Comparative Analysis of Mouse Decidualization Models at the Molecular Level
Abstract
1. Introduction
2. Materials and Methods
2.1. Mouse Models of Decidualization
2.2. RNA-seq
2.3. Validation by Quantitative RT-PCR
2.4. Gene Ontology (GO) Analysis
2.5. Pathway Analysis
2.6. Gene Network Analysis
3. Results
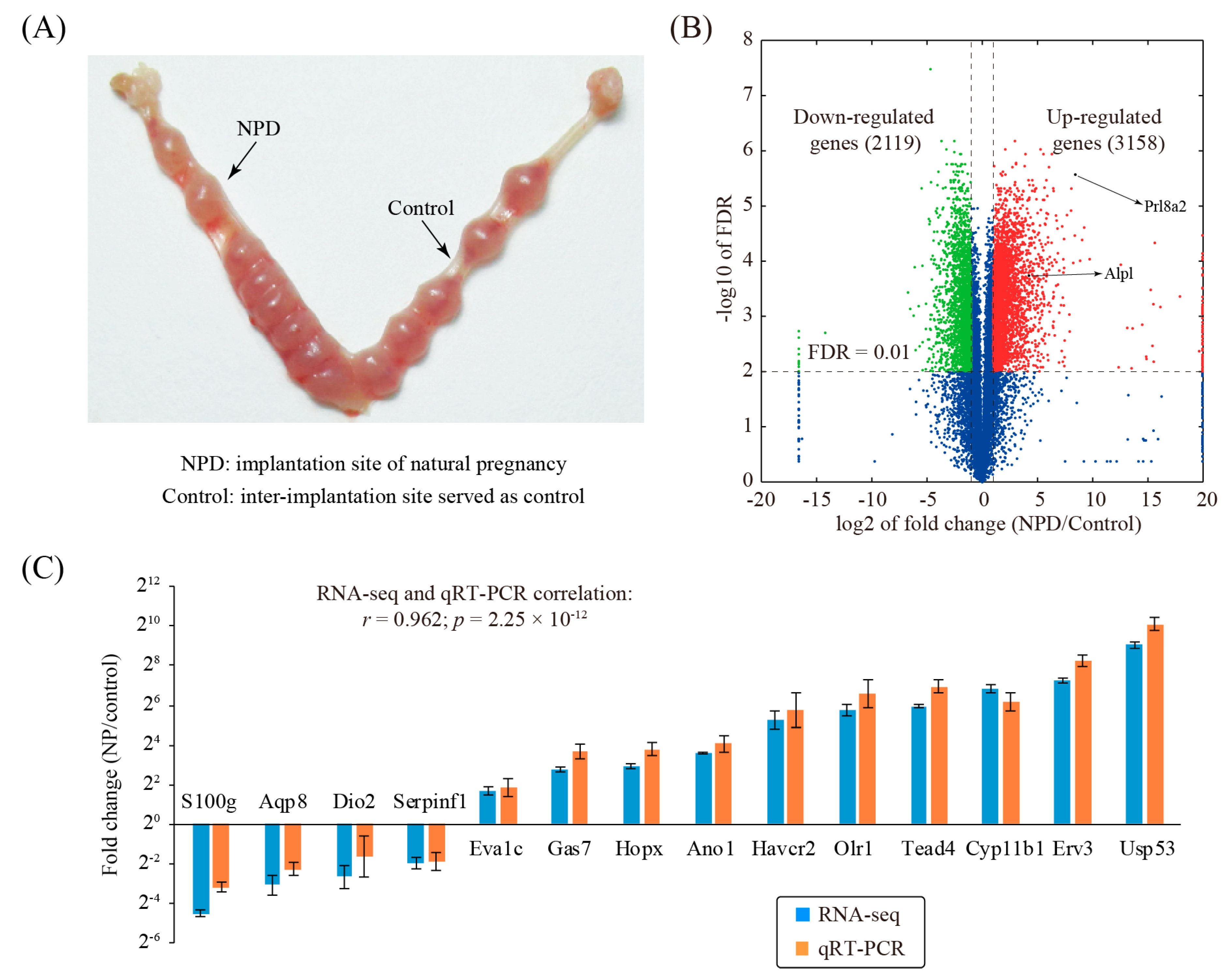
3.1. Identification of Changed Genes Associated with Decidualization During Natural Pregnancy
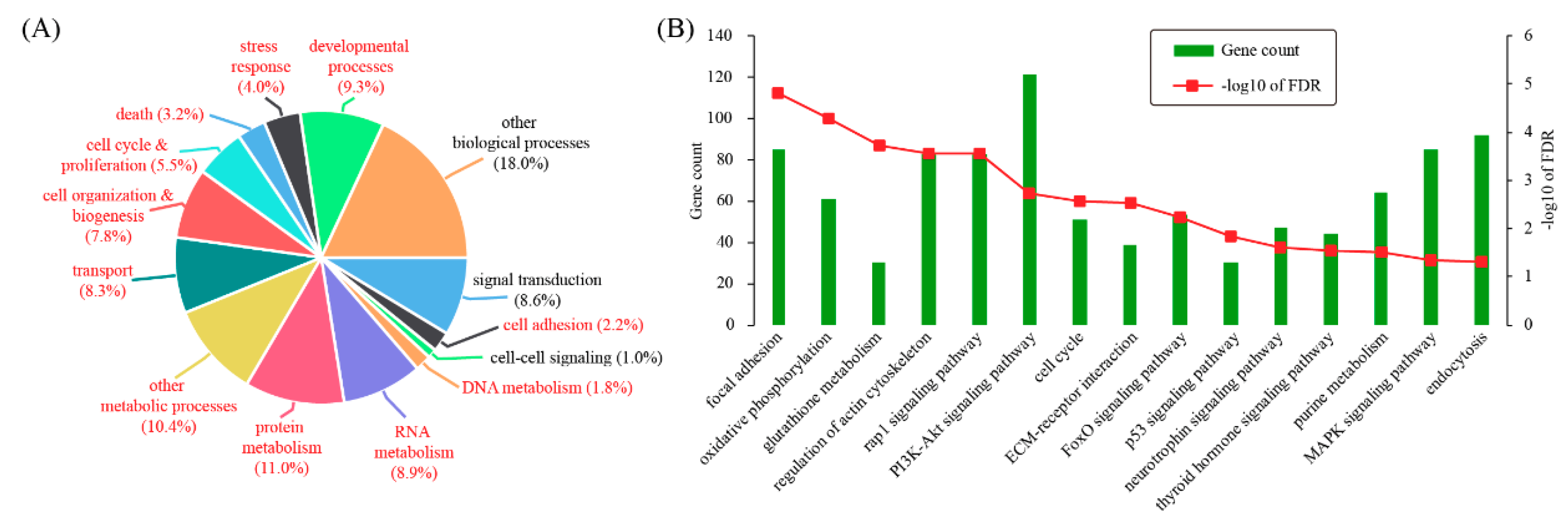
3.2. Characterizing Differentially Expressed Genes by Gene Ontology and Pathway Analysis
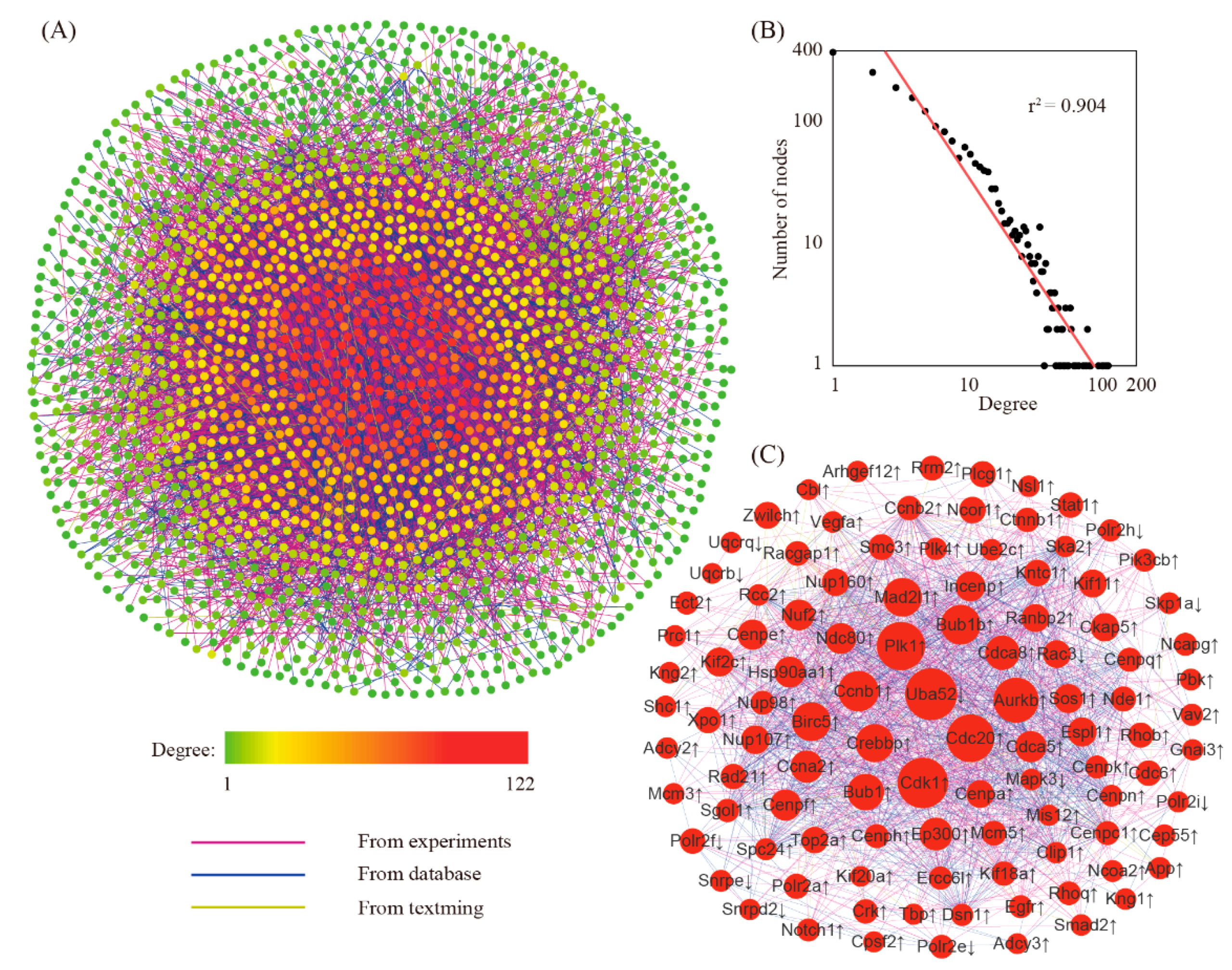
3.3. Searching for Hub Genes Through Network Analysis
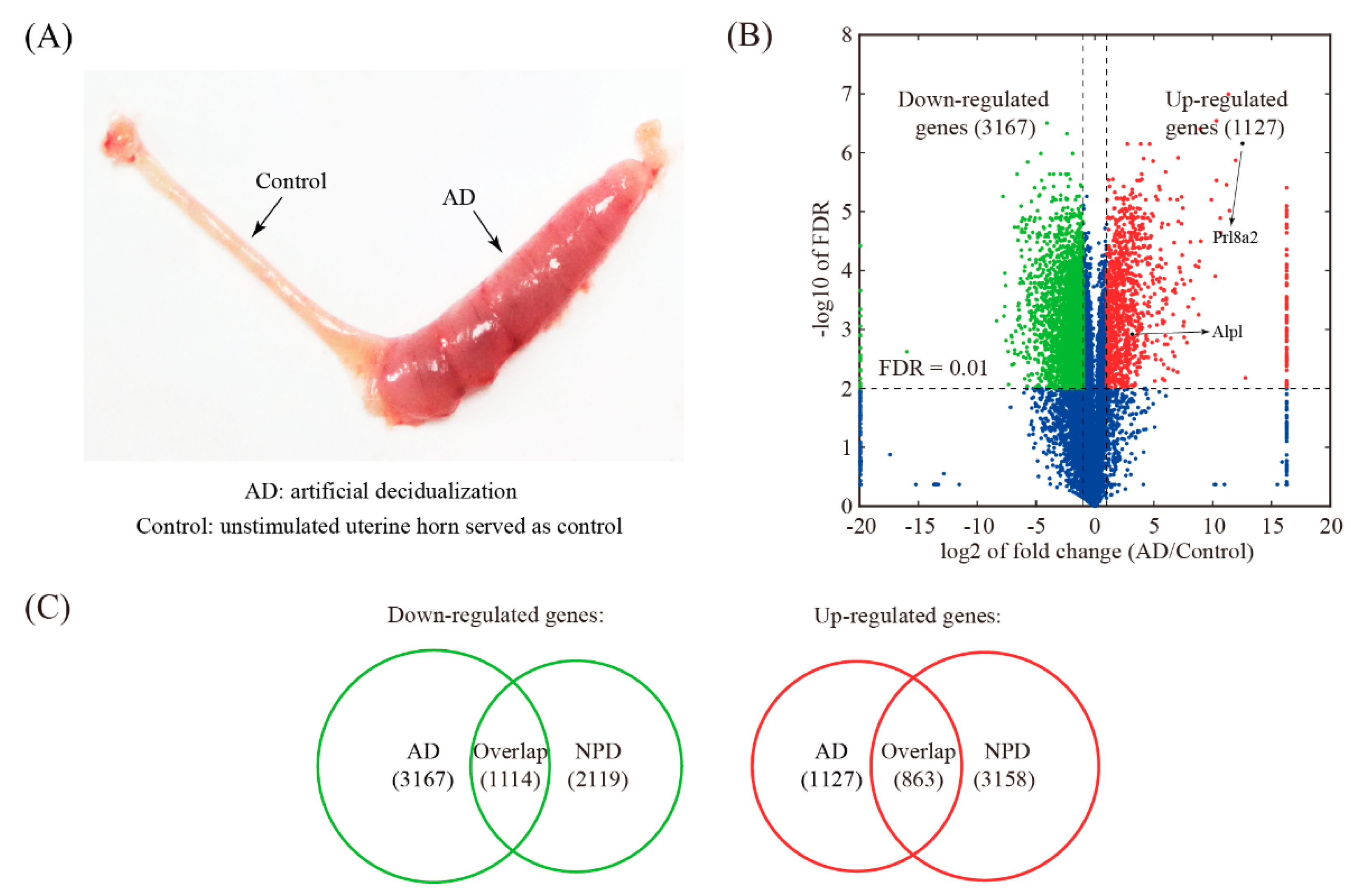
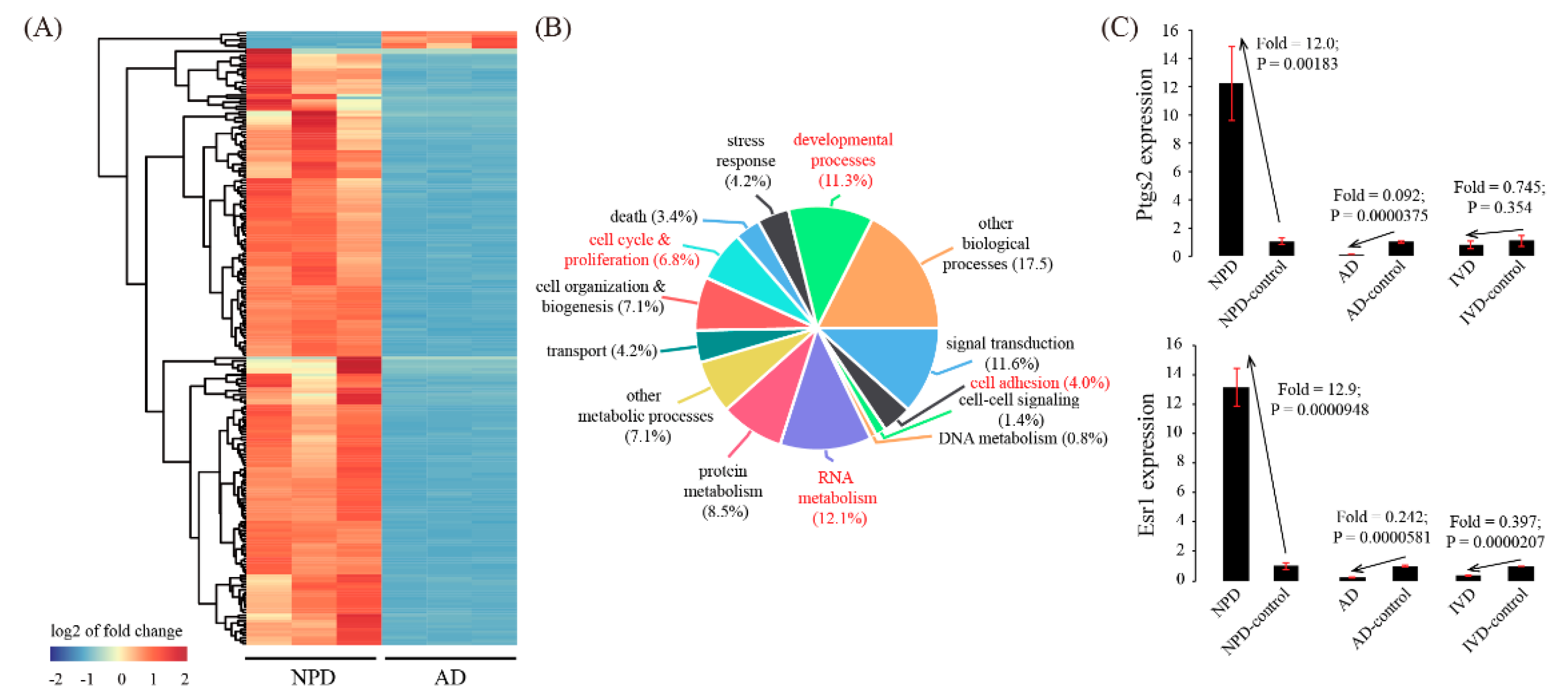
3.4. Global Comparison with Artificial Decidualization (AD) Model
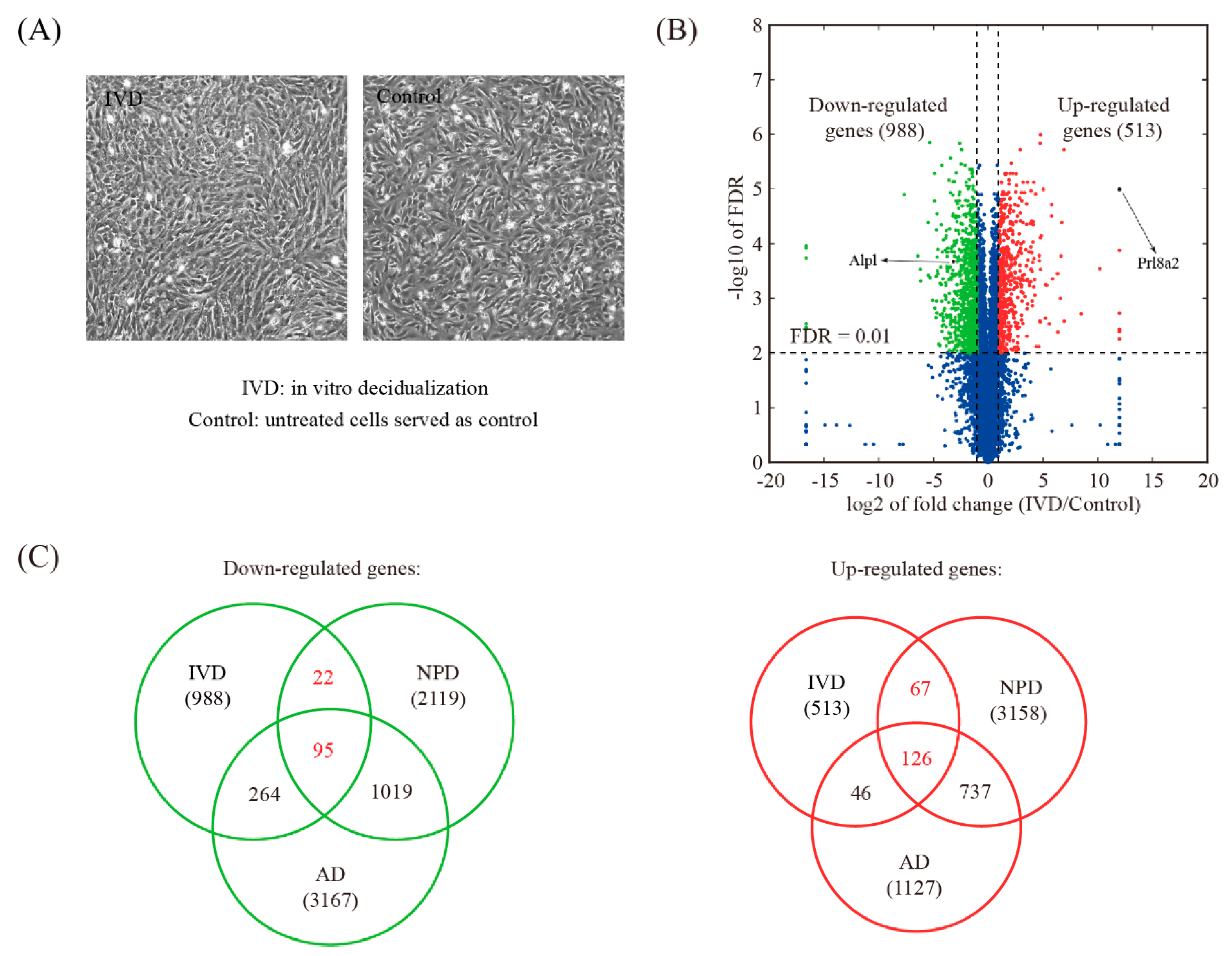
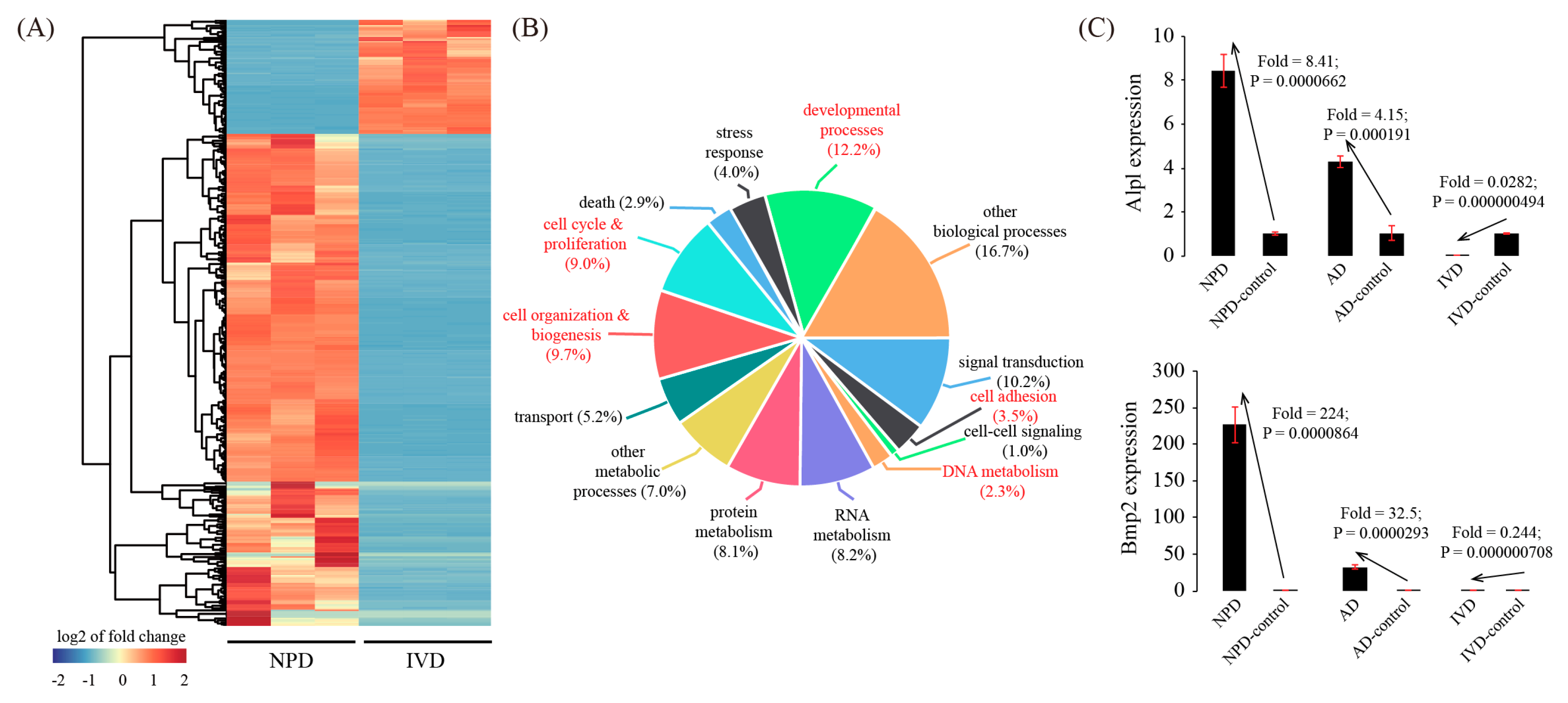
3.5. Global Comparison with In Vitro Decidualization (IVD) Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, H.; Dey, S.K. Roadmap to embryo implantation: Clues from mouse models. Nat. Rev. Genet. 2006, 7, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Ramathal, C.Y.; Bagchi, I.C.; Taylor, R.N.; Bagchi, M.K. Endometrial decidualization: Of mice and men. Semin. Reprod. Med. 2010, 28, 17–26. [Google Scholar] [CrossRef]
- Christian, M.; Mak, I.; White, J.O.; Brosens, J.J. Mechanisms of decidualization. Reprod. Biomed. Online 2002, 4 (Suppl. 3), 24–30. [Google Scholar] [CrossRef]
- Liu, J.L.; Wang, T.S. Systematic analysis of the molecular mechanism underlying decidualization using a text mining approach. PLoS ONE 2015, 10, e0134585. [Google Scholar] [CrossRef] [PubMed]
- Gellersen, B.; Brosens, J.J. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr. Rev. 2014, 35, 851–905. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.K.; Lim, H.; Das, S.K.; Reese, J.; Paria, B.C.; Daikoku, T.; Wang, H. Molecular cues to implantation. Endocr. Rev. 2004, 25, 341–373. [Google Scholar] [CrossRef] [PubMed]
- Deb, K.; Reese, J.; Paria, B.C. Methodologies to study implantation in mice. Methods Mol. Med. 2006, 121, 9–34. [Google Scholar]
- Kimura, F.; Takakura, K.; Takebayashi, K.; Ishikawa, H.; Kasahara, K.; Goto, S.; Noda, Y. Messenger ribonucleic acid for the mouse decidual prolactin is present and induced during in vitro decidualization of endometrial stromal cells. Gynecol. Endocrinol. Off. J. Int. Soc. Gynecol. Endocrinol. 2001, 15, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. Rna-seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by rna-seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Garber, M.; Grabherr, M.G.; Guttman, M.; Trapnell, C. Computational methods for transcriptome annotation and quantification using rna-seq. Nat. Methods 2011, 8, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Wang, T.S.; Zhao, M. Genome-wide association mapping for female infertility in inbred mice. G3 2016, 6, 2929–2935. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.Y.; Zhang, W.Q.; Zhao, M.; Zhu, C.; He, J.P.; Liu, J.L. Assessment of embryo-induced transcriptomic changes in hamster uterus using rna-seq. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 46, 1868–1878. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. Tophat: Discovering splice junctions with rna-seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by rna-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Liu, J.L.; Zuo, R.J.; Peng, Y.; Fu, Y.S. The impact of multiparity on uterine gene expression and decidualization in mice. Reprod. Sci. 2016, 23, 687–694. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. David bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. String v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Assenov, Y.; Ramirez, F.; Schelhorn, S.E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef]
- Kashiwagi, A.; DiGirolamo, C.M.; Kanda, Y.; Niikura, Y.; Esmon, C.T.; Hansen, T.R.; Shioda, T.; Pru, J.K. The postimplantation embryo differentially regulates endometrial gene expression and decidualization. Endocrinology 2007, 148, 4173–4184. [Google Scholar] [CrossRef] [PubMed]
- McConaha, M.E.; Eckstrum, K.; An, J.; Steinle, J.J.; Bany, B.M. Microarray assessment of the influence of the conceptus on gene expression in the mouse uterus during decidualization. Reproduction 2011, 141, 511–527. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, I.; Das, S.K.; Wang, J.; Dey, S.K. Developmental expression of the cyclo-oxygenase-1 and cyclo-oxygenase-2 genes in the peri-implantation mouse uterus and their differential regulation by the blastocyst and ovarian steroids. J. Mol. Endocrinol. 1996, 16, 107–122. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Paria, B.C.; Das, S.K.; Dinchuk, J.E.; Langenbach, R.; Trzaskos, J.M.; Dey, S.K. Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell 1997, 91, 197–208. [Google Scholar] [CrossRef]
- Wang, H.; Ma, W.G.; Tejada, L.; Zhang, H.; Morrow, J.D.; Das, S.K.; Dey, S.K. Rescue of female infertility from the loss of cyclooxygenase-2 by compensatory up-regulation of cyclooxygenase-1 is a function of genetic makeup. J. Biol. Chem. 2004, 279, 10649–10658. [Google Scholar] [CrossRef]
- Curtis Hewitt, S.; Goulding, E.H.; Eddy, E.M.; Korach, K.S. Studies using the estrogen receptor alpha knockout uterus demonstrate that implantation but not decidualization-associated signaling is estrogen dependent. Biol. Reprod. 2002, 67, 1268–1277. [Google Scholar] [CrossRef]
- Hewitt, S.C.; Kissling, G.E.; Fieselman, K.E.; Jayes, F.L.; Gerrish, K.E.; Korach, K.S. Biological and biochemical consequences of global deletion of exon 3 from the er alpha gene. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2010, 24, 4660–4667. [Google Scholar]
- Dupont, S.; Krust, A.; Gansmuller, A.; Dierich, A.; Chambon, P.; Mark, M. Effect of single and compound knockouts of estrogen receptors alpha (eralpha) and beta (erbeta) on mouse reproductive phenotypes. Development 2000, 127, 4277–4291. [Google Scholar]
- Pawar, S.; Laws, M.J.; Bagchi, I.C.; Bagchi, M.K. Uterine epithelial estrogen receptor-alpha controls decidualization via a paracrine mechanism. Mol. Endocrinol. 2015, 29, 1362–1374. [Google Scholar] [CrossRef]
- Lee, K.Y.; Jeong, J.W.; Wang, J.; Ma, L.; Martin, J.F.; Tsai, S.Y.; Lydon, J.P.; DeMayo, F.J. Bmp2 is critical for the murine uterine decidual response. Mol. Cell. Biol. 2007, 27, 5468–5478. [Google Scholar] [CrossRef]
- Li, Q.; Kannan, A.; Wang, W.; Demayo, F.J.; Taylor, R.N.; Bagchi, M.K.; Bagchi, I.C. Bone morphogenetic protein 2 functions via a conserved signaling pathway involving wnt4 to regulate uterine decidualization in the mouse and the human. J. Biol. Chem. 2007, 282, 31725–31732. [Google Scholar] [CrossRef] [PubMed]
- Franco, H.L.; Dai, D.; Lee, K.Y.; Rubel, C.A.; Roop, D.; Boerboom, D.; Jeong, J.W.; Lydon, J.P.; Bagchi, I.C.; Bagchi, M.K.; et al. Wnt4 is a key regulator of normal postnatal uterine development and progesterone signaling during embryo implantation and decidualization in the mouse. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2011, 25, 1176–1187. [Google Scholar] [CrossRef] [PubMed]
- Large, M.J.; Wetendorf, M.; Lanz, R.B.; Hartig, S.M.; Creighton, C.J.; Mancini, M.A.; Kovanci, E.; Lee, K.F.; Threadgill, D.W.; Lydon, J.P.; et al. The epidermal growth factor receptor critically regulates endometrial function during early pregnancy. PLoS Genet. 2014, 10, e1004451. [Google Scholar] [CrossRef] [PubMed]
- Whirledge, S.D.; Oakley, R.H.; Myers, P.H.; Lydon, J.P.; DeMayo, F.; Cidlowski, J.A. Uterine glucocorticoid receptors are critical for fertility in mice through control of embryo implantation and decidualization. Proc. Natl. Acad. Sci. USA 2015, 112, 15166–15171. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Bian, F.; Ma, X.; Kalinichenko, V.V.; Das, S.K. Control of regional decidualization in implantation: Role of foxm1 downstream of hoxa10 and cyclin d3. Sci. Rep. 2015, 5, 13863. [Google Scholar] [CrossRef]
- Herington, J.L.; Bany, B.M. The conceptus increases secreted phosphoprotein 1 gene expression in the mouse uterus during the progression of decidualization mainly due to its effects on uterine natural killer cells. Reproduction 2007, 133, 1213–1221. [Google Scholar] [CrossRef]
- Simon, L.; Spiewak, K.A.; Ekman, G.C.; Kim, J.; Lydon, J.P.; Bagchi, M.K.; Bagchi, I.C.; DeMayo, F.J.; Cooke, P.S. Stromal progesterone receptors mediate induction of indian hedgehog (ihh) in uterine epithelium and its downstream targets in uterine stroma. Endocrinology 2009, 150, 3871–3876. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Zhao, M.; Zhang, W.-Q.; Huang, M.-Y.; Zhu, C.; He, J.-P.; Liu, J.-L. Comparative Analysis of Mouse Decidualization Models at the Molecular Level. Genes 2020, 11, 935. https://doi.org/10.3390/genes11080935
Wang C, Zhao M, Zhang W-Q, Huang M-Y, Zhu C, He J-P, Liu J-L. Comparative Analysis of Mouse Decidualization Models at the Molecular Level. Genes. 2020; 11(8):935. https://doi.org/10.3390/genes11080935
Chicago/Turabian StyleWang, Chong, Miao Zhao, Wen-Qian Zhang, Ming-Yu Huang, Can Zhu, Jia-Peng He, and Ji-Long Liu. 2020. "Comparative Analysis of Mouse Decidualization Models at the Molecular Level" Genes 11, no. 8: 935. https://doi.org/10.3390/genes11080935
APA StyleWang, C., Zhao, M., Zhang, W.-Q., Huang, M.-Y., Zhu, C., He, J.-P., & Liu, J.-L. (2020). Comparative Analysis of Mouse Decidualization Models at the Molecular Level. Genes, 11(8), 935. https://doi.org/10.3390/genes11080935






