Evidence of Interspecific Chromosomal Diversification in Rainbowfishes (Melanotaeniidae, Teleostei)
Abstract
1. Introduction
2. Materials and Methods
2.1. Studied Material
2.2. Chromosome Preparation and Staining
2.3. Fluorescence In Situ Hybridisation (FISH) with Telomeric and rRNA Genes Probes
2.4. Microscopy and Image Analyses
3. Results
3.1. Karyotypes
3.2. rDNA Chromosome Mapping
3.3. Telomere Mapping
3.4. CMA3/DAPI Staining
3.5. C Banding
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Biemont, C.; Vieira, C. Genetics—Junk DNA as an evolutionary force. Nature 2006, 443, 521–524. [Google Scholar] [CrossRef] [PubMed]
- Gornung, E. Twenty years of physical mapping of major ribosomal RNA genes across the Teleosts: A review of research. Cytogenet. Genome Res. 2013. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.B.; Camacho, J.P.M.; Bertollo, L.A.C. Repetitive DNAs and differentiation of sex chromosomes in neotropical fishes. Cytogenet. Genome Res. 2011, 132, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Martins, C. Chromosomes and Repetitive DNAs: A Contribution to the Knowledge of the Fish Genome. Fish Cytogenet. 2007, 421, 452. [Google Scholar]
- Allen, G.R.; Midgley, S.H.; Allen, M. Field Guide to the Freshwater Fishes of Australia; Western Australian Museum: Perth, Australia, 2002. [Google Scholar]
- Fricke, R.; Eschmeyer, W.N.; van der Laan, R. Eschmeyer’s Catalog of Fishes: Genera, Species, Rederences. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 26 May 2020).
- Unmack, P.J.; Allen, G.R.; Johnson, J.B. Phylogeny and biogeography of rainbowfishes (Melanotaeniidae) from Australia and New Guinea. Mol. Phylogenet. Evol. 2013, 67, 15–27. [Google Scholar] [CrossRef]
- Zhu, D.; Jamieson, B.G.; Hugall, A.; Moritz, C. Sequence evolution and phylogenetic signal in control-region and cytochrome b sequences of rainbow fishes (Melanotaeniidae). Mol. Biol. Evol. 1994, 11, 672–683. [Google Scholar] [CrossRef]
- McGuigan, K.; Zhu, D.; Allen, G.R.; Moritz, C. Phylogenetic relationships and historical biogeography of melanotaeniid fishes in Australia and New Guinea. Mar. Freshw. Res. 2000, 51, 713–723. [Google Scholar] [CrossRef]
- Campanella, D.; Hughes, L.C.; Unmack, P.J.; Bloom, D.D.; Piller, K.R.; Ortí, G. Multi-locus fossil-calibrated phylogeny of Atheriniformes (Teleostei, Ovalentaria). Mol. Phylogenet. Evol. 2015, 86, 8–23. [Google Scholar] [CrossRef]
- Tappin, A.R. Rainbowfishes, Their Care and Keeping in Captivity, 2nd ed.; Available online: http://rainbowfish.angfaqld.org.au (accessed on 26 May 2020).
- Allen, G.R. Rainbowfishes: In Nature and In the Aquarium; Tetra-Verlag: Velten, Germany, 1995; ISBN 978-1-56465-149-5. [Google Scholar]
- Molina, W.F.; da Costa, G.W.W.F.; de Bello Cioffi, M.; Bertollo, L.A.C. Chromosomal differentiation and speciation in sister-species of Grammatidae (Perciformes) from the Western Atlantic. Helgol. Mar. Res. 2012, 66, 363–370. [Google Scholar] [CrossRef]
- Hinegardner, R.; Rosen, D.E. Cellular DNA Content and the Evolution of Teleostean Fishes. Am. Nat. 1972, 106, 621–644. [Google Scholar] [CrossRef]
- Scheel, J.J. Rivuline Karyotypes and their evolution (Rivulinae, Cyprinodontidae, Pisces). J. Zool. Syst. Evol. Res. 1972, 10, 180–209. [Google Scholar] [CrossRef]
- Arai, R. Chromosomes of two species of Atherinoid fishes. Bull. Natl. Sci. Mus. Ser. A (Zool) 1978, 4, 147–150. [Google Scholar]
- Carey, G.; Mather, P. Karyotypes of four Australian fish species Melanotaenia duboulayi; Bidyanus bidyanus, Macquaria novemaculeata and Lates calcarifer. Cytobios 1999, 100, 137–146. [Google Scholar]
- Said, D.S. Kekerab atan beberapa spesies Ikan Pelangi Irian (Famili Melanotaeniidae) berdasarkan karyotipe [The closely related of some Rainbow Fishes (Melanotaeniidae) from Irian based of caryotipe]. J. Iktiologi Indones. 2005, 5, 31–38. [Google Scholar] [CrossRef]
- Bertollo, L.A.C.; de Cioffi, M.B.; Moreira-Filho, O. Direct Chromosome Preparation from Freshwater Teleost Fishes. In Fish Cytogenetic Techniques; Ozouf-Costaz, C., Pisano, E., Foresti, F., Foresti de Almeida-Toledo, L., Eds.; CRC Press, Inc.: Enfield, NH, USA, 2015; pp. 21–26. [Google Scholar]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Pokorná, M.; Rens, W.; Rovatsos, M.; Kratochvíl, L. A ZZ/ZW sex chromosome system in the thick-tailed Gecko (Underwoodisaurus milii; Squamata: Gekkota: Carphodactylidae), a member of the ancient gecko lineage. Cytogenet. Genome Res. 2014, 142, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Sola, L.; Rossi, A.R.; Iaselli, V.; Rasch, E.M.; Monaco, P.J. Cytogenetics of bisexual/unisexual species of Poecilia. II. Analysis of heterochromatin and nucleolar organizer regions in Poecilia mexicana mexicana by C-banding and DAPI, quinacrine, chromomycin A3, and silver staining. Cytogenet. Cell Genet. 1992, 60, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Komiya, H.; Takemura, S. Nucleotide sequence of 5S ribosomal RNA from rainbow trout (Salmo gairdnerii) liver. J. Biochem. 1979, 86, 1067–1080. [Google Scholar] [CrossRef]
- Zhang, Q.; Cooper, R.K.; Tiersch, T.R. Chromosomal location of the 28S ribosomal RNA gene of channel catfish by in situ polymerase chain reaction. J. Fish Biol. 2000, 56, 388–397. [Google Scholar] [CrossRef]
- Symonová, R.; Sember, A.; Majtánová, Z.; Ráb, P. Characterization of fish genomes by GISH and CGH. In Fish Cytogenetic Techniques; Ozouf-Costaz, C., Pisano, E., Foresti, F., de Almeida, L., Eds.; CRC Press: Boca Raton, FL, USA, 2015; pp. 118–131. ISBN 978-1-4822-1198-6. [Google Scholar]
- McGuigan, K.; Franklin, C.E.; Moritz, C.; Blows, M.W. Adaptation of rainbow fish to lake and stream habitats. Evolution 2003, 57, 104–118. [Google Scholar] [CrossRef]
- Page, T.J.; Sharma, S.; Hughes, J.M. Deep phylogenetic structure has conservation implications for ornate rainbowfish (Melanotaeniidae: Rhadinocentrus ornatus) in Queensland, eastern Australia. Mar. Freshw. Res. 2004, 55, 165–172. [Google Scholar] [CrossRef]
- Colléter, M.; Brown, C. Personality traits predict hierarchy rank in male rainbowfish social groups. Anim. Behav. 2011, 81, 1231–1237. [Google Scholar] [CrossRef]
- Dion-Côté, A.-M.; Symonová, R.; Lamaze, F.C.; Pelikánová, Š.; Ráb, P.; Bernatchez, L. Standing chromosomal variation in Lake Whitefish species pairs: The role of historical contingency and relevance for speciation. Mol. Ecol. 2016, 26, 178–192. [Google Scholar] [CrossRef]
- Unmack, P.J. Update on the running river rainbowfish. Fishes Sahul 2016, 1025–1032. [Google Scholar]
- Mank, J.E.; Avise, J.C. Phylogenetic conservation of chromosome numbers in Actinopterygiian fishes. Genetica 2006, 127, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Arai, R. Fish Karyotypes: A Check List; 2011 edition; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2011; ISBN 978-4-431-53876-9. [Google Scholar]
- Barby, F.F.; Bertollo, L.A.C.; de Oliveira, E.A.; Yano, C.F.; Hatanaka, T.; Ráb, P.; Sember, A.; Ezaz, T.; Artoni, R.F.; Liehr, T.; et al. Emerging patterns of genome organization in Notopteridae species (Teleostei, Osteoglossiformes) as revealed by Zoo-FISH and Comparative Genomic Hybridization (CGH). Sci. Rep. 2019, 9, 1112. [Google Scholar] [CrossRef]
- Moy, K.G.; Unmack, P.J.; Lintermans, M.; Duncan, R.P.; Brown, C. Barriers to hybridisation and their conservation implications for a highly threatened Australian fish species. Ethology 2019, 125, 142–152. [Google Scholar] [CrossRef]
- Unmack, P.J. Historical Biogeography and a Priori Hypotheses Based on Freshwater Fishes; Arizona State University: Tempe, AZ, USA, 2005. [Google Scholar]
- Mayr, B.; Rab, P.; Kalat, M. Localization of NORs and counterstain-enhanced fluorescence studies in Perca fluviatilis (Pisces, Percidae). Genetica 1985, 67, 51–56. [Google Scholar] [CrossRef]
- Amemiya, C.T.; Gold, J.R. Chromomycin A3 stains nucleolus organizer regions of fish chromosomes. Copeia 1986, 1986, 226–231. [Google Scholar] [CrossRef]
- Schmid, M.; Guttenbach, M. Evolutionary diversity of reverse (R) fluorescent chromosome bands in vertebrates. Chromosoma 1988, 97, 101–114. [Google Scholar] [CrossRef]
- Gromicho, M.; Ozouf-Costaz, C.; Collares-Pereira, M.J. Lack of correspondence between CMA3-, Ag-positive signals and 28S rDNA loci in two Iberian minnows (Teleostei, Cyprinidae) evidenced by sequential banding. Cytogenet. Genome Res. 2005, 109, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Sola, L.; Gornung, E.; Naoi, H.; Gunji, R.; Sato, C.; Kawamura, K.; Arai, R.; Ueda, T. FISH-mapping of 18S ribosomal RNA genes and telomeric sequences in the Japanese bitterlings Rhodeus ocellatus kurumeus and Tanakia limbata (Pisces, Cyprinidae) reveals significant cytogenetic differences in morphologically similar karyotypes. Genetica 2003, 119, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Meyne, J.; Ratliff, R.L.; Moyzis, R.K. Conservative of the human telomere sequences (TTAGGG)n among vertebartes. Proc. Natl. Acad. Sci. USA 1989, 86, 7049–7053. [Google Scholar] [CrossRef] [PubMed]
- Ocalewicz, K. Telomeres in Fishes. Cytogen. Genome Res. 2013, 141, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Nanda, I.; Schneider-Rasp, S.; Winking, H.; Schmid, M. Loss of telomeric sites in the chromosomes of Mus musculus domesticus (Rodentia: Muridae) during Robertsonian rearrangements. Chromosom. Res. 1995, 3, 399–409. [Google Scholar] [CrossRef]
- Mayr, E.; White, M.J.D. Modes of Speciation. Syst. Biol. 1978, 27, 478–482. [Google Scholar] [CrossRef]
- King, M. Species Evolution: The Role of Chromosome Change; Cambridge University Press: Cambridge, UK, 1995; ISBN 978-0-521-48454-1. [Google Scholar]



| Species | No. of Individuals | No. of Metaphases Examined | ||||
|---|---|---|---|---|---|---|
| Giemsa | C Banding | CMA3 | Telomeres | FISH | ||
| Melanotaenia sp. | 3 ; 3 ; 3 | 50 | 13 | 31 | 10 | 15 |
| Glossolepis incisus | 2 ; 2 ; 2 | 22 | 21 | 13 | 12 | 16 |
| Iriatherina werneri | 2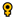 ; 5 ; 5 | 25 | 35 | 30 | 35 | 28 |
| Rhadinocentrus ornatus | 2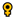 ; 2 ; 2 | 40 | 14 | 10 | 59 | 22 |
| Cairnsichthys rhombosomoides | 2 ; 2 ; 2 | 80 | 40 | 15 | 14 | 28 |
| Species | 2n | Karyotype Composition | NF | 28S rDNA Clusters | 5S rDNA Clusters | Telomeres | AT-Rich/GC-Rich Regions | Heterochromatin Regions |
|---|---|---|---|---|---|---|---|---|
| Melanotaenia sp. | 48 | 48st/a | 48 | 2 | 4 | no ITS | 2 pericenromeric | 32 centromeric |
| Glossolepis incisus | 48 | 48st/a | 48 | 2 | 4 | no ITS | 2 pericenromeric | 14 centromeric |
| Iriatherina werneri | 48 | 48st/a | 48 | 2 | 4 | no ITS | 2 pericenromeric | 36 centromeric 4 interstitial |
| Rhadinocentrus ornatus | 48 | 12sm + 36st/a | 60 | 2 | 4 | no ITS | 2 telomeric | 28 centromeric 2telomeric |
| Cairnsichthys rhombosomoides | 48 | 8sm + 40st/a | 56 | 2 | 8 | no ITS | 2 pericenromeric | 8 centromeric 4 interstitial 2 large clusters |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majtánová, Z.; Unmack, P.J.; Prasongmaneerut, T.; Shams, F.; Srikulnath, K.; Ráb, P.; Ezaz, T. Evidence of Interspecific Chromosomal Diversification in Rainbowfishes (Melanotaeniidae, Teleostei). Genes 2020, 11, 818. https://doi.org/10.3390/genes11070818
Majtánová Z, Unmack PJ, Prasongmaneerut T, Shams F, Srikulnath K, Ráb P, Ezaz T. Evidence of Interspecific Chromosomal Diversification in Rainbowfishes (Melanotaeniidae, Teleostei). Genes. 2020; 11(7):818. https://doi.org/10.3390/genes11070818
Chicago/Turabian StyleMajtánová, Zuzana, Peter J. Unmack, Tulyawat Prasongmaneerut, Foyez Shams, Kornsorn Srikulnath, Petr Ráb, and Tariq Ezaz. 2020. "Evidence of Interspecific Chromosomal Diversification in Rainbowfishes (Melanotaeniidae, Teleostei)" Genes 11, no. 7: 818. https://doi.org/10.3390/genes11070818
APA StyleMajtánová, Z., Unmack, P. J., Prasongmaneerut, T., Shams, F., Srikulnath, K., Ráb, P., & Ezaz, T. (2020). Evidence of Interspecific Chromosomal Diversification in Rainbowfishes (Melanotaeniidae, Teleostei). Genes, 11(7), 818. https://doi.org/10.3390/genes11070818






