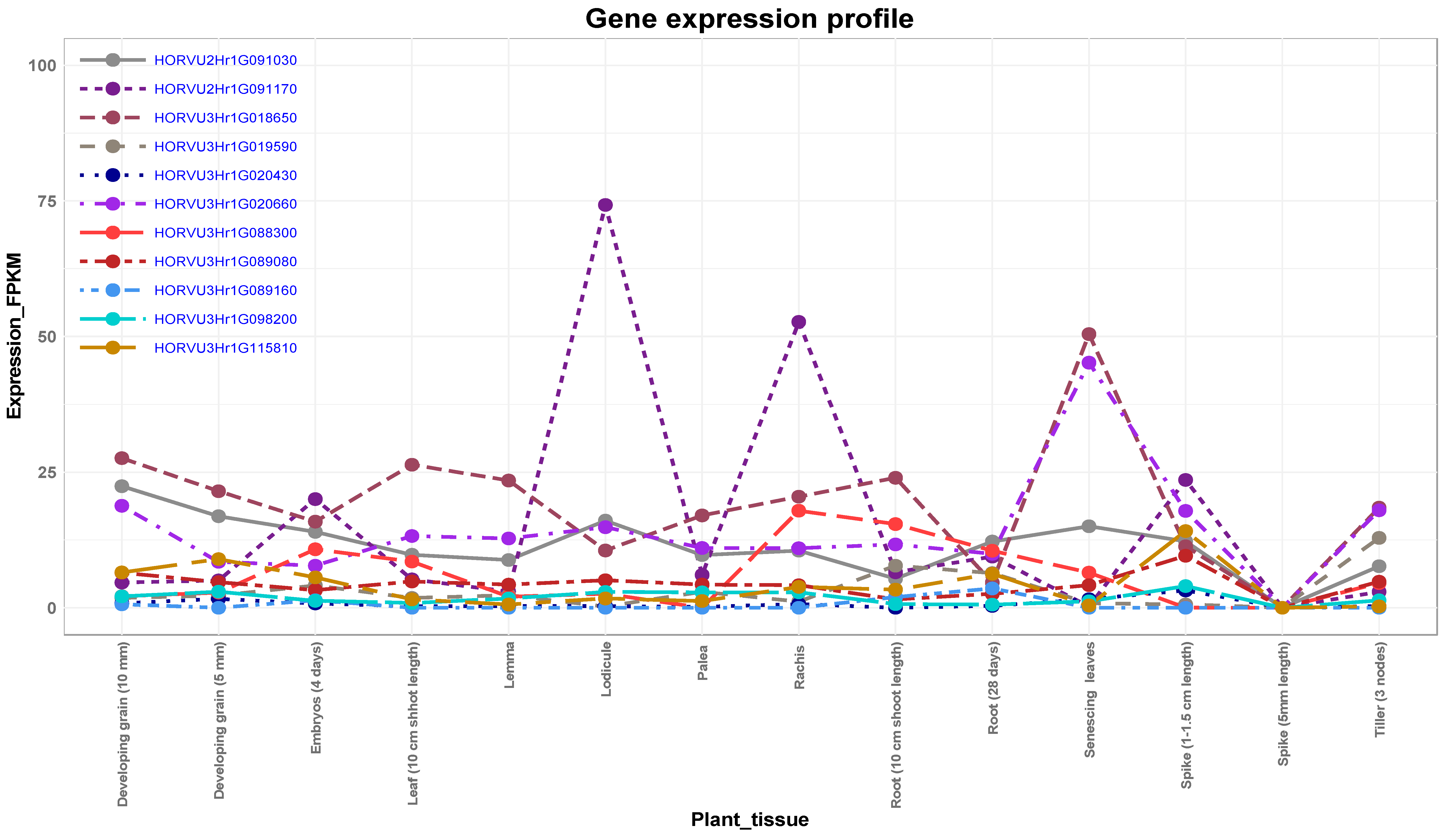
3.5. Expression Analysis of Candidate Genes
4. Discussion
The Role of Putative Candidate Genes in Drought Tolerance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lobell, D.B.; Schlenker, W.; Costa-Roberts, J. Climate trends and global crop production since 1980. Science 2011, 333, 616–620. [Google Scholar] [CrossRef] [Green Version]
- Samarah, N.H.; Alqudah, A.M.; Amayreh, J.A.; McAndrews, G.M. The effect of late-terminal drought stress on yield components of four barley cultivars. J. Agron. Crop Sci. 2009, 195, 427–441. [Google Scholar] [CrossRef]
- Thabet, S.G.; Alqudah, A.M. Crops and drought. In eLS; John Wiley & Sons, Ltd., Ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2019; pp. 1–8. [Google Scholar] [CrossRef]
- Sallam, A.; Alqudah, A.M.; Dawood, M.F.A.; Baenziger, P.S.; Borner, A. Drought stress tolerance in wheat and barley: Advances in physiology, breeding and genetics research. Int. J. Mol. Sci. 2019, 20, 3137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shavrukov, Y.; Kurishbayev, A.; Jatayev, S.; Shvidchenko, V.; Zotova, L.; Koekemoer, F.; de Groot, S.; Soole, K.; Langridge, P. Early flowering as a drought escape mechanism in plants: How can it aid wheat production? Front. Plant Sci. 2017, 8, 1950. [Google Scholar] [CrossRef] [PubMed]
- Alqudah, A.M.; Samarah, N.H.; Mullen, R.E. Drought stress effect on crop pollination, seed set, yield and quality. In Alternative Farming Systems, Biotechnology, Drought Stress and Ecological Fertilization; Springer: Dordrecht, The Netherlands, 2011; pp. 193–213. [Google Scholar] [CrossRef]
- Thabet, S.G.; Moursi, Y.S.; Karam, M.A.; Graner, A.; Alqudah, A.M. Genetic basis of drought tolerance during seed germination in barley. PLoS ONE 2018, 13, e0206682. [Google Scholar] [CrossRef]
- Fischer, R.; Turner, N.C. Plant productivity in the arid and semiarid zones. Annu. Rev. Plant Physiol. 1978, 29, 277–317. [Google Scholar] [CrossRef]
- Barnabas, B.; Jager, K.; Feher, A. The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 2008, 31, 11–38. [Google Scholar] [CrossRef]
- Samarah, N.; Alqudah, A. Effects of late-terminal drought stress on seed germination and vigor of barley (Hordeum vulgareL.). Arch. Agron. Soil Sci. 2011, 57, 27–32. [Google Scholar] [CrossRef]
- Botwright, T.; Condon, A.; Rebetzke, G.; Richards, R. Field evaluation of early vigour for genetic improvement of grain yield in wheat. Aust. J. Agric. Res. 2002, 53, 1137–1145. [Google Scholar] [CrossRef]
- Cowley, R.; Luckett, D.; Moroni, J.; Diffey, S. Use of remote sensing to determine the relationship of early vigour to grain yield in canola (Brassica napus L.) germplasm. Crop Pasture Sci. 2014, 65, 1288–1299. [Google Scholar] [CrossRef]
- Alqudah, A.M.; Sallam, A.; Stephen Baenziger, P.; Borner, A. GWAS: Fast-forwarding gene identification and characterization in temperate Cereals: Lessons from Barley—A review. J. Adv. Res. 2020, 22, 119–135. [Google Scholar] [CrossRef] [PubMed]
- Reig-Valiente, J.L.; Marques, L.; Talon, M.; Domingo, C. Genome-wide association study of agronomic traits in rice cultivated in temperate regions. BMC Genom. 2018, 19, 706. [Google Scholar] [CrossRef] [PubMed]
- Mazaheri, M.; Heckwolf, M.; Vaillancourt, B.; Gage, J.L.; Burdo, B.; Heckwolf, S.; Barry, K.; Lipzen, A.; Ribeiro, C.B.; Kono, T.J.Y.; et al. Genome-wide association analysis of stalk biomass and anatomical traits in maize. BMC Plant Biol. 2019, 19, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teulat, B.; Merah, O.; Sirault, X.; Borries, C.; Waugh, R.; This, D. QTLs for grain carbon isotope discrimination in field-grown barley. Theor. Appl. Genet. 2002, 106, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Zhu, G.; Li, Y.; Li, W.; Fu, J.; Niu, E.; Li, L.; Zhang, D.; Guo, W. Genome-wide association studies reveal genetic variation and candidate genes of drought stress related traits in cotton (Gossypium hirsutum L.). Front. Plant Sci. 2018, 9, 1276. [Google Scholar] [CrossRef] [PubMed]
- De Ronde, J.A.; Cress, W.A.; Kruger, G.H.; Strasser, R.J.; Van Staden, J. Photosynthetic response of transgenic soybean plants, containing an Arabidopsis P5CR gene, during heat and drought stress. J. Plant Physiol. 2004, 161, 1211–1224. [Google Scholar] [CrossRef]
- Haberle, J.; Svoboda, P.; Raimanová, I. The effect of post-anthesis water supply on grain nitrogen concentration and grain nitrogen šeld of winter wheat. Plant Soil Environ. 2018, 54, 304–312. [Google Scholar] [CrossRef] [Green Version]
- VSN-International. VSN GenStat for Windows, 18th ed.; VSN International: Hemel Hempstead, UK, 2016; Available online: https://genstat.kb.vsni.co.uk/knowledge-base/hcitegen/ (accessed on 15 August 2019).
- RStudio-Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2015; Available online: http://www.rstudio.com/ (accessed on 15 August 2019).
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [Green Version]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef] [Green Version]
- Al-Ajlouni, Z.; Al-Abdallat, A.; Al-Ghzawi, A.; Ayad, J.; Abu Elenein, J.; Al-Quraan, N.; Baenziger, P. Impact of pre-anthesis water deficit on yield and yield components in barley (Hordeum vulgare L.) plants grown under controlled conditions. Agronomy 2016, 6, 33. [Google Scholar] [CrossRef] [Green Version]
- Hossain, A.; Teixeira da Silva, J.A.; Lozovskaya, M.V.; Zvolinsky, V.P. High temperature combined with drought affect rainfed spring wheat and barley in South-Eastern Russia: I. Phenology and growth. Saudi J. Biol. Sci. 2012, 19, 473–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Liu, S.; Liu, Y.; Liu, Y.; You, J.; Deng, M.; Ma, J.; Chen, G.; Wei, Y.; Liu, C.; et al. Mapping and validation of major quantitative trait loci for kernel length in wild barley (Hordeum vulgare ssp. spontaneum). BMC Genet. 2016, 17, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Sharma, R.; Tondelli, A.; Russell, J.; Comadran, J.; Schnaithmann, F.; Pillen, K.; Kilian, B.; Cattivelli, L.; Thomas, W.T.B.; et al. Genome-wide association analysis of grain yield-associated traits in a pan-european barley cultivar collection. Plant Genome 2018, 11, 0073. [Google Scholar] [CrossRef]
- Sharafi, S.; Ghassemi-Golezani, K.; Mohammadi, S.; Lak, S.; Sorkhy, B. Evaluation of drought tolerance and yield potential in winter barley (Hordeum vulgare) genotypes. J. Food Agric. Environ. 2011, 9, 419–422. [Google Scholar]
- Perlikowski, D.; Kosmala, A.; Rapacz, M.; Koscielniak, J.; Pawlowicz, I.; Zwierzykowski, Z. Influence of short-term drought conditions and subsequent re-watering on the physiology and proteome of Lolium multiflorum/Festuca arundinacea introgression forms, with contrasting levels of tolerance to long-term drought. Plant Biol. Stuttg 2014, 16, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Koiwa, H.; Barb, A.W.; Xiong, L.; Li, F.; McCully, M.G.; Lee, B.H.; Sokolchik, I.; Zhu, J.; Gong, Z.; Reddy, M.; et al. C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc. Natl. Acad. Sci. USA 2002, 99, 10893–10898. [Google Scholar] [CrossRef] [Green Version]
- Thatcher, L.F.; Foley, R.; Casarotto, H.J.; Gao, L.L.; Kamphuis, L.G.; Melser, S.; Singh, K.B. The Arabidopsis RNA Polymerase II Carboxyl Terminal Domain (CTD) Phosphatase-Like1 (CPL1) is a biotic stress susceptibility gene. Sci. Rep. 2018, 8, 13454. [Google Scholar] [CrossRef]
- Ji, H.; Kim, S.R.; Kim, Y.H.; Kim, H.; Eun, M.Y.; Jin, I.D.; Cha, Y.S.; Yun, D.W.; Ahn, B.O.; Lee, M.C.; et al. Inactivation of the CTD phosphatase-like gene OsCPL1 enhances the development of the abscission layer and seed shattering in rice. Plant J. 2010, 61, 96–106. [Google Scholar] [CrossRef]
- Wu, Y.; Thorne, E.T.; Sharp, R.E.; Cosgrove, D.J. Modification of expansin transcript levels in the maize primary root at low water potentials. Plant Physiol. 2001, 126, 1471–1479. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Zhao, J.; Li, X.; Qin, L.; Yan, X.; Liao, H. A soybean beta-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J. 2011, 66, 541–552. [Google Scholar] [CrossRef]
- Dai, F.; Nevo, E.; Wu, D.; Comadran, J.; Zhou, M.; Qiu, L.; Chen, Z.; Beiles, A.; Chen, G.; Zhang, G. Tibet is one of the centers of domestication of cultivated barley. Proc. Natl. Acad. Sci. USA 2012, 109, 16969–16973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, P.; Kang, M.; Jiang, X.; Dai, F.; Gao, J.; Zhang, C. RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta 2013, 237, 1547–1559. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, C.; Yi, J.; Yang, Y.; Lei, C.; Gong, M. Transcriptome response to drought, rehydration and re-dehydration in potato. Int. J. Mol. Sci. 2019, 21, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Maury, L.; Marguerat, S.; Bahler, J. Tuning gene expression to changing environments: From rapid responses to evolutionary adaptation. Nat. Rev. Genet. 2008, 9, 583–593. [Google Scholar] [CrossRef]
- Kürsteiner, O.; Dupuis, I.; Kuhlemeier, C. The pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia but not other environmental stresses. Plant Physiol. 2003, 132, 968–978. [Google Scholar] [CrossRef] [Green Version]
- Uga, Y.; Kitomi, Y.; Ishikawa, S.; Yano, M. Genetic improvement for root growth angle to enhance crop production. Breed Sci. 2015, 65, 111–119. [Google Scholar] [CrossRef] [Green Version]
- Dolferus, R.; Ellis, M.; De Bruxelles, G.; Trevaskis, B.; Hoeren, F.; Dennis, E.; Peacock, W. Strategies of gene action in Arabidopsis during hypoxia. Ann. Bot. 1997, 79, 21–31. [Google Scholar] [CrossRef] [Green Version]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Jaradat, M.R.; Feurtado, J.A.; Huang, D.; Lu, Y.; Cutler, A.J. Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 2013, 13, 192. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.-H.; Fujii, H.; Zheng, X.; Zhu, J.-K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahaie, M.; Xue, G.P.; Naghavi, M.R.; Alizadeh, H.; Schenk, P.M. A MYB gene from wheat (Triticum aestivum L.) is up-regulated during salt and drought stresses and differentially regulated between salt-tolerant and sensitive genotypes. Plant Cell Rep. 2010, 29, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, G.; Jia, J.; Liu, X.; Kong, X. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J. Exp. Bot. 2012, 63, 203–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, T.; Fujisawa, M.; Shima, Y.; Ito, Y. The AP2/ERF transcription factor SlERF52 functions in flower pedicel abscission in tomato. J. Exp. Bot. 2014, 65, 3111–3119. [Google Scholar] [CrossRef] [Green Version]
- Koyama, T.; Nii, H.; Mitsuda, N.; Ohta, M.; Kitajima, S.; Ohme-Takagi, M.; Sato, F. A regulatory cascade involving class II ETHYLENE RESPONSE FACTOR transcriptional repressors operates in the progression of leaf senescence. Plant Physiol. 2013, 162, 991–1005. [Google Scholar] [CrossRef] [Green Version]
- Mehrnia, M.; Balazadeh, S.; Zanor, M.I.; Mueller-Roeber, B. EBE, an AP2/ERF transcription factor highly expressed in proliferating cells, affects shoot architecture in Arabidopsis. Plant Physiol. 2013, 162, 842–857. [Google Scholar] [CrossRef] [Green Version]
- Houston, K.; McKim, S.M.; Comadran, J.; Bonar, N.; Druka, I.; Uzrek, N.; Cirillo, E.; Guzy-Wrobelska, J.; Collins, N.C.; Halpin, C.; et al. Variation in the interaction between alleles of HvAPETALA2 and microRNA172 determines the density of grains on the barley inflorescence. Proc. Natl. Acad. Sci. USA 2013, 110, 16675–16680. [Google Scholar] [CrossRef] [Green Version]
- Johansson, I.; Karlsson, M.; Johanson, U.; Larsson, C.; Kjellbom, P. The role of aquaporins in cellular and whole plant water balance. Biochim. Biophys. Acta 2000, 1465, 324–342. [Google Scholar] [CrossRef] [Green Version]
- Maurel, C.; Verdoucq, L.; Luu, D.T.; Santoni, V. Plant aquaporins: Membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 2008, 59, 595–624. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Ban, L.; Wen, H.; Wang, Z.; Dzyubenko, N.; Chapurin, V.; Gao, H.; Wang, X. An aquaporin protein is associated with drought stress tolerance. Biochem. Biophys. Res. Commun. 2015, 459, 208–213. [Google Scholar] [CrossRef] [Green Version]
- Afzal, Z.; Howton, T.C.; Sun, Y.; Mukhtar, M.S. The roles of aquaporins in plant stress responses. J. Dev. Biol. 2016, 4, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, X.; Ling, H.-Q. Current advances in genome sequencing of common wheat and its ancestral species. Crop J. 2018, 6, 15–21. [Google Scholar] [CrossRef]
- Hedden, P. The genes of the green revolution. Trends Genet. 2003, 19, 5–9. [Google Scholar] [CrossRef]
- Borner, A.; Worland, A.; Plaschke, J.; Schumann, E.; Law, C. Pleiotropic effects of genes for reduced height (Rht) and day-length insensitivity (Ppd) on yield and its components for wheat grown in middle Europe. Plant Breed. 1993, 111, 204–216. [Google Scholar] [CrossRef]
- Youssefian, S.; Kirby, E.; Gale, M. Pleiotropic effects of the GA-insensitive Rht dwarfing genes in wheat. 2. Effects on leaf, stem, ear and floret growth. Field Crops Res. 1992, 28, 191–210. [Google Scholar] [CrossRef]
- Geisel, N. Constitutive versus responsive gene expression strategies for growth in changing environments. PLoS ONE 2011, 6, e27033. [Google Scholar] [CrossRef] [Green Version]
- Blum, A. Drought resistance and its improvement. In Plant Breeding for Water-Limited Environments; Springer: Berlin/Heidelberg, Germany, 2011; pp. 53–152. [Google Scholar]
- Dawe, R.K.; Reed, L.M.; Yu, H.-G.; Muszynski, M.G.; Hiatt, E.N. A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 1999, 11, 1227–1238. [Google Scholar] [CrossRef]
- Zhong, C.X.; Marshall, J.B.; Topp, C.; Mroczek, R.; Kato, A.; Nagaki, K.; Birchler, J.A.; Jiang, J.; Dawe, R.K. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 2002, 14, 2825–2836. [Google Scholar] [CrossRef]
- Cheeseman, I.M.; Desai, A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell. Biol. 2008, 9, 33–46. [Google Scholar] [CrossRef]
- Zheng, H.; Wu, H.; Pan, X.; Jin, W.; Li, X. Aberrant meiotic modulation partially contributes to the lower germination rate of pollen grains in maize (Zea mays L.) under low nitrogen supply. Plant Cell Physiol. 2017, 58, 342–353. [Google Scholar] [CrossRef]
- Li, X.; Dawe, R.K. Fused sister kinetochores initiate the reductional division in meiosis I. Nat. Cell Biol. 2009, 11, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Powers, A.F.; Franck, A.D.; Gestaut, D.R.; Cooper, J.; Gracyzk, B.; Wei, R.R.; Wordeman, L.; Davis, T.N.; Asbury, C.L. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell 2009, 136, 865–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabbari, M.; Fakheri, B.A.; Aghnoum, R.; Mahdi Nezhad, N.; Ataei, R. GWAS analysis in spring barley (Hordeum vulgare L.) for morphological traits exposed to drought. PLoS ONE 2018, 13, e0204952. [Google Scholar] [CrossRef]
- Varshney, R.K.; Paulo, M.J.; Grando, S.; van Eeuwijk, F.A.; Keizer, L.C.P.; Guo, P.; Ceccarelli, S.; Kilian, A.; Baum, M.; Graner, A. Genome wide association analyses for drought tolerance related traits in barley (Hordeum vulgare L.). Field Crops Res. 2012, 126, 171–180. [Google Scholar] [CrossRef] [Green Version]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.; Gampala, S.S.L.; Lynch, T.J.; Thomas, T.L.; Rock, C.D. Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol. 2005, 59, 253–267. [Google Scholar] [CrossRef]
- Mittal, A.; Gampala, S.S.L.; Ritchie, G.L.; Payton, P.; Burke, J.J.; Rock, C.D. Related to ABA-Insensitive3(ABI3)/Viviparous1 and AtABI5 transcription factor coexpression in cotton enhances drought stress adaptation. Plant Biotechnol. J. 2014, 12, 578–589. [Google Scholar] [CrossRef] [Green Version]
- Tang, N.; Zhang, H.; Li, X.; Xiao, J.; Xiong, L. Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 2012, 158, 1755–1768. [Google Scholar] [CrossRef] [Green Version]



| Trait | Abbreviation | Description | |
|---|---|---|---|
| Control Drought | |||
| Awn Tipping | AT_C | AT_D | The number of days from planting up to awn tipping. |
| Spike Heading | SH_C | SH_D | The number of days from planting up to spike heading |
| Anther Extrusion | AE_C | AE_D | The number of days from planting up to anther extrusion. |
| Plant Height | PH_C | PH_D | The distance between the ground level to the tip of the highest spikelet (excluding awns) in cm. |
| Spike Length | SL_C | SL_D | Distance from the base of the spike to the tip of the highest spikelet (excluding awns) in cm. |
| Awn Length | AL_C | AL_D | Distance from the tip of the spike to the end of the awn in cm. |
| No of Spikelets per Spike | NSS_C | NSS_D | The actual count of the number of spikelets. |
| No of Grains per Spike | NGS_C | NGS_D | The actual count of the number of the grains. |
| Thousand Grain Weight | TGW_C | TGW_D | The weight of 1000 grains randomly taken from each plot in gram (g). |
| Drought tolerance index (Awn Tipping) | DTI_AT | DTI | |
| Drought tolerance index (Spike Heading) | DTI_SH | DTI | |
| Drought tolerance index (Anther Extrusion) | DTI_AE | DTI | |
| Drought tolerance index (Plant Height) | DTI_PH | DTI | |
| Drought tolerance index (Spike Length) | DTI_SL | DTI | |
| Drought tolerance index (Awn Length) | DTI_AL | DTI | |
| Drought tolerance index (No of Spikelet per Spike) | DTI_NSS | DTI | |
| Drought tolerance index (No of Grain per Spike) | DTI_NGS | DTI | |
| Drought tolerance index (Thousand Grain Weight) | DTI_TGW | DTI | |
| Trait | Control | H2 | Drought | H2 | ||||
|---|---|---|---|---|---|---|---|---|
| T | G | T × G | T | G | T × G | |||
| Awn Tipping | ns | *** | ** | 0.72 | ns | ** | *** | 0.84 |
| Spike Heading | ns | *** | ** | 0.80 | ns | ** | *** | 0.78 |
| Anther Extrusion | ns | *** | * | 0.79 | ns | * | *** | 0.81 |
| Plant Height | *** | *** | *** | 0.75 | *** | *** | *** | 0.71 |
| Spike Length | *** | *** | *** | 0.74 | *** | *** | *** | 0.69 |
| Awn Length | *** | *** | *** | 0.79 | *** | *** | *** | 0.76 |
| No of Spikelet per Spike | *** | *** | *** | 0.76 | *** | *** | *** | 0.65 |
| No of Grain per Spike | *** | *** | *** | 0.77 | *** | *** | *** | 0.68 |
| Thousand Grain Weight | *** | *** | *** | 0.72 | *** | *** | *** | 0.64 |
| Awn Tipping_DTI | ̶ | *** | ̶ | 0.78 | ||||
| Spike Heading_DTI | ̶ | *** | ̶ | 0.71 | ||||
| Anther Extrusion_DTI | ̶ | *** | ̶ | 0.68 | ||||
| Plant Height_DTI | ̶ | *** | ̶ | 0.73 | ||||
| Spike Length_DTI | ̶ | *** | ̶ | 0.74 | ||||
| Awn Length_DTI | ̶ | *** | ̶ | 0.71 | ||||
| No of Spikelet per Spike _DTI | ̶ | *** | ̶ | 0.74 | ||||
| No of Grain per Spike _DTI | ̶ | *** | ̶ | 0.68 | ||||
| Thousand Grain Weight_DTI | ̶ | *** | ̶ | 0.77 | ||||
| Trait | Marker | Chr | Pos. | SNP Pos. | Gene | Gene Pos. | GO ID | Annotation in Barley | Orthologs |
|---|---|---|---|---|---|---|---|---|---|
| NGS_C NGS_D NSS_D | SCRI_RS_166540 | 2H | 75.56 | 646934425 | HORVU2Hr1G091030 | 646930069-646939693 | None | RNA polymerase II C-terminal domain phosphatase-like 1 | AT4G21670 |
| NGS_D NSS_D | SCRI_RS_157347 | 2H | 75.56 | 647255135 | HORVU2Hr1G091170 | 647252087-647258342 | None | expansin B3 | AT4G28250 |
| AE_C NGS_C NSS_C | SCRI_RS_229693 | 3H | 44.26 | 48643412 | HORVU3Hr1G018650 | 48643412-48647662 | GO:0000287 GO:0003824 GO:0030976 | pyruvate decarboxylase-2 | AT5G54960 |
| AE_C NGS_C NSS_C | BOPA2_12_30737 | 3H | 45.55 | 63626447 | HORVU3Hr1G020430 | 63623393-63623651 | None | Protein HASTY 1 | AT3G05040 |
| AE_C NGS_C NSS_C | BOPA1_7045-950 | 3H | 45.55 | 64618663 | HORVU3Hr1G020660 | 64617802-64621872 | GO:0005515 | Chromosome 3B, genomic scaffold, cultivar Chinese Spring | AT2G36270 |
| AE_C NGS_C NSS_D | BOPA1_2391-566 | 3H | 46.25 | 55590274 | HORVU3Hr1G019590 | 55915087-55917096 | GO:0003677 | myb domain protein 37 | AT5G23000 |
| AE_C SH_C | SCRI_RS_230075 | 3H | 103.27 | 624309908 | HORVU3Hr1G088300 | 624309585-624311169 | None | Chromosome 3B, genomic scaffold, cultivar Chinese Spring | AT2G36270 |
| AE_C SH_C | BOPA2_12_30223 | 3H | 104.32 | 627767267 | HORVU3Hr1G089160 | 627749990-627754917 | GO:0003677 GO:0003700 GO:0006355 | AP2-like ethylene-responsive transcription factor | AT2G28550 |
| AE_C SH_C | SCRI_RS_192360 | 3H | 104.26 | 627260224 | HORVU3Hr1G089080 | 627258327-627265762 | None | undescribed protein | |
| AE_DTI AL_DTI AE_C AL_C SH_C SL_C TGW_C TGW_D SH_DTI AT_DTI | SCRI_RS_177313 | 3H | 126.69 | 657713242 | HORVU3Hr1G098200 | 657712024-657713975 | GO:0005515 | Chromosome 3B, genomic scaffold, cultivar Chinese Spring | AT2G36270 |
| AE_DTI SH_DTI | BOPA2_12_10981 | 3H | 154.15 | 696452271 | HORVU3Hr1G116790 | 696450874-696453390 | GO:0005215 GO:0006810 GO:0016020 | Aquaporin-like superfamily protein | AT2G36830 |
| AE_DTI SH_DTI | SCRI_RS_237738 | 3H | 154.6 | 694105354 | HORVU3Hr1G115810 | 694103839-694106776 | None | Kinetochore protein spc25 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thabet, S.G.; Moursi, Y.S.; Karam, M.A.; Börner, A.; Alqudah, A.M. Natural Variation Uncovers Candidate Genes for Barley Spikelet Number and Grain Yield under Drought Stress. Genes 2020, 11, 533. https://doi.org/10.3390/genes11050533
Thabet SG, Moursi YS, Karam MA, Börner A, Alqudah AM. Natural Variation Uncovers Candidate Genes for Barley Spikelet Number and Grain Yield under Drought Stress. Genes. 2020; 11(5):533. https://doi.org/10.3390/genes11050533
Chicago/Turabian StyleThabet, Samar G., Yasser S. Moursi, Mohamed A. Karam, Andreas Börner, and Ahmad M. Alqudah. 2020. "Natural Variation Uncovers Candidate Genes for Barley Spikelet Number and Grain Yield under Drought Stress" Genes 11, no. 5: 533. https://doi.org/10.3390/genes11050533
APA StyleThabet, S. G., Moursi, Y. S., Karam, M. A., Börner, A., & Alqudah, A. M. (2020). Natural Variation Uncovers Candidate Genes for Barley Spikelet Number and Grain Yield under Drought Stress. Genes, 11(5), 533. https://doi.org/10.3390/genes11050533






