Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Ergosterol Synthesis, Uptake and Detoxification in S. cerevisiae
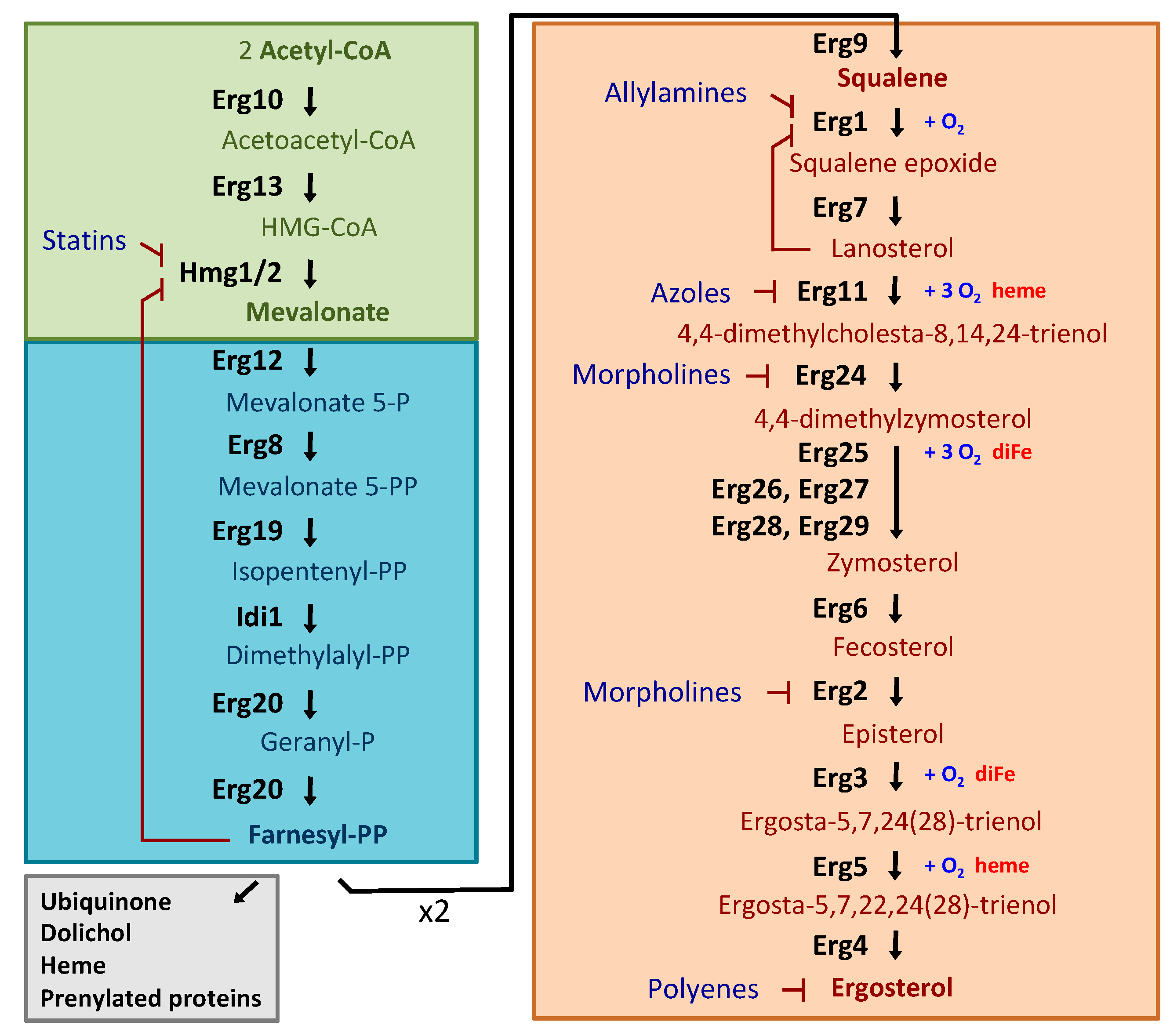
2.1. Ergosterol Biosynthesis in S. cerevisiae
2.2. Sterol Acquisition and Transport in S. cerevisiae
2.3. Sterol Detoxification
3. Regulation of Ergosterol Biosynthesis
3.1. Subcellular Localization of Ergosterol Biosynthesis Enzymes
3.2. Post-Translational Feedback Regulation
3.3. Transcriptional Regulation
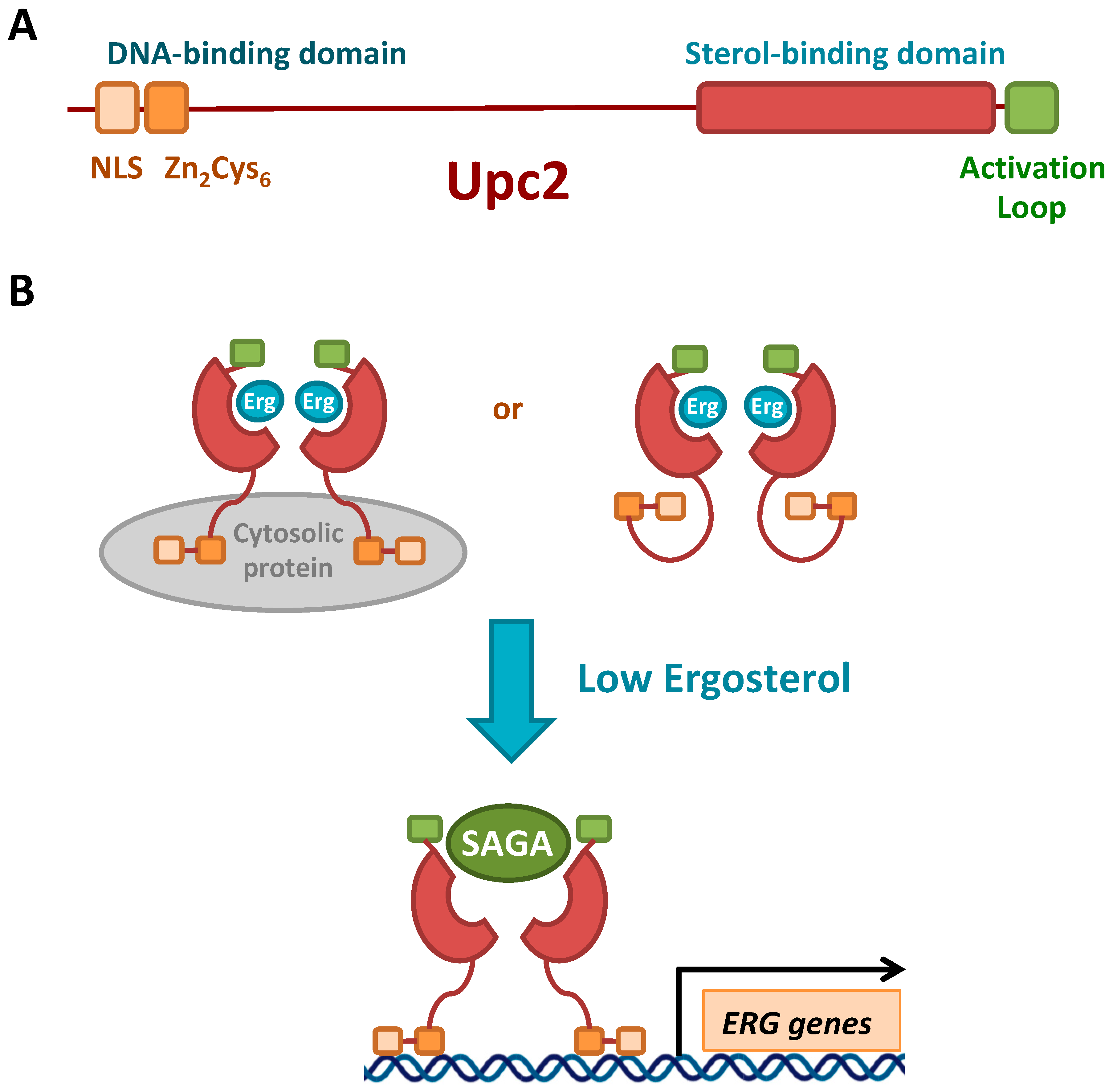
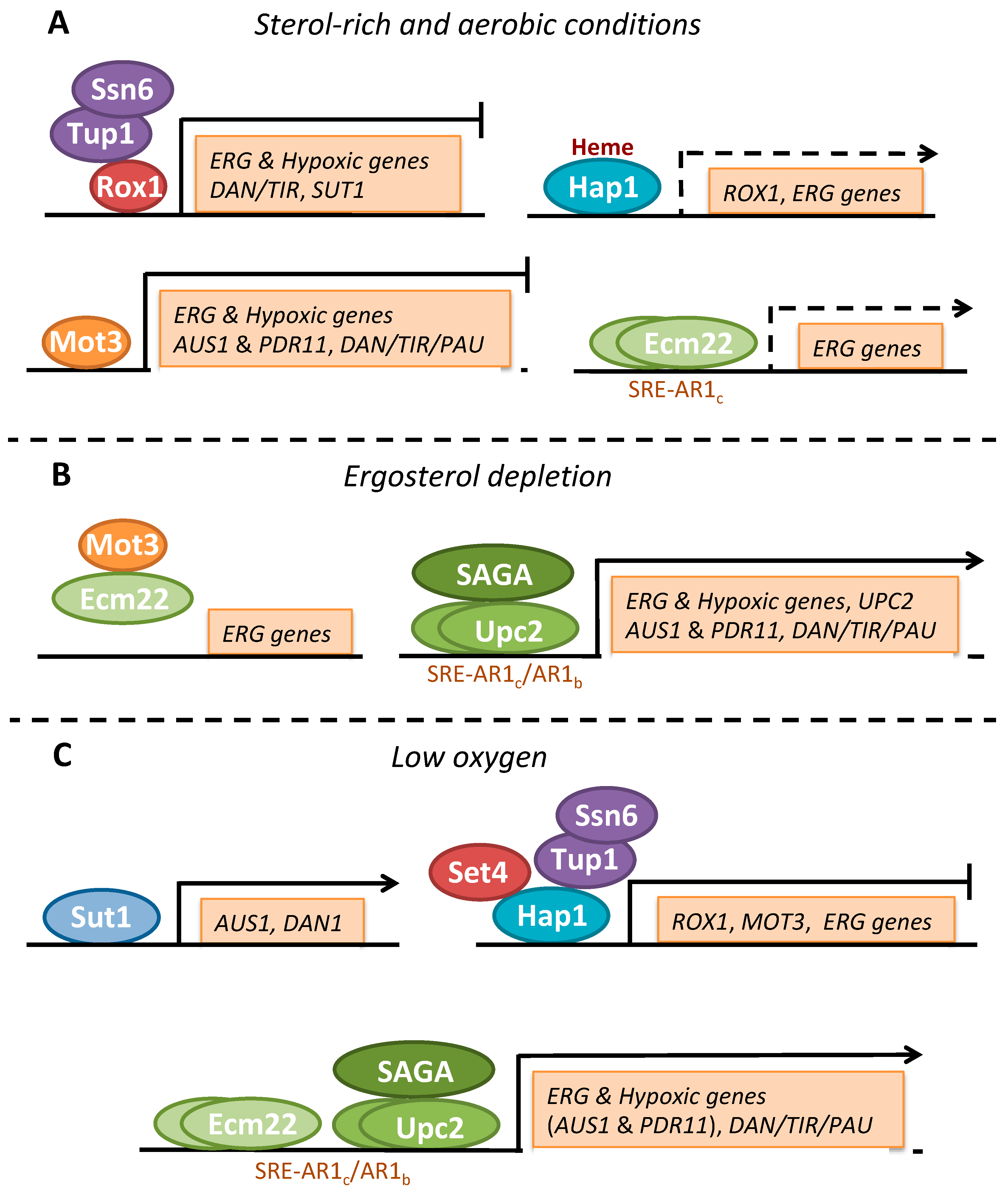
3.3.1. Transcriptional Regulation by Sterols
3.3.2. Transcriptional Regulation by Oxygen
3.3.3. Transcriptional Regulation by Osmotic Stress
3.4. Regulation by Iron Bioavailability
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maxfield, F.R.; Tabas, I. Role of cholesterol and lipid organization in disease. Nature 2005, 438, 612–621. [Google Scholar] [CrossRef] [PubMed]
- Tarkowska, D.; Strnad, M. Plant ecdysteroids: Plant sterols with intriguing distributions, biological effects and relations to plant hormones. Planta 2016, 244, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.L. The Multifunctional Fungal Ergosterol. mBio 2018, 9. [Google Scholar] [CrossRef]
- Cirigliano, A.; Macone, A.; Bianchi, M.M.; Oliaro-Bosso, S.; Balliano, G.; Negri, R.; Rinaldi, T. Ergosterol reduction impairs mitochondrial DNA maintenance in S. cerevisiae. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 290–303. [Google Scholar] [CrossRef]
- Gerhold, J.M.; Cansiz-Arda, S.; Lohmus, M.; Engberg, O.; Reyes, A.; van Rennes, H.; Sanz, A.; Holt, I.J.; Cooper, H.M.; Spelbrink, J.N. Human Mitochondrial DNA-Protein Complexes Attach to a Cholesterol-Rich Membrane Structure. Sci. Rep. 2015, 5, 15292. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; He, B.; Ma, L.; Sun, Y.; Niu, Y.; Zeng, B. Recent Advances in Ergosterol Biosynthesis and Regulation Mechanisms in Saccharomyces cerevisiae. Indian J. Microbiol. 2017, 57, 270–277. [Google Scholar] [CrossRef]
- Montanes, F.M.; Pascual-Ahuir, A.; Proft, M. Repression of ergosterol biosynthesis is essential for stress resistance and is mediated by the Hog1 MAP kinase and the Mot3 and Rox1 transcription factors. Mol. Microbiol. 2011, 79, 1008–1023. [Google Scholar] [CrossRef]
- Liu, J.F.; Xia, J.J.; Nie, K.L.; Wang, F.; Deng, L. Outline of the biosynthesis and regulation of ergosterol in yeast. World J. Microbiol. Biotechnol. 2019, 35, 98. [Google Scholar] [CrossRef]
- Shakoury-Elizeh, M.; Protchenko, O.; Berger, A.; Cox, J.; Gable, K.; Dunn, T.M.; Prinz, W.A.; Bard, M.; Philpott, C.C. Metabolic response to iron deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 2010, 285, 14823–14833. [Google Scholar] [CrossRef]
- Zhang, Y.; Nielsen, J.; Liu, Z. Engineering yeast metabolism for production of terpenoids for use as perfume ingredients, pharmaceuticals and biofuels. FEMS Yeast Res. 2017, 17. [Google Scholar] [CrossRef]
- Vil, V.A.; Gloriozova, T.A.; Poroikov, V.V.; Terent’ev, A.O.; Savidov, N.; Dembitsky, V.M. Peroxy steroids derived from plant and fungi and their biological activities. Appl. Microbiol. Biotechnol. 2018, 102, 7657–7667. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Pan, M.; Liu, H.; Tian, H.; Ye, Q.; Liu, H. Ergosterol peroxide inhibits ovarian cancer cell growth through multiple pathways. Onco Targets Ther. 2017, 10, 3467–3474. [Google Scholar] [CrossRef] [PubMed]
- Ward, D.M.; Chen, O.S.; Li, L.; Kaplan, J.; Bhuiyan, S.A.; Natarajan, S.K.; Bard, M.; Cox, J.E. Altered sterol metabolism in budding yeast affects mitochondrial iron-sulfur (Fe-S) cluster synthesis. J. Biol. Chem. 2018, 293, 10782–10795. [Google Scholar] [CrossRef]
- Zinser, E.; Paltauf, F.; Daum, G. Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J. Bacteriol. 1993, 175, 2853–2858. [Google Scholar] [CrossRef]
- Joshua, I.M.; Hofken, T. From Lipid Homeostasis to Differentiation: Old and New Functions of the Zinc Cluster Proteins Ecm22, Upc2, Sut1 and Sut2. Int. J. Mol. Sci. 2017, 18, 772. [Google Scholar] [CrossRef] [PubMed]
- Johnston, E.J.; Moses, T.; Rosser, S.J. The wide-Ranging phenotypes of ergosterol biosynthesis mutants, and implications for microbial cell factories. Yeast 2020, 37, 27–44. [Google Scholar] [CrossRef] [PubMed]
- Heese-Peck, A.; Pichler, H.; Zanolari, B.; Watanabe, R.; Daum, G.; Riezman, H. Multiple functions of sterols in yeast endocytosis. Mol. Biol. Cell 2002, 13, 2664–2680. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, S.S.; Trushina, N.I.; Severin, F.F.; Knorre, D.A. Ergosterol Turnover in Yeast: An Interplay between Biosynthesis and Transport. Biochemistry (Mosc) 2019, 84, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Esquivel, B.D.; White, T.C. Overexpression or Deletion of Ergosterol Biosynthesis Genes Alters Doubling Time, Response to Stress Agents, and Drug Susceptibility in Saccharomyces cerevisiae. mBio 2018, 9. [Google Scholar] [CrossRef]
- Ruckenstuhl, C.; Lang, S.; Poschenel, A.; Eidenberger, A.; Baral, P.K.; Kohut, P.; Hapala, I.; Gruber, K.; Turnowsky, F. Characterization of squalene epoxidase of Saccharomyces cerevisiae by applying terbinafine-sensitive variants. Antimicrob. Agents Chemother. 2007, 51, 275–284. [Google Scholar] [CrossRef]
- Berkow, E.L.; Lockhart, S.R. Fluconazole resistance in Candida species: A current perspective. Infect Drug Resist 2017, 10, 237–245. [Google Scholar] [CrossRef]
- Kelly, S.L.; Lamb, D.C.; Kelly, D.E.; Manning, N.J.; Loeffler, J.; Hebart, H.; Schumacher, U.; Einsele, H. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol delta5,6-desaturation. FEBS Lett. 1997, 400, 80–82. [Google Scholar] [CrossRef]
- Sanglard, D.; Ischer, F.; Parkinson, T.; Falconer, D.; Bille, J. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob. Agents Chemother. 2003, 47, 2404–2412. [Google Scholar] [CrossRef]
- White, T.C.; Marr, K.A.; Bowden, R.A. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 1998, 11, 382–402. [Google Scholar] [CrossRef] [PubMed]
- Serhan, G.; Stack, C.M.; Perrone, G.G.; Morton, C.O. The polyene antifungals, amphotericin B and nystatin, cause cell death in Saccharomyces cerevisiae by a distinct mechanism to amphibian-derived antimicrobial peptides. Ann. Clin. Microbiol. Antimicrob. 2014, 13, 18. [Google Scholar] [CrossRef] [PubMed]
- Espenshade, P.J.; Hughes, A.L. Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet 2007, 41, 401–427. [Google Scholar] [CrossRef]
- Kodedova, M.; Sychrova, H. Changes in the Sterol Composition of the Plasma Membrane Affect Membrane Potential, Salt Tolerance and the Activity of Multidrug Resistance Pumps in Saccharomyces cerevisiae. PLoS ONE 2015, 10, e0139306. [Google Scholar] [CrossRef]
- Lorenz, R.T.; Parks, L.W. Regulation of ergosterol biosynthesis and sterol uptake in a sterol-auxotrophic yeast. J. Bacteriol. 1987, 169, 3707–3711. [Google Scholar] [CrossRef]
- Hughes, A.L.; Todd, B.L.; Espenshade, P.J. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 2005, 120, 831–842. [Google Scholar] [CrossRef]
- Zavrel, M.; Hoot, S.J.; White, T.C. Comparison of sterol import under aerobic and anaerobic conditions in three fungal species, Candida albicans, Candida glabrata, and Saccharomyces cerevisiae. Eukaryot Cell 2013, 12, 725–738. [Google Scholar] [CrossRef]
- Kohut, P.; Wustner, D.; Hronska, L.; Kuchler, K.; Hapala, I.; Valachovic, M. The role of ABC proteins Aus1p and Pdr11p in the uptake of external sterols in yeast: Dehydroergosterol fluorescence study. Biochem. Biophys. Res. Commun. 2011, 404, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Prinz, W.A. ATP-Binding cassette (ABC) transporters mediate nonvesicular, raft-Modulated sterol movement from the plasma membrane to the endoplasmic reticulum. J. Biol. Chem. 2004, 279, 45226–45234. [Google Scholar] [CrossRef] [PubMed]
- Gulati, S.; Balderes, D.; Kim, C.; Guo, Z.A.; Wilcox, L.; Area-Gomez, E.; Snider, J.; Wolinski, H.; Stagljar, I.; Granato, J.T.; et al. ATP-Binding cassette transporters and sterol O-acyltransferases interact at membrane microdomains to modulate sterol uptake and esterification. FASEB J. 2015, 29, 4682–4694. [Google Scholar] [CrossRef] [PubMed]
- Georgiev, A.G.; Sullivan, D.P.; Kersting, M.C.; Dittman, J.S.; Beh, C.T.; Menon, A.K. Osh proteins regulate membrane sterol organization but are not required for sterol movement between the ER and PM. Traffic 2011, 12, 1341–1355. [Google Scholar] [CrossRef]
- Kentala, H.; Weber-Boyvat, M.; Olkkonen, V.M. OSBP-Related Protein Family: Mediators of Lipid Transport and Signaling at Membrane Contact Sites. Int. Rev. Cell Mol. Biol. 2016, 321, 299–340. [Google Scholar]
- Tian, S.; Ohta, A.; Horiuchi, H.; Fukuda, R. Oxysterol-Binding protein homologs mediate sterol transport from the endoplasmic reticulum to mitochondria in yeast. J. Biol. Chem. 2018, 293, 5636–5648. [Google Scholar] [CrossRef]
- Gatta, A.T.; Wong, L.H.; Sere, Y.Y.; Calderon-Norena, D.M.; Cockcroft, S.; Menon, A.K.; Levine, T.P. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. Elife 2015, 4. [Google Scholar] [CrossRef]
- Tong, J.; Manik, M.K.; Im, Y.J. Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites. Proc. Natl. Acad. Sci. USA 2018, 115, E856–E865. [Google Scholar] [CrossRef]
- Tinkelenberg, A.H.; Liu, Y.; Alcantara, F.; Khan, S.; Guo, Z.; Bard, M.; Sturley, S.L. Mutations in yeast ARV1 alter intracellular sterol distribution and are complemented by human ARV1. J. Biol. Chem. 2000, 275, 40667–40670. [Google Scholar] [CrossRef]
- Kajiwara, K.; Watanabe, R.; Pichler, H.; Ihara, K.; Murakami, S.; Riezman, H.; Funato, K. Yeast ARV1 is required for efficient delivery of an early GPI intermediate to the first mannosyltransferase during GPI assembly and controls lipid flow from the endoplasmic reticulum. Mol. Biol. Cell 2008, 19, 2069–2082. [Google Scholar] [CrossRef]
- Georgiev, A.G.; Johansen, J.; Ramanathan, V.D.; Sere, Y.Y.; Beh, C.T.; Menon, A.K. Arv1 regulates PM and ER membrane structure and homeostasis but is dispensable for intracellular sterol transport. Traffic 2013, 14, 912–921. [Google Scholar] [CrossRef] [PubMed]
- Demuyser, L.; Swinnen, E.; Fiori, A.; Herrera-Malaver, B.; Vestrepen, K.; Van Dijck, P. Mitochondrial Cochaperone Mge1 Is Involved in Regulating Susceptibility to Fluconazole in Saccharomyces cerevisiae and Candida Species. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Munoz, A.J.; Giusiano, G.; Ezkurra, P.A.; Quindos, G. Antifungal agents: Mode of action in yeast cells. Rev. Esp. Quimioter. 2006, 19, 130–139. [Google Scholar] [PubMed]
- Zweytick, D.; Leitner, E.; Kohlwein, S.D.; Yu, C.; Rothblatt, J.; Daum, G. Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 2000, 267, 1075–1082. [Google Scholar] [CrossRef]
- Koffel, R.; Tiwari, R.; Falquet, L.; Schneiter, R. The Saccharomyces cerevisiae YLL012/YEH1, YLR020/YEH2, and TGL1 genes encode a novel family of membrane-anchored lipases that are required for steryl ester hydrolysis. Mol. Cell Biol. 2005, 25, 1655–1668. [Google Scholar] [CrossRef] [PubMed]
- Koffel, R.; Schneiter, R. Yeh1 constitutes the major steryl ester hydrolase under heme-deficient conditions in Saccharomyces cerevisiae. Eukaryot. Cell 2006, 5, 1018–1025. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jandrositz, A.; Petschnigg, J.; Zimmermann, R.; Natter, K.; Scholze, H.; Hermetter, A.; Kohlwein, S.D.; Leber, R. The lipid droplet enzyme Tgl1p hydrolyzes both steryl esters and triglycerides in the yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta 2005, 1735, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Sorger, D.; Athenstaedt, K.; Hrastnik, C.; Daum, G. A yeast strain lacking lipid particles bears a defect in ergosterol formation. J. Biol. Chem. 2004, 279, 31190–31196. [Google Scholar] [CrossRef]
- Arthington-Skaggs, B.A.; Crowell, D.N.; Yang, H.; Sturley, S.L.; Bard, M. Positive and negative regulation of a sterol biosynthetic gene (ERG3) in the post-squalene portion of the yeast ergosterol pathway. FEBS Lett. 1996, 392, 161–165. [Google Scholar] [CrossRef]
- Ploier, B.; Korber, M.; Schmidt, C.; Koch, B.; Leitner, E.; Daum, G. Regulatory link between steryl ester formation and hydrolysis in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 2015, 1851, 977–986. [Google Scholar] [CrossRef]
- Tiwari, R.; Koffel, R.; Schneiter, R. An acetylation/deacetylation cycle controls the export of sterols and steroids from S. cerevisiae. EMBO J. 2007, 26, 5109–5119. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, V.; Schneiter, R. Pathogen-Related Yeast (PRY) proteins and members of the CAP superfamily are secreted sterol-binding proteins. Proc. Natl. Acad. Sci. USA 2012, 109, 16882–16887. [Google Scholar] [CrossRef] [PubMed]
- Mo, C.; Bard, M. A systematic study of yeast sterol biosynthetic protein-protein interactions using the split-Ubiquitin system. Biochim. Biophys. Acta 2005, 1737, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Mullner, H.; Zweytick, D.; Leber, R.; Turnowsky, F.; Daum, G. Targeting of proteins involved in sterol biosynthesis to lipid particles of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 2004, 1663, 9–13. [Google Scholar] [CrossRef]
- Kristan, K.; Rizner, T.L. Steroid-Transforming enzymes in fungi. J. Steroid Biochem. Mol. Biol. 2012, 129, 79–91. [Google Scholar] [CrossRef]
- Caldas, H.; Herman, G.E. NSDHL, an enzyme involved in cholesterol biosynthesis, traffics through the Golgi and accumulates on ER membranes and on the surface of lipid droplets. Hum. Mol. Genet. 2003, 12, 2981–2991. [Google Scholar] [CrossRef]
- Leber, R.; Landl, K.; Zinser, E.; Ahorn, H.; Spok, A.; Kohlwein, S.D.; Turnowsky, F.; Daum, G. Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol. Biol. Cell 1998, 9, 375–386. [Google Scholar] [CrossRef]
- Mukherjee, D.; Gao, M.; O’Connor, J.P.; Raijmakers, R.; Pruijn, G.; Lutz, C.S.; Wilusz, J. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002, 21, 165–174. [Google Scholar] [CrossRef]
- Mo, C.; Valachovic, M.; Bard, M. The ERG28-Encoded protein, Erg28p, interacts with both the sterol C-4 demethylation enzyme complex as well as the late biosynthetic protein, the C-24 sterol methyltransferase (Erg6p). Biochim. Biophys. Acta 2004, 1686, 30–36. [Google Scholar] [CrossRef]
- Mo, C.; Milla, P.; Athenstaedt, K.; Ott, R.; Balliano, G.; Daum, G.; Bard, M. In yeast sterol biosynthesis the 3-keto reductase protein (Erg27p) is required for oxidosqualene cyclase (Erg7p) activity. Biochim. Biophys. Acta 2003, 1633, 68–74. [Google Scholar] [CrossRef]
- Layer, J.V.; Barnes, B.M.; Yamasaki, Y.; Barbuch, R.; Li, L.; Taramino, S.; Balliano, G.; Bard, M. Characterization of a mutation that results in independence of oxidosqualene cyclase (Erg7) activity from the downstream 3-ketoreductase (Erg27) in the yeast ergosterol biosynthetic pathway. Biochim. Biophys. Acta 2013, 1831, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Burg, J.S.; Espenshade, P.J. Regulation of HMG-CoA reductase in mammals and yeast. Prog. Lipid Res. 2011, 50, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Hampton, R.Y.; Gardner, R.G.; Rine, J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell 1996, 7, 2029–2044. [Google Scholar] [CrossRef] [PubMed]
- Hampton, R.Y.; Bhakta, H. Ubiquitin-mediated regulation of 3-hydroxy-3-methylglutaryl-CoA reductase. Proc. Natl. Acad. Sci. USA 1997, 94, 12944–12948. [Google Scholar] [CrossRef]
- Foresti, O.; Ruggiano, A.; Hannibal-Bach, H.K.; Ejsing, C.S.; Carvalho, P. Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4. Elife 2013, 2, e00953. [Google Scholar] [CrossRef]
- Chang, Y.C.; Bien, C.M.; Lee, H.; Espenshade, P.J.; Kwon-Chung, K.J. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol. Microbiol. 2007, 64, 614–629. [Google Scholar] [CrossRef]
- Radhakrishnan, A.; Sun, L.P.; Kwon, H.J.; Brown, M.S.; Goldstein, J.L. Direct binding of cholesterol to the purified membrane region of SCAP: Mechanism for a sterol-Sensing domain. Mol. Cell 2004, 15, 259–268. [Google Scholar] [CrossRef]
- Vik, A.; Rine, J. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell Biol. 2001, 21, 6395–6405. [Google Scholar] [CrossRef]
- Maguire, S.L.; Wang, C.; Holland, L.M.; Brunel, F.; Neuveglise, C.; Nicaud, J.M.; Zavrel, M.; White, T.C.; Wolfe, K.H.; Butler, G. Zinc finger transcription factors displaced SREBP proteins as the major Sterol regulators during Saccharomycotina evolution. PLoS Genet. 2014, 10, e1004076. [Google Scholar] [CrossRef]
- MacPherson, S.; Akache, B.; Weber, S.; De Deken, X.; Raymond, M.; Turcotte, B. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 2005, 49, 1745–1752. [Google Scholar] [CrossRef]
- Yang, H.; Tong, J.; Lee, C.W.; Ha, S.; Eom, S.H.; Im, Y.J. Structural mechanism of ergosterol regulation by fungal sterol transcription factor Upc2. Nat. Commun. 2015, 6, 6129. [Google Scholar] [CrossRef] [PubMed]
- Marie, C.; Leyde, S.; White, T.C. Cytoplasmic localization of sterol transcription factors Upc2p and Ecm22p in S. cerevisiae. Fungal Genet Biol. 2008, 45, 1430–1438. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, L.J.; Balderes, D.A.; Wharton, B.; Tinkelenberg, A.H.; Rao, G.; Sturley, S.L. Transcriptional profiling identifies two members of the ATP-Binding cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 2002, 277, 32466–32472. [Google Scholar] [CrossRef] [PubMed]
- Davies, B.S.; Wang, H.S.; Rine, J. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: Similar activation/regulatory domains but different response mechanisms. Mol. Cell Biol. 2005, 25, 7375–7385. [Google Scholar] [CrossRef] [PubMed]
- Davies, B.S.; Rine, J. A role for sterol levels in oxygen sensing in Saccharomyces cerevisiae. Genetics 2006, 174, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Dewhurst-Maridor, G.; Abegg, D.; David, F.P.A.; Rougemont, J.; Scott, C.C.; Adibekian, A.; Riezman, H. The SAGA complex, together with transcription factors and the endocytic protein Rvs167p, coordinates the reprofiling of gene expression in response to changes in sterol composition in Saccharomyces cerevisiae. Mol. Biol. Cell 2017, 28, 2637–2649. [Google Scholar] [CrossRef]
- Vasicek, E.M.; Berkow, E.L.; Flowers, S.A.; Barker, K.S.; Rogers, P.D. UPC2 is universally essential for azole antifungal resistance in Candida albicans. Eukaryot Cell 2014, 13, 933–946. [Google Scholar] [CrossRef]
- Gallo-Ebert, C.; Donigan, M.; Liu, H.Y.; Pascual, F.; Manners, M.; Pandya, D.; Swanson, R.; Gallagher, D.; Chen, W.; Carman, G.M.; et al. The yeast anaerobic response element AR1b regulates aerobic antifungal drug-dependent sterol gene expression. J. Biol. Chem. 2013, 288, 35466–35477. [Google Scholar] [CrossRef]
- Kennedy, M.A.; Barbuch, R.; Bard, M. Transcriptional regulation of the squalene synthase gene (ERG9) in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1999, 1445, 110–122. [Google Scholar] [CrossRef]
- Tamura, K.; Gu, Y.; Wang, Q.; Yamada, T.; Ito, K.; Shimoi, H. A hap1 mutation in a laboratory strain of Saccharomyces cerevisiae results in decreased expression of ergosterol-related genes and cellular ergosterol content compared to sake yeast. J. Biosci. Bioeng. 2004, 98, 159–166. [Google Scholar] [CrossRef]
- MacIsaac, K.D.; Wang, T.; Gordon, D.B.; Gifford, D.K.; Stormo, G.D.; Fraenkel, E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinform. 2006, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- Bergenholm, D.; Liu, G.; Holland, P.; Nielsen, J. Reconstruction of a Global Transcriptional Regulatory Network for Control of Lipid Metabolism in Yeast by Using Chromatin Immunoprecipitation with Lambda Exonuclease Digestion. mSystems 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Valachovic, M.; Bareither, B.M.; Shah Alam Bhuiyan, M.; Eckstein, J.; Barbuch, R.; Balderes, D.; Wilcox, L.; Sturley, S.L.; Dickson, R.C.; Bard, M. Cumulative mutations affecting sterol biosynthesis in the yeast Saccharomyces cerevisiae result in synthetic lethality that is suppressed by alterations in sphingolipid profiles. Genetics 2006, 173, 1893–1908. [Google Scholar] [CrossRef]
- Rice, C.; Cooke, M.; Treloar, N.; Vollbrecht, P.; Stukey, J.; McDonough, V. A role for MGA2, but not SPT23, in activation of transcription of ERG1 in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2010, 403, 293–297. [Google Scholar] [CrossRef]
- Burr, R.; Stewart, E.V.; Espenshade, P.J. Coordinate Regulation of Yeast Sterol Regulatory Element-binding Protein (SREBP) and Mga2 Transcription Factors. J. Biol. Chem. 2017, 292, 5311–5324. [Google Scholar] [CrossRef] [PubMed]
- Abramova, N.E.; Cohen, B.D.; Sertil, O.; Kapoor, R.; Davies, K.J.; Lowry, C.V. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics 2001, 157, 1169–1177. [Google Scholar]
- Kwast, K.E.; Lai, L.C.; Menda, N.; James, D.T.; Aref, S.; Burke, P.V. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: Functional roles of Rox1 and other factors in mediating the anoxic response. J. Bacteriol. 2002, 184, 250–265. [Google Scholar] [CrossRef]
- Ter Linde, J.J.; Steensma, H.Y. A microarray-Assisted screen for potential Hap1 and Rox1 target genes in Saccharomyces cerevisiae. Yeast 2002, 19, 825–840. [Google Scholar] [CrossRef]
- Sertil, O.; Kapoor, R.; Cohen, B.D.; Abramova, N.; Lowry, C.V. Synergistic repression of anaerobic genes by Mot3 and Rox1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2003, 31, 5831–5837. [Google Scholar] [CrossRef][Green Version]
- Klinkenberg, L.G.; Mennella, T.A.; Luetkenhaus, K.; Zitomer, R.S. Combinatorial repression of the hypoxic genes of Saccharomyces cerevisiae by DNA binding proteins Rox1 and Mot3. Eukaryot Cell 2005, 4, 649–660. [Google Scholar] [CrossRef]
- Abramova, N.; Sertil, O.; Mehta, S.; Lowry, C.V. Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J. Bacteriol. 2001, 183, 2881–2887. [Google Scholar] [CrossRef]
- Hongay, C.; Jia, N.; Bard, M.; Winston, F. Mot3 is a transcriptional repressor of ergosterol biosynthetic genes and is required for normal vacuolar function in Saccharomyces cerevisiae. EMBO J. 2002, 21, 4114–4124. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; van Vuuren, H.J.J. Functional analyses of PAU genes in Saccharomyces cerevisiae. Microbiology 2009, 155, 4036–4049. [Google Scholar] [CrossRef] [PubMed]
- Kwast, K.E.; Burke, P.V.; Staahl, B.T.; Poyton, R.O. Oxygen sensing in yeast: Evidence for the involvement of the respiratory chain in regulating the transcription of a subset of hypoxic genes. Proc. Natl. Acad. Sci. USA 1999, 96, 5446–5451. [Google Scholar] [CrossRef]
- Cohen, B.D.; Sertil, O.; Abramova, N.E.; Davies, K.J.; Lowry, C.V. Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res. 2001, 29, 799–808. [Google Scholar] [CrossRef]
- Hickman, M.J.; Spatt, D.; Winston, F. The Hog1 mitogen-activated protein kinase mediates a hypoxic response in Saccharomyces cerevisiae. Genetics 2011, 188, 325–338. [Google Scholar] [CrossRef]
- Jiang, Y.; Vasconcelles, M.J.; Wretzel, S.; Light, A.; Martin, C.E.; Goldberg, M.A. MGA2 is involved in the low-oxygen response element-dependent hypoxic induction of genes in Saccharomyces cerevisiae. Mol. Cell Biol. 2001, 21, 6161–6169. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hickman, M.J.; Winston, F. Heme levels switch the function of Hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Mol. Cell Biol. 2007, 27, 7414–7424. [Google Scholar] [CrossRef] [PubMed]
- Serratore, N.D.; Baker, K.M.; Macadlo, L.A.; Gress, A.R.; Powers, B.L.; Atallah, N.; Westerhouse, K.M.; Hall, M.C.; Weake, V.M.; Briggs, S.D. A Novel Sterol-Signaling Pathway Governs Azole Antifungal Drug Resistance and Hypoxic Gene Repression in Saccharomyces cerevisiae. Genetics 2018, 208, 1037–1055. [Google Scholar] [CrossRef]
- Bendjilali, N.; MacLeon, S.; Kalra, G.; Willis, S.D.; Hossian, A.K.; Avery, E.; Wojtowicz, O.; Hickman, M.J. Time-Course Analysis of Gene Expression During the Saccharomyces cerevisiae Hypoxic Response. G3 (Bethesda) 2017, 7, 221–231. [Google Scholar] [CrossRef]
- Becerra, M.; Lombardia-Ferreira, L.J.; Hauser, N.C.; Hoheisel, J.D.; Tizon, B.; Cerdan, M.E. The yeast transcriptome in aerobic and hypoxic conditions: Effects of hap1, rox1, rox3 and srb10 deletions. Mol. Microbiol. 2002, 43, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.C.; Kosorukoff, A.L.; Burke, P.V.; Kwast, K.E. Dynamical remodeling of the transcriptome during short-term anaerobiosis in Saccharomyces cerevisiae: Differential response and role of Msn2 and/or Msn4 and other factors in galactose and glucose media. Mol. Cell Biol. 2005, 25, 4075–4091. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.C.; Kosorukoff, A.L.; Burke, P.V.; Kwast, K.E. Metabolic-state-dependent remodeling of the transcriptome in response to anoxia and subsequent reoxygenation in Saccharomyces cerevisiae. Eukaryot Cell 2006, 5, 1468–1489. [Google Scholar] [CrossRef]
- Rosenfeld, E.; Beauvoit, B. Role of the non-Respiratory pathways in the utilization of molecular oxygen by Saccharomyces cerevisiae. Yeast 2003, 20, 1115–1144. [Google Scholar] [CrossRef]
- Alimardani, P.; Regnacq, M.; Moreau-Vauzelle, C.; Ferreira, T.; Rossignol, T.; Blondin, B.; Berges, T. SUT1-promoted sterol uptake involves the ABC transporter Aus1 and the mannoprotein Dan1 whose synergistic action is sufficient for this process. Biochem. J. 2004, 381, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, D.P.; Georgiev, A.; Menon, A.K. Tritium suicide selection identifies proteins involved in the uptake and intracellular transport of sterols in Saccharomyces cerevisiae. Eukaryot Cell 2009, 8, 161–169. [Google Scholar] [CrossRef]
- Shianna, K.V.; Dotson, W.D.; Tove, S.; Parks, L.W. Identification of a UPC2 homolog in Saccharomyces cerevisiae and its involvement in aerobic sterol uptake. J. Bacteriol. 2001, 183, 830–834. [Google Scholar] [CrossRef]
- Saito, H.; Posas, F. Response to hyperosmotic stress. Genetics 2012, 192, 289–318. [Google Scholar] [CrossRef]
- Proft, M.; Gibbons, F.D.; Copeland, M.; Roth, F.P.; Struhl, K. Genomewide identification of Sko1 target promoters reveals a regulatory network that operates in response to osmotic stress in Saccharomyces cerevisiae. Eukaryot Cell 2005, 4, 1343–1352. [Google Scholar] [CrossRef]
- Martinez-Montanes, F.; Rienzo, A.; Poveda-Huertes, D.; Pascual-Ahuir, A.; Proft, M. Activator and repressor functions of the Mot3 transcription factor in the osmostress response of Saccharomyces cerevisiae. Eukaryot Cell 2013, 12, 636–647. [Google Scholar] [CrossRef]
- Craven, R.J.; Mallory, J.C.; Hand, R.A. Regulation of iron homeostasis mediated by the heme-binding protein Dap1 (damage resistance protein 1) via the P450 protein Erg11/Cyp51. J. Biol. Chem. 2007, 282, 36543–36551. [Google Scholar] [CrossRef]
- Mallory, J.C.; Crudden, G.; Johnson, B.L.; Mo, C.; Pierson, C.A.; Bard, M.; Craven, R.J. Dap1p, a heme-Binding protein that regulates the cytochrome P450 protein Erg11p/Cyp51p in Saccharomyces cerevisiae. Mol Cell Biol 2005, 25, 1669–1679. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.M.; Reddi, A.R.; Shi, X.; Goldbeck, R.A.; Moenne-Loccoz, P.; Gibney, B.R.; Holman, T.R. Measurement of the heme affinity for yeast dap1p, and its importance in cellular function. Biochemistry 2007, 46, 14629–14637. [Google Scholar] [CrossRef]
- Li, L.; Kaplan, J. Characterization of yeast methyl sterol oxidase (ERG25) and identification of a human homologue. J. Biol. Chem. 1996, 271, 16927–16933. [Google Scholar] [CrossRef] [PubMed]
- Moretti-Almeida, G.; Netto, L.E.; Monteiro, G. The essential gene YMR134W from Saccharomyces cerevisiae is important for appropriate mitochondrial iron utilization and the ergosterol biosynthetic pathway. FEBS Lett. 2013, 587, 3008–3013. [Google Scholar] [CrossRef] [PubMed]
- Puig, S.; Askeland, E.; Thiele, D.J. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 2005, 120, 99–110. [Google Scholar] [CrossRef]
- Hausmann, A.; Samans, B.; Lill, R.; Muhlenhoff, U. Cellular and mitochondrial remodeling upon defects in iron-sulfur protein biogenesis. J. Biol. Chem. 2008, 283, 8318–8330. [Google Scholar] [CrossRef]
- Puig, S.; Vergara, S.V.; Thiele, D.J. Cooperation of two mRNA-Binding proteins drives metabolic adaptation to iron deficiency. Cell Metab. 2008, 7, 555–564. [Google Scholar] [CrossRef]
- Pedro-Segura, E.; Vergara, S.V.; Rodriguez-Navarro, S.; Parker, R.; Thiele, D.J.; Puig, S. The Cth2 ARE-binding protein recruits the Dhh1 helicase to promote the decay of succinate dehydrogenase SDH4 mRNA in response to iron deficiency. J. Biol. Chem. 2008, 283, 28527–28535. [Google Scholar] [CrossRef]
- Ramos-Alonso, L.; Romero, A.M.; Soler, M.A.; Perea-Garcia, A.; Alepuz, P.; Puig, S.; Martinez-Pastor, M.T. Yeast Cth2 protein represses the translation of ARE-containing mRNAs in response to iron deficiency. PLoS Genet 2018, 14, e1007476. [Google Scholar] [CrossRef]
- Romero, A.M.; Ramos-Alonso, L.; Montella-Manuel, S.; Garcia-Martinez, J.; de la Torre-Ruiz, M.A.; Perez-Ortin, J.E.; Martinez-Pastor, M.T.; Puig, S. A genome-Wide transcriptional study reveals that iron deficiency inhibits the yeast TORC1 pathway. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 194414. [Google Scholar] [CrossRef] [PubMed]
- Singh, P. Budding Yeast: An Ideal Backdrop for In vivo Lipid Biochemistry. Front Cell Dev. Biol. 2017, 4, 156. [Google Scholar] [CrossRef] [PubMed]
- Sanglard, D. Emerging Threats in Antifungal-Resistant Fungal Pathogens. Front Med (Lausanne) 2016, 3, 11. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jordá, T.; Puig, S. Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae. Genes 2020, 11, 795. https://doi.org/10.3390/genes11070795
Jordá T, Puig S. Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae. Genes. 2020; 11(7):795. https://doi.org/10.3390/genes11070795
Chicago/Turabian StyleJordá, Tania, and Sergi Puig. 2020. "Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae" Genes 11, no. 7: 795. https://doi.org/10.3390/genes11070795
APA StyleJordá, T., & Puig, S. (2020). Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae. Genes, 11(7), 795. https://doi.org/10.3390/genes11070795





