Increased Accumulation of Medium-Chain Fatty Acids by Dynamic Degradation of Long-Chain Fatty Acids in Mucor circinelloides
Abstract
:1. Introduction
2. Material and Methods
2.1. Strains, Plasmids, Cultivation, and Transformation Conditions
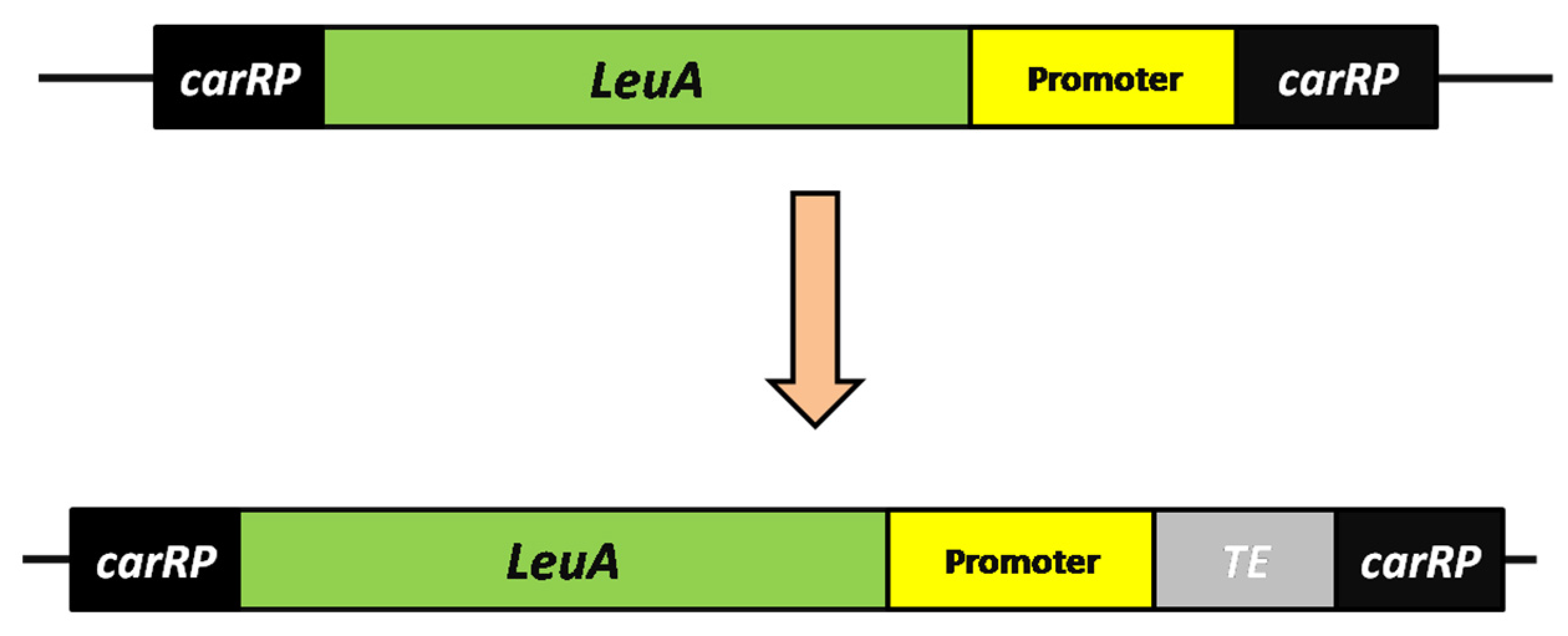
2.2. Construction of the Thioesterase (TE) Over-Expressing Plasmid
2.3. Construction of a Green pyrF Marker
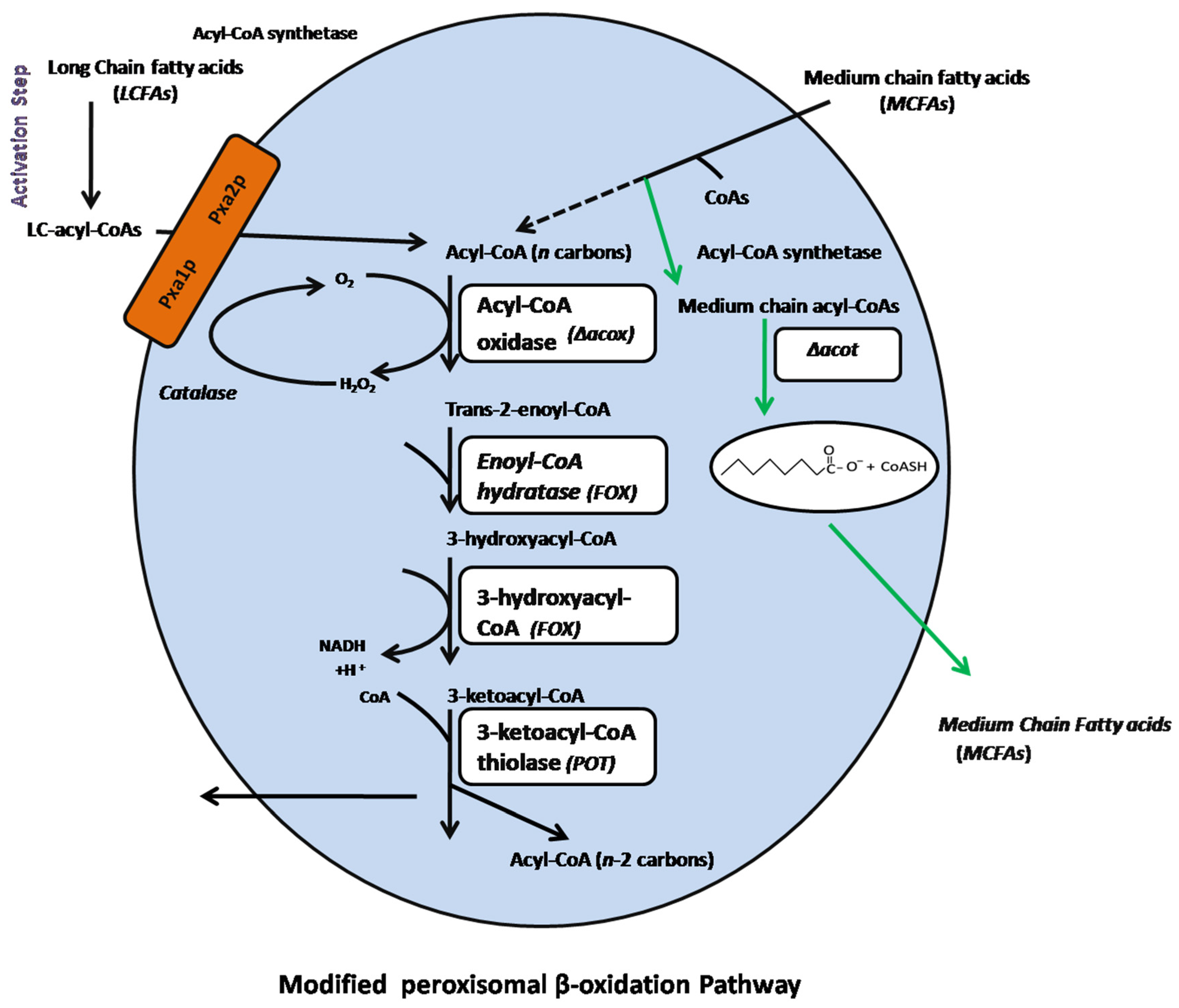
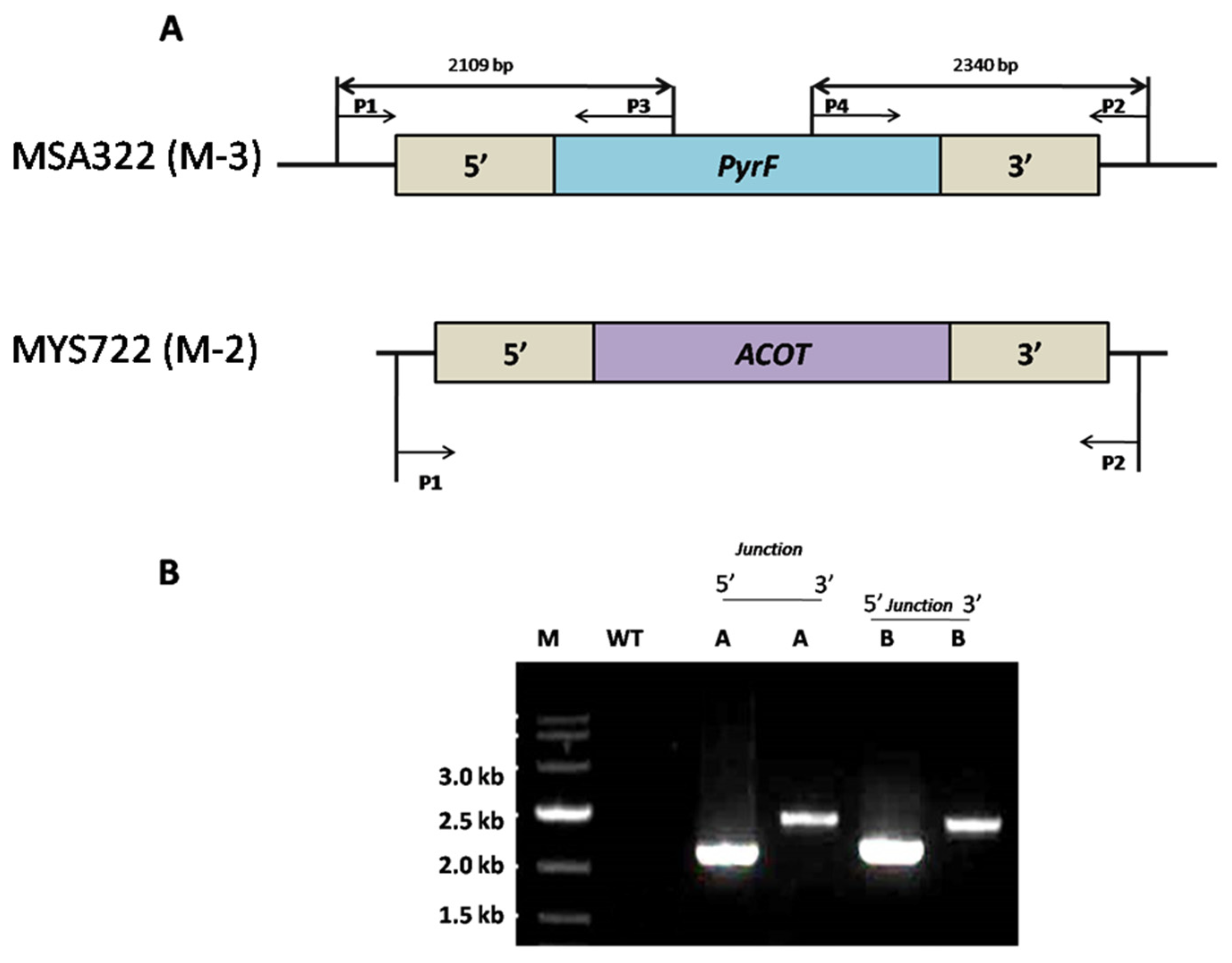
2.4. Disruption of the ACOX and ACOT Genes
2.5. Excision of the pyrF Gene from the pyrF Blaster Marker
2.6. Preparation for Genomic DNA and Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) Analysis
2.7. Determination of Cell Dry Weight (CDW) and Lipid Accumulation
2.8. Determination of Glucose and Nitrogen Concentration in the Culture Medium
2.9. Separation of Lipid Classes
2.10. Statistical Analysis
3. Results
3.1. Generation of Heterologous Acyl-ACP Thioesterase (TE) Gene-Over-Expressing Strains
3.2. Generation of the pyrF Blaster Marker and Disruption of the Acyl-CoA Oxidase (ACOX) and Acyl-CoA Thioesterase (ACOT) Genes in the TE-Over-Expressing and acoxΔ::dpl237 Mutant Strains
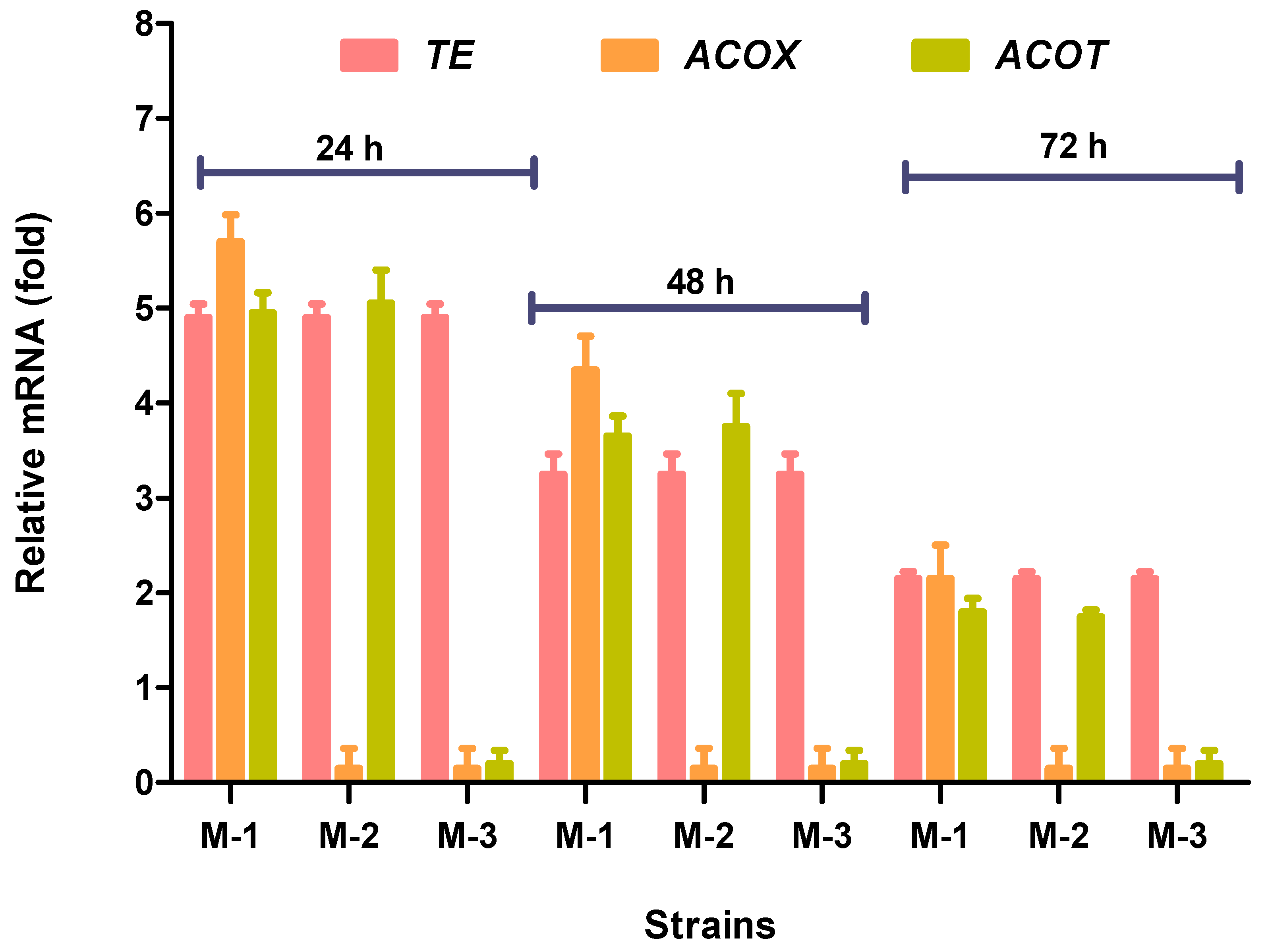
3.3. Expression Levels of Acyl-ACP-Thioesterase (TE), Acyl-CoA-Oxidase (ACOX), and Acyl-CoA Thioesterase (ACOT) Genes in Mutant Fungal Strains
3.4. Fungal Cell Growth and Lipid Yield from Mutant Fungal Strains
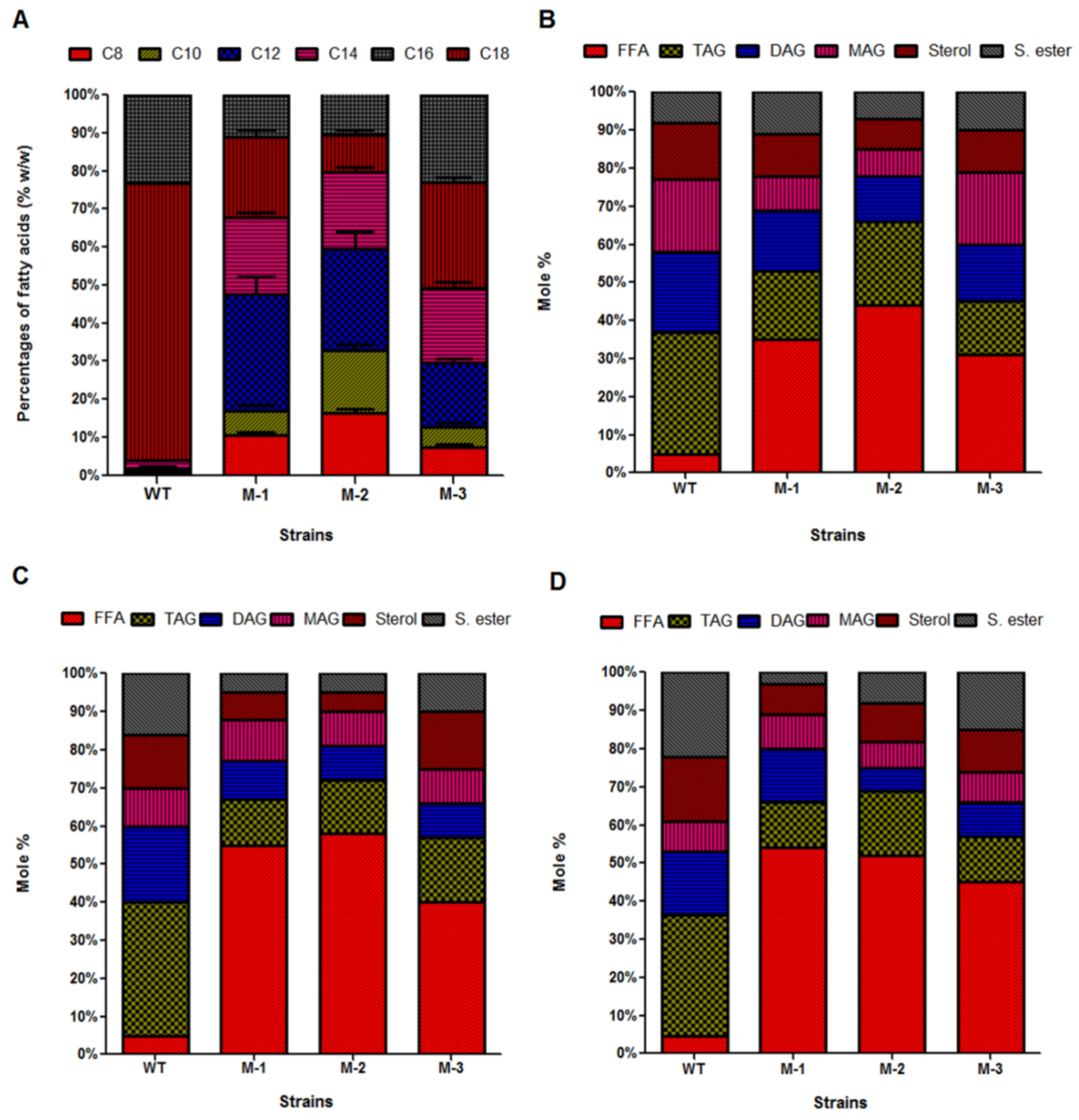
3.5. Incorporation of De Novo Medium-Chain Fatty Acids (MCFAs) into Diverse Lipid Classes
4. Discussion
4.1. The Impact of Heterologous Thioesterase (TE) Protein Over-Expression on Total Lipids and Fatty Acid Composition in the Mutant Fungal Strains
4.2. Combined Impact of TEOver-Expression and ACOX (and ACOT) Gene Disruption on Lipid Accumulation and the Fatty Acid Portfolio in the Mutant Fungal Strains
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MCFAs | medium-chain fatty acids |
| LCFAs | long chain fatty acids |
| TE | Thioesterase |
| CDW | cell dry weight |
| M.C | Mucor circinelloides |
| FFA | free fatty acid |
| HR | homologous recombination |
| TLC | total lipid contents |
| ACP | acyl-carrier protein |
| FAME | fatty acid methyl esters |
| ACOX | acyl-CoA oxidase |
| ACOT | acyl-CoA thioesterase |
| FAS | fatty acid synthase |
| Vvm | volume of air per volume of medium per min |
| TAG | Triacylglycerol |
| DAG | Diacylglycerol |
| MAG | Monoacylglycerol |
| SE | steryl ester |
| MMC | minimal media with casamino acids |
| YPG | yeast peptone glucose |
| GC | gas chromatography |
References
- Fortman, J.; Chhabra, S.; Mukhopadhyay, A.; Chou, H.; Lee, T.S.; Steen, E.; Keasling, J.D. Biofuel alternatives to ethanol: Pumping the microbial well. Trend Biotechnol. 2008, 26, 375–381. [Google Scholar] [CrossRef] [Green Version]
- Allouche, Y.; Cameleyre, X.; Guillouet, S.; Hulin, S.; Thevenieau, F.; Akomia, L.; Molina-Jouve, C. ProBio3 Project: How to Achieve Scientific and Technological Challenges to Boost the Sustainable Microbial Production of Lipids as Biojet Fuel and Chemical CompoundsOCL: Oilseeds Fats Crop. Lipid. 2013, 20, 6D605. [Google Scholar] [CrossRef]
- Runguphan, W.; Keasling, J.D. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 2014, 21, 103–113. [Google Scholar] [CrossRef]
- Hussain, S.A.; Hameed, A.; Khan, M.A.K.; Zhang, Y.; Zhang, H.; Garre, V.; Song, Y. Engineering of Fatty Acid Synthases (FASs) to Boost the Production of Medium-Chain Fatty Acids (MCFAs) in Mucor circinelloides. Int. J. Mol. Sci. 2019, 20, 786. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Qiao, K.; Ahn, W.S.; Stephanopoulos, G. Engineering Yarrowia lipolytica as a Platform for Synthesis of Drop-in Transportation Fuels and Oleochemicals. Proc. Natl. Acad. Sci. USA 2016, 113, 10848–10853. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Li, L.; Zhang, F.; Stephanopoulos, G.; Koffas, M. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc. Natl. Acad. Sci. USA 2014, 111, 11299–11304. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Zhao, L.; Chen, H.; Chen, Y.Q.; Chen, W.; Song, Y.; Ratledge, C. Complete Genome Sequence of a High Lipid-Producing Strain of Mucor circinelloides WJ11 and Comparative Genome Analysis with a Low Lipid-Producing Strain CBS 277.49. PLoS ONE 2015, 10, e0137543. [Google Scholar] [CrossRef] [Green Version]
- Naz, T.; Nosheen, S.; Li, S.; Nazir, Y.; Mustafa, K.; Liu, Q.; Garre, V.; Song, Y. Comparative Analysis of β-Carotene Production by Mucor circinelloides Strains CBS 277.49 and WJ11 under Light and Dark Conditions. Metabolites 2020, 10, 38. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Luan, X.; Zhang, H.; Garre, V.; Song, Y.; Ratledge, C. Improved γ-linolenic acid production in Mucor circinelloides by homologous overexpressing of delta-12 and delta-6 desaturases. Microb. Cell Fact. 2017, 16, 113. [Google Scholar] [CrossRef]
- Sakuradani, E.; Kobayashi, M.; Shimizu, S. Delta6-fatty acid desaturase from an arachidonic acid-producing Mortierella fungus. Gene cloning and its heterologous expression in a fungus, Aspergillus. Gene 1999, 238, 445–453. [Google Scholar] [CrossRef]
- Kelder, B.; Mukeji, P.; Kirchner, S.; Hovanec, G.; Leonard, A.E.; Chuang, L.T.; Kopchick, J.J.; Huang, Y.S. Expression of fungal desaturase genes in cultured mammalian cells. Mol. Cell Biochem. 2001, 219, 7–11. [Google Scholar] [CrossRef]
- Michinak, Y.; Aki, T.; Shimauchi, T.; Nakajima, T.; Kawamoto, S.; Shigeta, S.; Suzuki, O.; Ono, K. Differential response to low temperature of two Delta6 fatty acid desaturases from Mucor circinelloides. Appl. Microbiol. Biotechnol. 2003, 62, 362–368. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, H.; Chen, H.; Chen, Y.Q.; Song, Y. Effects of 20 standard amino acids on the growth, total fatty acids production, and gamma-linolenic acid yield in Mucor circinelloides. Curr. Microbiol. 2014, 69, 899–908. [Google Scholar] [CrossRef]
- Carvalho, A.K.F.; Rivaldi, J.D.; Barbosa, J.C.; Castro, H.F.D. Biosynthesis, characterization and enzymatic transesterification of single cell oil of Mucor circinelloides—A sustainable pathway for biofuel production. Bioresour. Technol. 2015, 181, 47–53. [Google Scholar] [CrossRef]
- Carvalho, A.K.F.; Conceicao, L.R.V.D.; Silva, J.P.V.; Perez, V.H.; Castro, H.F.D. Biodiesel production from Mucor circinelloides using ethanol and heteropolyacid in one and two-step transesterification. Fuel 2017, 202, 503–511. [Google Scholar] [CrossRef]
- Khan, M.A.K.; Yang, J.; Hussain, S.A.; Zhang, H.; Garre, V.; Song, Y. Genetic modification of Mucor circinelloides to construct stearidonic acid producing cell factory. Int. J. Mol. Sci. 2019, 20, 1683. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.A.K.; Yang, J.; Hussain, S.A.; Zhang, H.; Garre, V.; Song, Y. Construction of DGLA producing cell factory by genetic modification of Mucor circinelloides. Microb. Cell Fact. 2019, 18, 64. [Google Scholar] [CrossRef] [Green Version]
- Sarria, S.; Kruyer, N.S.; Yahya, P.P. Microbial synthesis of medium-chain chemicals from renewable. Nat. Biotechnol. 2017, 35, 1158–1166. [Google Scholar] [CrossRef]
- Rigouin, C.; Gueroult, M.; Croux, C.; Dubosis, G.; Borsenberger, V.; Barbe, S.; Marty, A.; Daboussi, F.; André, I.; Bordes, F. Production of Medium Chain Fatty Acids by Yarrowia lipolytica: Combining Molecular Design and TALEN to Engineer the Fatty Acid Synthase. ACS Synth. Biol. 2017, 6, 1870–1879. [Google Scholar] [CrossRef]
- Ruter, C.D.; Zhang, S.; Rao, C.V. Engineering Yarrowia lipolytica for production of medium-chain fatty acids. Appl. Microbiol. Biotechnol. 2015, 99, 7359–7368. [Google Scholar] [CrossRef]
- Camões, F.; Islinger, M.; Guimarães, S.C.; Kilaru, S.; Schuster, M.; Godinho, L.F.; Steinberg, G.; Schrader, M. New insights into the peroxisomal protein inventory: Acyl-CoA oxidases and dehydrogenases are an ancient feature of peroxisomes. Biochim. Biophy. ActaMol. Cell Res. 2015, 1853, 111–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodhi, I.J.; Semenkovich, C.F. Peroxisomes: A nexus for lipid metabolism and cellular signaling. Cell Metab. 2014, 19, 380–392. [Google Scholar] [CrossRef] [Green Version]
- Gabriel, F.; Accoceberry, I.; Bessoule, J.J.; Salin, B.; Lucas-Guérin, M.; Manon, S.; Dementhon, K.; Noël, T. A Fox2-Dependent Fatty Acid β-Oxidation Pathway Coexists both in Peroxisomes and Mitochondria of the Ascomycete Yeast Candida lusitaniae. PLoS ONE 2014, 9, e114531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Zhang, J.; Chen, W.N. Engineering the Saccharomyces cerevisiae β-Oxidation Pathway to Increase Medium Chain Fatty Acid Production as Potential Biofuel. PLoS ONE 2014, 9, e84853. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.J.; Buijs, N.A.; Zhu, Z.; Qin, J.; Siewers, V.; Nielsen, J. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat. Commun. 2016, 7, 11709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lian, J.; Zhao, H. Reversal of the β-oxidation cycle in Saccharomyces cerevisiae for production of fuels and chemicals. ACS Synth. Biol. 2015, 4, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Clomburg, J.M.; Gonzalez, R. Synthesis of medium-chain length (C6-C10) fuels and chemicals via β-oxidation reversal in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2015, 42, 465–475. [Google Scholar] [CrossRef]
- Goh, E.B.; Baidoo, E.E.K.; Keasling, J.D.; Beller, H.R. Engineering of bacterial methyl ketone synthesis for biofuels. Appl. Environ. Microbiol. 2012, 78, 70–80. [Google Scholar] [CrossRef] [Green Version]
- Dellomonaco, C.; Clomburg, J.M.; Miller, E.N.; Gonzalez, R. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature 2011, 476, 355–359. [Google Scholar] [CrossRef]
- Cronan, J.E.; Rock, C.O. Biosynthesis of Membrane Lipids Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd ed.; ASM Press: Washington, DC, USA, 1996; pp. 612–636. [Google Scholar]
- Harwood, J.L. Fatty acid metabolism. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1988, 39, 101–138. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhou, Y.J.; Krivoruchko, A.; Grininger, M.; Zhao, Z.K.; Nielsen, J. Expanding the product portfolio of fungal type I fatty acid synthases. Nat. Chem. Biol. 2017, 13, 360–362. [Google Scholar] [CrossRef] [PubMed]
- Gajewski, J.; Pavlovic, R.; Fischer, M.; Boles, E.; Grininger, M. Engineering fungal de novo fatty acid synthesis for short chain fatty acid production. Nat. Commun. 2017, 8, 14650. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hicks, W.M.; Silver, P.A.; Way, J.C. Engineering acyl carrier protein to enhance production of shortened fatty acids. Biotechnol. Biofuel. 2016, 9, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabrales, L.; Calderon, K.; Hinojosa, I.; Valencia, F.; Abidi, N. Synthesis and characterization of polyesters derived from sebacic acid, hexanediol, and hydroquinone. Int. J. Polym. Anal. Charact. 2016, 21, 718–727. [Google Scholar] [CrossRef]
- Leber, C.; Da-Silva, N.A. Engineering of Saccharomyces cerevisiae for the synthesis of short chain fatty acids. Biotechnol. Bioeng. 2014, 111, 347–358. [Google Scholar] [CrossRef]
- McMahon, M.D.; Prather, K.L. Functional screening and in vitro analysis reveal thioesterases with enhanced substrate specificity profiles that improve short-chain fatty acid production in Escherichia coli. Appl. Environ. Microbiol. 2014, 80, 1042–1050. [Google Scholar] [CrossRef] [Green Version]
- Torella, J.P.; Ford, T.J.; Kim, S.N.; Chen, A.M.; Way, J.C.; Silver, P.A. Tailored fatty acid synthesis via dynamic control of fatty acid elongation. Proc. Natl. Acad. Sci. USA 2013, 110, 11290–11295. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.J.; Lee, S.Y. Microbial production of short-chain alkanes. Nature 2013, 502, 571–574. [Google Scholar] [CrossRef]
- Honda-Malca, S.; Scheps, D.; Kühnel, L.; Venegas-Venegas, E.; Seifert, A.; Nestl, B.M.; Hauer, B. Bacterial CYP153A monooxygenases for the synthesis of omega-hydroxylated fatty acids. Chem. Commun. 2012, 48, 5115–5117. [Google Scholar] [CrossRef]
- Nagao, K.; Yanagita, T. Medium-chain fatty acids: Functional lipids for the prevention and treatment of the metabolic syndrome. Pharm. Res. 2010, 61, 208–212. [Google Scholar] [CrossRef]
- Saerens, S.M.G.; Verstrepen, K.J.; Dijck, P.V.; Thevelein, J.M.; Delvaux, F.R. Parameters affecting ethyl ester production by Saccharomyces cerevisiae during fermentation. Appl. Environ. Microbiol. 2008, 74, 454–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knothe, G. “Designer” biodiesel: Optimizing fatty ester composition to improve fuel properties. Energy Fuels 2008, 22, 1358–1364. [Google Scholar] [CrossRef]
- Lynd, L.R.; Zyl, V.W.H.; McBride, J.E.; Laser, M. Consolidated bioprocessing of cellulosic biomass: An update. Curr. Opin. Biotech. 2005, 16, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Li, A.; Calo, S.; Heitman, J. Calcineur in plays key roles in the dimorphic transition and virulence of the human pathogenic zygomycete Mucor circinelloides. PLoS Pathog. 2013, 9, e1003625. [Google Scholar]
- Torres-Martinez, S.; Ruiz-Vázquez, R.M.; Garre, V.; López-García, S.; Navarro, E.; Vila, A. Molecular tools for carotenogenesis analysis in the zygomycete Mucor circinelloides. Method. Mol. Biol. 2012, 898, 85–107. [Google Scholar]
- Nicolas, F.E.; de-Haro, J.P.; Torres-Martínez, S.; Ruiz-Vázquez, R.M. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet. Biol. 2007, 44, 504–516. [Google Scholar] [CrossRef]
- Bartnicki-García, S.; Nickerson, W.J. Nutrition, growth and morphogenesis of Mucor rouxii. J. Bacteriol. 1962, 84, 841–858. [Google Scholar] [CrossRef] [Green Version]
- Garcia, A.; Adedoyin, G.; Heitman, J.; Lee, S.C. Construction of a Recyclable Genetic Marker and Serial Gene Deletions in the Human Pathogenic Mucorales Mucor circinelloides. G3 Genes Genomes Genet. 2017, 7, 2047–2054. [Google Scholar]
- Hameed, A.; Hussain, S.A.; Yang, J.; Ijaz, M.U.; Liu, Q.; Suleria, H.A.R.; Song, Y. Antioxidants Potential of the Filamentous Fungi (Mucor circinelloides). Nutrients 2017, 9, 1101. [Google Scholar] [CrossRef] [Green Version]
- Hussain, S.A.; Nazir, Y.; Hameed, A.; Yang, W.; Mustafa, K.; Song, Y. Optimization of Diverse Carbon Sources and Cultivation Conditions for Enhanced Growthand Lipid and Medium-Chain Fatty Acid (MCFAs) Production by Mucor circinelloides. Fermentation 2019, 5, 35. [Google Scholar] [CrossRef] [Green Version]
- Hameed, A.; Hussain, S.A.; Nosheen, S.; Muhammad, Z.; Wu, Y.; Ullah, S.; Suleria, H.A.R.; Song, Y. Microencapsulation of microbial antioxidants from Mucor circinelloides, their physico-chemical characterization, in vitro digestion and releasing behaviors in food. Appl. Biol. Chem. 2020, 63, 28. [Google Scholar] [CrossRef]
- Hameed, A.; Hussain, S.A.; Ijaz, M.U.; Ullah, S.; Muhammad, Z.; Suleria, H.A.R.; Song, Y. Antioxidant activity of polyphenolic extracts of filamentous fungus Mucor circinelloides (WJ11): Extraction, characterization and storage stability of food emulsions. Food Bioscience 2020, 34, 100525. [Google Scholar] [CrossRef]
- Vellanki, S.; Navarro-Mendoza, M.I.; Garcia, A.; Murcia, L.; Perez-Arques, C.; Garre, V.; Nicolas, F.E.; Lee, S.C. Mucor circinelloides: Growth, maintenance, and genetic manipulation. Curr. Prot. Micro. 2018, 49, e53. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, M.; Wang, J.; Kronstad, J.W. Peroxisomal and Mitochondrial β-Oxidation Pathways Influence the Virulence of the Pathogenic Fungus Cryptococcus neoformans. Eukar. Cell. 2012, 11, 1042–1054. [Google Scholar] [CrossRef] [Green Version]
- Froman, B.E.; Edwards, P.C.; Bursch, A.G.; Dehesh, K. ACX3, a novel medium-chain acyl-coenzyme A oxidase from Arabidopsis. Plant Physiol. 2000, 123, 733–742. [Google Scholar] [CrossRef] [Green Version]
- Hunt, M.C.; Alexson, S.E. Novel functions of acyl-CoA thioesterases and acyltransferases as auxiliary enzymes in peroxisomal lipid metabolism. Prog. Lipid Res. 2008, 47, 405–421. [Google Scholar] [CrossRef] [Green Version]
- Hiltunen, J.K.; Mursula, A.M.; Rottensteiner, H.; Wierenga, R.K.; Kastaniotis, A.J.; Gurvitz, A. The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2003, 27, 35–64. [Google Scholar] [CrossRef] [Green Version]
- Folch, J.; Lees, M.; Stanley, G.H.S. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1956, 226, 497–509. [Google Scholar]
- Chaney, A.L.; Marbach, E.P. Modified reagents for determination of urea and ammonia. Clin. Chem. 1962, 8, 130–132. [Google Scholar] [CrossRef]
- Carroll, K.K. Separation of lipid classes by chromatography on Florisil. J. Lipid Res. 1961, 2, 135–141. [Google Scholar]
- Freeman, C.; West, D. Complete separation of lipid classes on a single thin-layer plate. J. Lipid Res. 1966, 7, 324–327. [Google Scholar] [PubMed]
- Salunke, D.; Mangalekar, R.; Kuvalekar, A.; Harsulkar, A. Bioconversion of alpha-linolenic acid into long chain polyunsaturated fatty acids by oleaginous fungi. Int. J. Phar. Biol. Sci. 2014, 5, 27–35. [Google Scholar]
- Wynn, J.P.; Hamid, A.A.; Li, Y.; Ratledge, C. Biochemical events leading to diversion of carbon into storage lipids in oleaginous fungi Mucor circinelloides and Mortierella alpina. Microbiology 2001, 147, 2857–2864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, F.; Zhao, L.; Nelson, M.D.Y.; Nikolau, B.J. Two distinct domains contribute to the substrate acyl chain length selectivity of plant acyl-ACP thioesterase. Nat. Commun. 2018, 9, 860. [Google Scholar] [CrossRef]
- Ohlrogge, J.B.; Jaworski, J.G. Regulation of fatty acid synthesis. Ann. Rev. Plant Biol. 1997, 48, 109–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, P.; Cronan, J. Inhibition of fatty acid synthesis in Escherichia coli in the absence of phospholipid synthesis and release of inhibition by thioesterase action. J. Bacteriol. 1994, 176, 2814–2821. [Google Scholar] [CrossRef] [Green Version]
- Sherkhanov, S.; Korman, T.P.; Bowie, J.U. Improving the tolerance of Escherichia coli to medium-chain fatty acid production. Metab. Eng. 2014, 25, 1–7. [Google Scholar] [CrossRef]
- Hunt, M.C.; Alexson, S.E. The role Acyl-CoA thioesterases play in mediating intracellular lipid metabolism. Prog. Lipid Res. 2002, 41, 99–130. [Google Scholar] [CrossRef]
- Stefan, A.; Hochkoeppler, A.; Ugolini, L.; Lazzeri, L.; Conte, E. The Expression of the Cuphea Palustris Thioesterase CpFatB2 in Yarrowia Lipolytica Triggers Oleic Acid Accumulation. Biotechnol. Prog. 2016, 32, 26–35. [Google Scholar] [CrossRef]
- Bardi, L.; Cocito, C.; Marzona, M. Saccharomyces cerevisiae cell fatty acid composition and release during fermentation without aeration and in absence of exogenous lipids. Int. J. Food Microbiol. 1999, 47, 133–140. [Google Scholar] [CrossRef]
- Kopka, J.; Ohlrogge, J.B.; Jaworski, J.G. Analysis of in Vivo Levels of Acyl- Thioesters with Gas Chromatography/Mass Spectrometry of the Butylamide Derivative. Anal. Biochem. 1995, 224, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A. Amoeba provides insight into the origin of virulence in pathogenic fungi. Adv. Exp. Med. Biol. 2012, 710, 1–10. [Google Scholar] [PubMed]
- Chrisman, C.J.; Alvarez, M.; Casadevall, A. Phagocytosis of Cryptococcus neoformans by, and nonlytic exocytosis from Acanthamoeba castellanii. Appl. Environ. Microbiol. 2010, 76, 6056–6062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, K.; De-Obaldia, A.L.; Heitman, J. Cryptococcus neoformans mates on pigeon guano: Implications for the realized ecological niche and globalization. Eukar. Cell. 2007, 6, 949–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litvintseva, A.P.; Carbone, I.; Rossouw, J.; Thakur, R.; Govender, N.P.; Mitchell, T.G. Evidence that the human pathogenic fungus Cryptococcus neoformans var. grubii may have evolved in Africa. PLoS ONE. 2011, 6, e19688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruenwai, R.; Cheevadhanarak, S.; Laoteng, K. Overexpression of acetyl-CoA carboxylase gene of Mucorrouxii enhanced fatty acid content in Hansenula polymorpha. Mol. Biotechnol. 2009, 42, 327–332. [Google Scholar] [CrossRef]
- Vorapreeda, T.; Thammarongtham, C.; Cheevadhanarak, S.; Laoteng, K. Alternative routes of acetyl-CoA synthesis identified by comparative genomic analysis: Involvement in the lipid production of oleaginous yeast and fungi. Microbiology 2012, 158, 217–228. [Google Scholar] [CrossRef] [Green Version]
- Zan, X.; Tang, X.; Chu, L.; Zhao, L.; Chen, H.; Chen, Y.Q.; Chen, W.; Song, Y. Lipase genes in Mucor circinelloides: Identification, sub-cellular location, phylogenetic analysis and expression profiling during growth and lipid accumulation. J. Ind. Microbiol. Biotechnol. 2016, 43, 1467–1480. [Google Scholar] [CrossRef]
- Li, Z.; Sun, H.; Mo, X.; Li, X.; Xu, B.; Tian, P. Overexpression of malic enzyme (ME) of Mucor circinelloides improved lipid accumulation in engineered Rhodotorula glutinis. Appl. Microbiol. Biotechnol. 2013, 97, 14927–14936. [Google Scholar] [CrossRef]



| Name | Genotype/Characteristics | References | |
|---|---|---|---|
| M. circinelloides | MU758 | LeuA-pyrF- | This work |
| MYS719 | LeuA+pyrF-TE + | This work | |
| MYS720 | LeuA+pyrF+TE + acoxΔ ::pyrF-dpl237 | This work | |
| MYS721 | LeuA+pyrF+TE + acoxΔ::dpl237 | This work | |
| MYS722 | LeuA+pyrF+TE + acoxΔ::dpl237 | This work | |
| MSA321 | LeuA+pyrF+TE + acoxΔ::dpl237 acotΔ::pyrF | This work | |
| MSA322 | LeuA+pyrF+TE + acoxΔ::dpl237acotΔ ::pyrF | This work | |
| Plasmids | pMAT1552-LeuA | AmpicillinR, LeuA Marker gene | (Zhang et al. [9]) |
| pMAT1552-LeuA TE | Contain LeuA Marker gene and thioesterase gene (TE) | This work | |
| pCR21-TOPO | Ampicillin R Kanamycin R | Invitrogen | |
| pTOPO-pyrF | 890-bp pyrF fragment cloned into pCR21-TOPO | This work | |
| pACOX-KO | ACOX disruption construct cloned into pCR21-TOPO | This work | |
| pACOT-KO | ACOT disruption construct cloned into pCR21-TOPO | This work | |
| pSL13 | pyrG blaster marker in pCR21-TOPO | (Garcia et al. [49]) | |
| pYS719 | PyrF blaster marker in pCR21-TOPO | This work |
| WT (Control) | M-1 | M-2 | M-3 | |
|---|---|---|---|---|
| CDW (g/L) | 13.28 ± 1.37 a | 11.14 ± 0.21 b | 10.45 ± 1.26 b | 07.35 ± 1.29 |
| %TLC (CDW) | 36.70 ± 0.21 | 54.85 ± 1.20 b | 68.85 ± 2.20 a | 47.01 ± 2.40 c |
| TLC (g/L) | 04.87 ± 0.23 | 06.11 ± 0.12 a | 07.19 ± 0.08 a | 03.46 ± 0.09 |
| % MCFAs | 02.25 ± 1.10 | 47.45 ± 1.80 b | 60.09 ± 2.10 a | 29.61 ± 0.80 c |
| MCFAs (g/L) | 0.11 ± 0.71 | 02.88 ± 0.04 b | 04.32 ± 0.09 a | 01.04 ± 0.05 |
| PTL | 1217.5 ± 0.11 c | 1527.5 ± 0.41 b | 1800.5 ± 1.2 a | 865.1 ± 0.25 |
| PMCFAs | 204 ± 0.45 | 722 ± 0.23 b | 1084 ± 0.17 a | 260 ± 0.21 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, S.A.; Garcia, A.; Khan, M.A.K.; Nosheen, S.; Zhang, Y.; Koffas, M.A.G.; Garre, V.; Lee, S.C.; Song, Y. Increased Accumulation of Medium-Chain Fatty Acids by Dynamic Degradation of Long-Chain Fatty Acids in Mucor circinelloides. Genes 2020, 11, 890. https://doi.org/10.3390/genes11080890
Hussain SA, Garcia A, Khan MAK, Nosheen S, Zhang Y, Koffas MAG, Garre V, Lee SC, Song Y. Increased Accumulation of Medium-Chain Fatty Acids by Dynamic Degradation of Long-Chain Fatty Acids in Mucor circinelloides. Genes. 2020; 11(8):890. https://doi.org/10.3390/genes11080890
Chicago/Turabian StyleHussain, Syed Ammar, Alexis Garcia, Md. Ahsanul Kabir Khan, Shaista Nosheen, Yao Zhang, Mattheos A. G. Koffas, Victoriano Garre, Soo Chan Lee, and Yuanda Song. 2020. "Increased Accumulation of Medium-Chain Fatty Acids by Dynamic Degradation of Long-Chain Fatty Acids in Mucor circinelloides" Genes 11, no. 8: 890. https://doi.org/10.3390/genes11080890
APA StyleHussain, S. A., Garcia, A., Khan, M. A. K., Nosheen, S., Zhang, Y., Koffas, M. A. G., Garre, V., Lee, S. C., & Song, Y. (2020). Increased Accumulation of Medium-Chain Fatty Acids by Dynamic Degradation of Long-Chain Fatty Acids in Mucor circinelloides. Genes, 11(8), 890. https://doi.org/10.3390/genes11080890







