Dramatic Increase in Content of Diverse Flavonoids Accompanied with Down-Regulation of F-Box Genes in a Chrysanthemum (Chrysanthemum × morifolium (Ramat.) Hemsl.) Mutant Cultivar Producing Dark-Purple Ray Florets
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Analysis of Flower Color
2.2. Extraction of Phenolic Compounds
2.3. HPLC-DAD-ESI-MS Analysis
2.4. Transcriptome Analysis
2.5. Annotation, Functional Analysis, and Expression Level Determination of Unigenes
2.6. Analysis of Differentially Expressed Genes
2.7. Quantitative Reverse Transcription PCR and Unigene PCR Amplification
3. Results
3.1. Analysis of Ray Floret Corolla Color of “Noble Wine” and “ARTI-Dark Chocolate”
3.2. Identification and Quantification of Anthocyanins and Other Flavonoids
3.3. Transcriptome Sequencing and Analysis of Flavonoid Biosynthetic Gene Expression
3.4. Functional Analysis of Differentially Expressed Genes (DEGs)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HPLC-DAD-ESI-MS | high-performance liquid chromatography coupled with photodiode array detection and electrospray ionization mass spectrometry |
| CHS | chalcone synthase |
| CHI | chalcone isomerase |
| bHLH | basic helix-loop-helix |
| KFB | Kelch motif-containing F-box |
| PAL | phenylalanine ammonia-lyase |
| FPKM | fragment per kilobase of transcripts per million mapped reads |
| C4H | cinnamate-4-hydroxylase |
| 4CL | 4-coumarate-CoA ligase |
| F3H | flavanone 3-hydroxylase |
| F3’H | flavanone 3’-hydroxylase |
| DFR | dihydroflavonol 4-reductase |
| ANS | anthocyanin synthase |
| 3-GT | UDP-glucose:anthocyanidin 3-O-glucosyltransferase |
| 3,5-GT | UDP-glucose:anthocyanin 5,3-O-glucosyltransferase |
| 3MAT | malonyl-coenzyme A:anthocyanin 3-O-glucoside-6”-O-malonyltransferase |
| UGAT | cyanidin-3-O-glucoside 2”-O-glucuronosyltransferase |
| FNS | flavone synthase |
| FLS | flavonol synthase |
| LAR | leucoanthocyanidin reductase |
| DEG | differentially expressed gene |
| GO | gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| KOG | eukaryotic orthologous groups |
| ELSD | evaporative light scattering detector |
| CDS | protein coding sequence |
References
- George, V.C.; Dellaire, G.; Rupasinghe, H.P.V. Plant flavonoids in cancer chmoprevention: Role in genome stability. J. Nutr. Biochem. 2014, 1, 26–31. [Google Scholar]
- Brunetti, C.; Fini, A.; Sebastiani, F.; Gori, A.; Tattini, M. Modulation of Phytohormone Signaling: A Primary Function of Flavonoids in Plant–Environment Interactions. Front. Plant Sci. 2018, 9, 1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treutter, D. Significance of Flavonoids in Plant Resistance and Enhancement of Their Biosynthesis. Plant Biol. 2005, 7, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Nabavi, S.F.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Chaves-Silva, S.; Dos Santos, A.L.; Chalfun-Júnior, A.; Zhao, J.; Peres, L.E.; Benedito, V.A. Understanding the genetic regulation of anthocyanin biosynthesis in plants—Tools for breeding purple varieties of fruits and vegetables. Phytochemistry 2018, 153, 11–27. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F.M. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [Green Version]
- Fornalè, S.; Lopez, E.; Salazar-Henao, J.E.; Nohales, P.F.; Rigau, J.; Caparros-Ruiz, D. AtMYB7, a New Player in the Regulation of UV-Sunscreens in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 507–516. [Google Scholar] [CrossRef]
- Matsui, K.; Umemura, Y.; Ohme-Takagi, M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008, 55, 954–967. [Google Scholar] [CrossRef]
- Luo, Q.-J.; Mittal, A.; Jia, F.; Rock, C.D. An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol. Biol. 2011, 80, 117–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gou, J.-Y.; De Felippes, F.F.; Liu, C.-J.; Weigel, D.; Wang, J.-W. Negative Regulation of Anthocyanin Biosynthesis in Arabidopsis by a miR156-Targeted SPL Transcription Factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Abrahan, C.; Colquhoun, T.A.; Liu, C.-J. A Proteolytic Regulator Controlling Chalcone Synthase Stability and Flavonoid Biosynthesis in Arabidopsis. Plant Cell 2017, 29, 1157–1174. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Gou, M.; Guo, C.; Yang, H.; Liu, C.-J. Down-regulation of Kelch domain-containing F-box protein in Arabidopsis enhances the production of (poly)phenols and tolerance to ultraviolet radiation. Plant Physiol. 2014, 167, 337–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Gou, M.; Liu, C.-J. Arabidopsis Kelch Repeat F-Box Proteins Regulate Phenylpropanoid Biosynthesis via Controlling the Turnover of Phenylalanine Ammonia-Lyase. Plant Cell 2013, 25, 4994–5010. [Google Scholar] [CrossRef] [Green Version]
- Del Pozo, J.C.; Estelle, M. F-box proteins and protein degradation: An emerging theme in cellular regulation. Plant Mol. Biol. 2000, 44, 123–128. [Google Scholar] [CrossRef]
- Zhou, L.-J.; Li, Y.-Y.; Zhang, R.-F.; Zhang, C.-L.; Xie, X.-B.; Zhao, C.; Hao, Y.-J. The small ubiquitin-like modifier E3 ligase MdSIZ1 promotes anthocyanin accumulation by sumoylating MdMYB1 under low-temperature conditions in apple. Plant Cell Environ. 2017, 40, 2068–2080. [Google Scholar] [CrossRef]
- Hanudin, H.; Budiarto, K.; Marwoto, B. Application of PGPR and Antagonist Fungi-based Biofungicide for White Rust Disease Control and its Economyc Analysis in Chrysanthemum Production. AGRIVITA J. Agric. Sci. 2017, 39, 266–278. [Google Scholar] [CrossRef] [Green Version]
- Ali, Y.; Park, S.; Park, B.-R.; Lee, J.-Y.; Lim, S.-H. Flavonoid Metabolic Engineering for Modification of Flower Color in Chrysanthemum. Korean J. Breed. Sci. 2018, 50, 351–363. [Google Scholar] [CrossRef]
- Nakayama, M.; Koshioka, M.; Shibata, M.; Hiradate, S.; Sugie, H.; Yamaguchi, M.-A. Identification of Cyanidin 3-O-(3″,6″-O-Dimalonyl-β-glucopyranoside) as a Flower Pigment of Chrysanthemum (Dendranthema grandiflorum). Biosci. Biotechnol. Biochem. 1997, 61, 1607–1608. [Google Scholar] [CrossRef]
- Schwinn, K.E.; Markham, K.R.; Giveno, N.K. Floral flavonoids and the potential for pelargonidin biosynthesis in commercial chrysanthemum cultivars. Phytochemistry 1993, 35, 145–150. [Google Scholar] [CrossRef]
- Hong, Y.; Tang, X.; Huang, H.; Zhang, Y.; Dai, S. Transcriptomic analyses reveal species-specific light-induced anthocyanin biosynthesis in chrysanthemum. BMC Genom. 2015, 16, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, J.; Zhu, C.; Zhou, Y.; Niu, X.; Miao, M.; Tang, X.; Chen, F.; Zhao, W.; Liu, Y. Transcriptome analysis of differentially expressed unigenes involved in flavonoid biosynthesis during flower development of Chrysanthemum morifolium ‘Chuju’. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Noda, N.; Aida, R.; Kishimoto, S.; Ishiguro, K.; Fukuchi-Mizutani, M.; Tanaka, Y.; Ohmiya, A. Genetic Engineering of Novel Bluer-Colored Chrysanthemums Produced by Accumulation of Delphinidin-Based Anthocyanins. Plant Cell Physiol. 2013, 54, 1684–1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noda, N.; Yoshioka, S.; Kishimoto, S.; Nakayama, M.; Douzono, M.; Tanaka, Y.; Aida, R. Generation of blue chrysanthemums by anthocyanin B-ring hydroxylation and glucosylation and its coloration mechanism. Sci. Adv. 2017, 3, e1602785. [Google Scholar] [CrossRef] [Green Version]
- Sung, S.Y.; Kim, S.H.; Velusamy, V.; Lee, Y.-M.; Ha, B.-K.; Kim, J.-B.; Kang, S.-Y.; Kim, H.G.; Kim, D.S. Comparative gene expression analysis in a highly anthocyanin pigmented mutant of colorless chrysanthemum. Mol. Biol. Rep. 2013, 40, 5177–5189. [Google Scholar] [CrossRef]
- Ryu, J.; Nam, B.; Kim, B.-R.; Kim, S.H.; Hong, M.J.; Ahn, J.-W.; Kim, J.-B.; Jin, C.-H.; Han, A.-R. Comparative Analysis of Phytochemical Composition of γ-Irradiated Mutant Cultivars of Chrysanthemum morifolium. Molecules 2019, 24, 3003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, Y.D.; Kim, Y.-S.; Ryu, J.; Choi, H.-I.; Kim, S.W.; Kang, H.S.; Ahn, J.-W.; Kim, J.-B.; Kang, S.-Y.; Kim, S.H. Deletion of carotenoid cleavage dioxygenase 4a (CmCCD4a) and global up-regulation of plastid protein-coding genes in a yellow-petal mutant cultivar of chrysanthemum. Sci. Hortic. 2016, 212, 49–59. [Google Scholar] [CrossRef]
- Han, A.-R.; Nam, B.; Kim, B.-R.; Lee, K.-C.; Song, B.-S.; Kim, S.H.; Kim, J.-B.; Jin, C.-H. Phytochemical Composition and Antioxidant Activities of Two Different Color Chrysanthemum Flower Teas. Molecules 2019, 24, 329. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.-M.; Li, C.-H.; Zhu, X.-R.; Deng, Y.-M.; Sun, W.; Wang, L.-S.; Chen, F.-D.; Zhang, Z. The identification of flavonoids and the expression of genes of anthocyanin biosynthesis in the chrysanthemum flowers. Biol. Plant. 2012, 56, 458–464. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Madan, A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Kadota, K.; Nishiyama, T.; Shimizu, K. A normalization strategy for comparing tag count data. Algorithms Mol. Biol. 2012, 7, 5. [Google Scholar] [CrossRef]
- WEGO 2.0 Home Page. Available online: http://wego.genomics.org.cn (accessed on 15 March 2017).
- KEGG Automatic Annotation Server Home Page. Available online: http://www.genome.jp/kegg/kaas/ (accessed on 15 March 2017).
- WebMGA Home Page. Available online: http://weizhong-lab.ucsd.edu/webMGA/server/kog/ (accessed on 23 March 2017).
- DAVID Bioinformatics Resources 6.7 Home Page. Available online: http://david-d.ncifcrf.gov (accessed on 15 May 2017).
- Lin, L.-Z.; Harnly, J.M. Identification of Hydroxycinnamoylquinic Acids of Arnica Flowers and Burdock Roots Using a Standardized LC-DAD-ESI/MS Profiling Method. J. Agric. Food Chem. 2008, 56, 10105–10114. [Google Scholar] [CrossRef] [PubMed]
- Benayad, Z.; Gómez-Cordovés, C.; Es-Safi, N. Characterization of Flavonoid Glycosides from Fenugreek (Trigonella foenum-graecum) Crude Seeds by HPLC–DAD–ESI/MS Analysis. Int. J. Mol. Sci. 2014, 15, 20668–20685. [Google Scholar] [CrossRef] [Green Version]
- Selenge, E.; Murata, T.; Tanaka, S.; Sasaki, K.; Batkhuu, J.; Yoshizaki, F. Monoterpene glycosides, phenylpropanoids, and acacetin glycosides from Dracocephalum foetidum. Phytochemistry 2014, 101, 91–100. [Google Scholar] [CrossRef]
- Barros, L.; Dueñas, M.; Dias, M.I.; Sousa, M.J.; Santos-Buelga, C.; Ferreira, I.C.F.R.; Dueñas, M. Phenolic profiles of in vivo and in vitro grown Coriandrum sativum L. Food Chem. 2012, 132, 841–848. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.-K. Faculty Opinions recommendation of Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Fac. Opin. Post-Publ. Peer Rev. Biomed. Lit. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Wang, X.; Guo, W. The cytochrome P450 superfamily: Key players in plant development and defense. J. Integr. Agric. 2015, 14, 1673–1686. [Google Scholar] [CrossRef] [Green Version]
- Jin, H.; Choi, S.-M.; Kang, M.-J.; Yun, S.-H.; Kwon, D.-J.; Noh, Y.-S.; Noh, B. Salicylic acid-induced transcriptional reprogramming by the HAC–NPR1–TGA histone acetyltransferase complex in Arabidopsis. Nucleic Acids Res. 2018, 46, 11712–11725. [Google Scholar] [CrossRef] [PubMed]
- Kawase, K.; Tsukamoto, Y.; Saito, N.; Osawa, Y. Studies on flower color in Chrysanthemum morifolium RAMAT. I. Anthocyanins. Plant Cell Physiol. 1970, 11, 349–353. [Google Scholar] [CrossRef]
- Thill, J.; Miosic, S.; Ahmed, R.; Schlangen, K.; Muster, G.; Stich, K.; Halbwirth, H. ‘Le Rouge et le Noir’: A decline in flavone formation correlates with the rare color of black dahlia (Dahlia variabilis hort.) flowers. BMC Plant Biol. 2012, 12, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, Q.; Liu, Y.; Qi, Y.; Jiao, S.; Tian, F.; Jiang, L.; Wang, Y. Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth. J. Exp. Bot. 2014, 65, 3157–3164. [Google Scholar] [CrossRef] [Green Version]
- Luo, P.; Ning, G.; Wang, Z.; Shen, Y.; Jin, H.; Li, P.; Huang, S.; Zhao, J.; Bao, M. Disequilibrium of Flavonol Synthase and Dihydroflavonol-4-Reductase Expression Associated Tightly to White vs. Red Color Flower Formation in Plants. Front. Plant Sci. 2016, 6. [Google Scholar] [CrossRef] [Green Version]
- Feder, A.; Burger, J.; Gao, S.; Lewinsohn, E.; Katzir, N.; Schaffer, A.A.; Meir, A.; Davidovich-Rikanati, R.; Portnoy, V.; Gal-On, A.; et al. A Kelch Domain-Containing F-Box Coding Gene Negatively Regulates Flavonoid Accumulation in Muskmelon. Plant Physiol. 2015, 169, 1714–1726. [Google Scholar] [CrossRef] [Green Version]
- Gu, Z.; Men, S.; Zhu, J.; Hao, Q.; Tong, N.; Liu, Z.-A.; Zhang, H.; Shu, Q.; Wang, L. Chalcone synthase is ubiquitinated and degraded via interactions with a RING-H2 protein in petals of Paeonia ‘He Xie’. J. Exp. Bot. 2019, 70, 4749–4762. [Google Scholar] [CrossRef]
- Hong, M.J.; Kim, D.Y.; Choi, H.-I.; Seo, Y.W.; Kim, J.-B. Isolation and characterization of kelch repeat-containing F-box proteins from colored wheat. Mol. Biol. Rep. 2020, 47, 1129–1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jo, Y.D.; Kim, J.-B. Frequency and Spectrum of Radiation-Induced Mutations Revealed by Whole-Genome Sequencing Analyses of Plants. Quantum Beam Sci. 2019, 3, 7. [Google Scholar] [CrossRef] [Green Version]
- Sung, P.-H.; Huang, F.-C.; Do, Y.-Y.; Huang, P.-L. Functional Expression of Geraniol 10-Hydroxylase Reveals Its Dual Function in the Biosynthesis of Terpenoid and Phenylpropanoid. J. Agric. Food Chem. 2011, 59, 4637–4643. [Google Scholar] [CrossRef]
- Long, L.; Liu, J.; Gao, Y.; Xu, F.-C.; Zhao, J.-R.; Li, B.; Gao, W. Flavonoid accumulation in spontaneous cotton mutant results in red coloration and enhanced disease resistance. Plant Physiol. Biochem. 2019, 143, 40–49. [Google Scholar] [CrossRef]

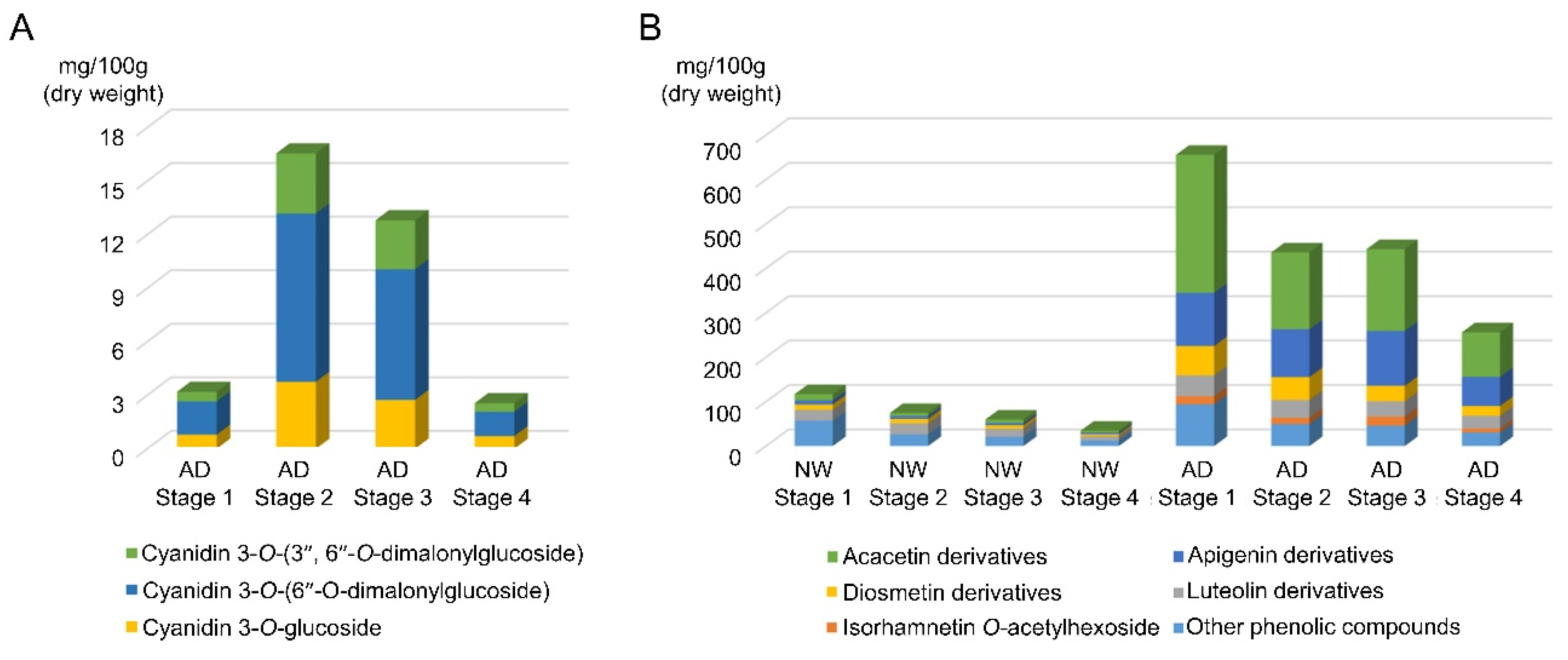
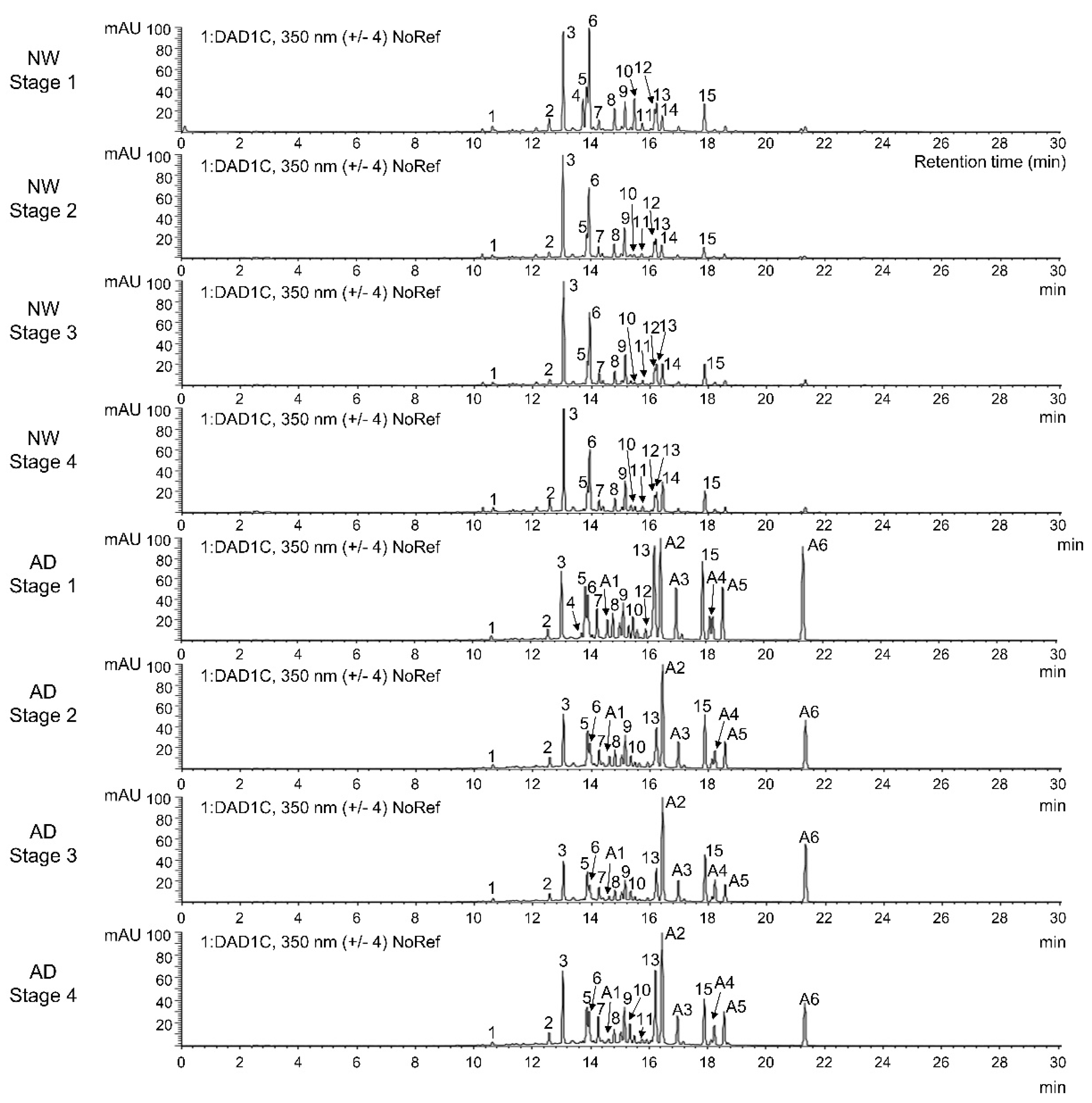





| Peak Number | tR (min) | M + H+/M + H− (m/z) | UV Λmax (nm) | Identification |
|---|---|---|---|---|
| 350 nm | ||||
| 1 | 10.74 | -/353 | 326 | 3-Caffeyolquinic acid [42] x |
| 2 | 12.67 | 595/563 | 338 | 6,8-C,C-diglucosylapigenin [42] x |
| 3 | 13.06 | 449/447 | 348 | Luteolin-7-O-glucoside [42] x |
| 4 | 13.84 | -/515 | 326 | 3,4-Di-caffeoylquinic acid [42] x |
| 5 | 13.88 | -/515 | 326 | 1,4-Di-caffeoylquinic acid [42] x |
| 6 | 13.96 | -/515 | 326 | 3,5-Di-caffeoylquinic acid [42] x |
| 7 | 14.04 | 433/431 | 332 | Apigenin-7-O-glucoside [42] x |
| A1 z | 14.36 | 463/461 | nd y | Trihydroxymethoxyflavone glucoside [42] x |
| 8 | 14.75 | 477/475 | 348 | Diosmetin-7-O-glucuronide [42] x |
| 9 | 15.17 | 549/547 | 348 | Diosmetin-7-O-6″-malonylglucoside [42] x |
| 10 | 15.50 | -/529 | nd | 4-Caffeoyl-5-feruloylquinic acid isomer [42] x |
| 11 | 15.76 | 287/285 | 348 | Luteolin [42] x |
| 12 | 16.16 | 271/269 | 334 | Apigenin [42] x |
| 13 | 16.17 | 533/531 | 334 | Acacetin-7-O-6″-malonylgactoside [42] x |
| A2 | 16.25 | 563/- | 338 | Apigenin-8-C-hexoside-7-O-pentoside [43] w |
| 14 | 16.26 | -/491 | nd | Caffeoyl-dihydroxyphenyllactoyl-tartaric acid [43] w |
| A3 | 17.06 | 533/- | 325 | Acacetin-malonylglucoside [44] x |
| 15 | 17.88 | 619/- | 326 | Acacetin-7-O-(3,6-O-dimalonyl)-β-D-glucopyranoside [30] w |
| A4 | 18.24 | -/519 | 358 | Isorhamnetin O-acetylhexoside [44] w |
| A5 | 18.66 | 547/- | 326 | Acacetin-7-O-(3-O-malonyl)-β-D-glucuronopyranoside [30] w |
| A6 | 21.35 | 285/283 | 330 | Acacetin [42] x |
| 515 nm | ||||
| A7 | 10.33 | 449/625 | 516 | Cyanidin 3-O-glucoside [45] x |
| A8 | 11.338 | 535/529 | 518 | Cyanidin 3-O-(6″-O-malonylglucoside) [45] x |
| A9 | 11.99 | 621/571 | 518 | Cyanidin 3-O-(3″6,″-O-dimalonylglucoside) [30] x |
| A10 | 12.251 | 549/571 | 347 | Chrysoeriol 7-O-malonylglucoside [30] x |
| A11 | 12.83 | 635/677 | 342 | Acetylated luteolin hexoxyl-rhamnoside [29] w |
| 6 | 13.96 | -/515 | 326 | 3,5-Di-caffeoylquinic acid [42] x |
| NW Stage 1 | NW Stage 2 | NW Stage 3 | NW Stage 4 | AD Stage 1 | AD Stage 2 | AD Stage 3 | AD Stage 4 | |
|---|---|---|---|---|---|---|---|---|
| 350 nm | ||||||||
| 1 y | 0.5 ± 0.001 ax | 0.3 ± 0.001 b | 0.3 ± 0.004 c | 0.3 ± 0.004 c | 1.0 ± 0.004 d | 0.6 ± 0.001 e | 0.6 ± 0.001 f | 0.4 ± 0.001 g |
| 2 | 2.3 ± 0.007 a | 0.8 ± 0.002 b | 0.7 ± 0.002 c | 1.2 ± 0.008 d | 8.0 ± 0.005 e | 8.4 ± 0.001 f | 7.9 ± 0.001 g | 5.3 ± 0.003 h |
| 3 | 22.4 ± 0.046 a | 23.6 ± 0.001 b | 17.5 ± 0.003 c | 9.5 ± 0.011 d | 47.0 ± 0.011 e | 39.8 ± 0.001 f | 34.3 ± 0.014 g | 26.7 ± 0.018 h |
| 4 | 12.7 ± 0.008 a | - | - | - | 3.1 ± 0.002 b | - | - | - |
| 5 | 9.3 ± 0.012 a | 5.2 ± 0.003 b | 3.6 ± 0.001 c | 2.3 ± 0.007 d | 39.3 ± 0.007 e | 26.4 ± 0.004 f | 25.5 ± 0.010 g | 14.0 ± 0.004 h |
| 6 | 23.3 ± 0.008 a | 16.5 ± 0.001 b | 12.6 ± 0.001 c | 5.8 ± 0.008 d | 33.1 ± 0.019 e | 18.4 ± 0.005 f | 14.4 ± 0.007 g | 12.2 ± 0.009 h |
| 7 | 2.1 ± 0.002 a | 2.4 ± 0.001 b | 1.8 ± 0.001 c | 1.0 ± 0.006 d | 24.8 ± 0.005 e | 15.0 ± 0.004 f | 12.4 ± 0.006 g | 11.7 ± 0.008 h |
| 8 | 5.3 ± 0.007 a | 3.1 ± 0.001 b | 2.4 ± 0.002 c | 1.3 ± 0.003 d | 20.8 ± 0.012 e | 15.0 ± 0.004 f | 10.4 ± 0.005 g | 6.8 ± 0.005 h |
| A1 | - | - | - | - | 16.1 ± 0.009 a | 10.1 ± 0.003 b | 3.9 ± 0.002 c | 2.4 ± 0.002 d |
| 9 | 6.6 ± 0.008 a | 7.1 ± 0.002 b | 5.2 ± 0.004 c | 2.9 ± 0.003 d | 29.2 ± 0.004 e | 26.7 ± 0.007 f | 20.6 ± 0.005 g | 13.2 ± 0.003 h |
| 10 | 7.6 ± 0.004 a | 0.7 ± 0.001 b | 0.3 ± 0.002 c | 0.4 ± 0.001 d | 17.6 ± 0.007 e | 4.1 ± 0.001 f | 5.2 ± 0.001 g | 3.9 ± 0.003 h |
| 11 | 2.1 ± 0.001 a | 0.9 ± 0.001 b | 0.6 ± 0.003 c | 0.5 ± 0.002 d | - | - | - | 1.9 ± 0.001 e |
| 12 | 4.9 ± 0.005 a | 3.5 ± 0.001 b | 2.5 ± 0.002 c | 1.5 ± 0.001 d | 8.1 ± 0.005 e | - | - | - |
| 13 | 6.5 ± 0.00 1 a | 4.5 ± 0.002 b | 3.7 ± 0.001 c | 1.9 ± 0.003 d | 90.5 ± 0.042 e | 41.9 ± 0.011 f | 37.2 ± 0.019 g | 33.8 ± 0.009 h |
| A2 | - | - | - | - | 78.9 ± 0.047 a | 84.1 ± 0.016 b | 103.1 ± 0.021 c | 48.6 ± 0.031 d |
| 14 | 3.7 ± 0.005 a | 3.5 ± 0.001 b | 4.3 ± 0.001 c | 3.4 ± 0.003 d | - | - | - | - |
| A3 | - | - | - | - | 40.8 ± 0.020 a | 21.2 ± 0.002 b | 20.5 ± 0.010 c | 12.0 ± 0.008 d |
| 15 | 7.4 ± 0.001 a | 2.4 ± 0.002 b | 4.3 ± 0.002 c | 2.4 ± 0.001 d | 59.9 ± 0.013 e | 44.2 ± 0.008 f | 46.4 ± 0.023 g | 19.9 ± 0.008 h |
| A4 | - | - | - | - | 17.8 ± 0.010 a | 14.2 ± 0.001 b | 20.7 ± 0.010 c | 8.7 ± 0.006 d |
| A5 | - | - | - | - | 38.8 ± 0.022 a | 21.4 ± 0.003 b | 16.5 ± 0.008 c | 14.1 ± 0.010 d |
| A6 | - | - | - | - | 80.7 ± 0.025 a | 44.5 ± 0.012 b | 63.7 ± 0.026 c | 20.5 ± 0.014 d |
| 515 nm | ||||||||
| A7 | - | - | - | - | 0.79 ± 0.005 a | 3.75 ± 0.007 b | 2.74 ± 0.005 c | 0.71 ± 0.011 d |
| A8 | - | - | - | - | 1.86 ± 0.036 a | 9.43 ± 0.152 b | 7.32± 0.020 c | 1.36 ± 0.008 d |
| A9 | - | - | - | - | 0.54 ± 0.013 a | 3.37 ± 0.022 b | 2.74 ± 0.018 c | 0.48 ± 0.004 a |
| A10 | - | - | - | - | 0.75 ± 0.011 a | 3.63 ± 0.004 b | 2.40 ± 0.018 c | 0.63 ± 0.016 d |
| A11 | - | - | - | - | 0.42 ± 0.001 a | 2.62 ± 0.007 b | 1.84 ± 0.009 c | 0.39 ± 0.013 a |
| 6 | - | - | - | - | 0.11 ± 0.001 a | 0.38 ± 0.014 b | 0.30 ± 0.002 c | 0.11 ± 0.005 a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jo, Y.D.; Ryu, J.; Kim, Y.-S.; Kang, K.-Y.; Hong, M.J.; Choi, H.-I.; Lim, G.-H.; Kim, J.-B.; Kim, S.H. Dramatic Increase in Content of Diverse Flavonoids Accompanied with Down-Regulation of F-Box Genes in a Chrysanthemum (Chrysanthemum × morifolium (Ramat.) Hemsl.) Mutant Cultivar Producing Dark-Purple Ray Florets. Genes 2020, 11, 865. https://doi.org/10.3390/genes11080865
Jo YD, Ryu J, Kim Y-S, Kang K-Y, Hong MJ, Choi H-I, Lim G-H, Kim J-B, Kim SH. Dramatic Increase in Content of Diverse Flavonoids Accompanied with Down-Regulation of F-Box Genes in a Chrysanthemum (Chrysanthemum × morifolium (Ramat.) Hemsl.) Mutant Cultivar Producing Dark-Purple Ray Florets. Genes. 2020; 11(8):865. https://doi.org/10.3390/genes11080865
Chicago/Turabian StyleJo, Yeong Deuk, Jaihyunk Ryu, Ye-Sol Kim, Kyung-Yun Kang, Min Jeong Hong, Hong-Il Choi, Gah-Hyun Lim, Jin-Baek Kim, and Sang Hoon Kim. 2020. "Dramatic Increase in Content of Diverse Flavonoids Accompanied with Down-Regulation of F-Box Genes in a Chrysanthemum (Chrysanthemum × morifolium (Ramat.) Hemsl.) Mutant Cultivar Producing Dark-Purple Ray Florets" Genes 11, no. 8: 865. https://doi.org/10.3390/genes11080865
APA StyleJo, Y. D., Ryu, J., Kim, Y.-S., Kang, K.-Y., Hong, M. J., Choi, H.-I., Lim, G.-H., Kim, J.-B., & Kim, S. H. (2020). Dramatic Increase in Content of Diverse Flavonoids Accompanied with Down-Regulation of F-Box Genes in a Chrysanthemum (Chrysanthemum × morifolium (Ramat.) Hemsl.) Mutant Cultivar Producing Dark-Purple Ray Florets. Genes, 11(8), 865. https://doi.org/10.3390/genes11080865







