Comparative Transcriptome Analysis of Pine Trees Treated with Resistance-Inducing Substances against the Nematode Bursaphelenchus xylophilus
Abstract
1. Introduction
2. Materials and Methods
2.1. PWN Source
2.2. Preparation of Chemical Formulations
2.3. In Vivo Pine Seedling Assay
2.4. RNA Isolation
2.5. RNA Sequencing
2.6. Genome Characterization
2.7. Transcriptome Analysis
2.8. DEG Analysis
2.9. Comparative Transcriptome and Functional Enrichment
2.10. Protein–Protein Interaction Network Analysis
2.11. Quantitative Real-Time PCR (qPCR) Analysis
3. Results
3.1. Inhibitory Effects of the ASM and MeSA Treatments against PWN Infection
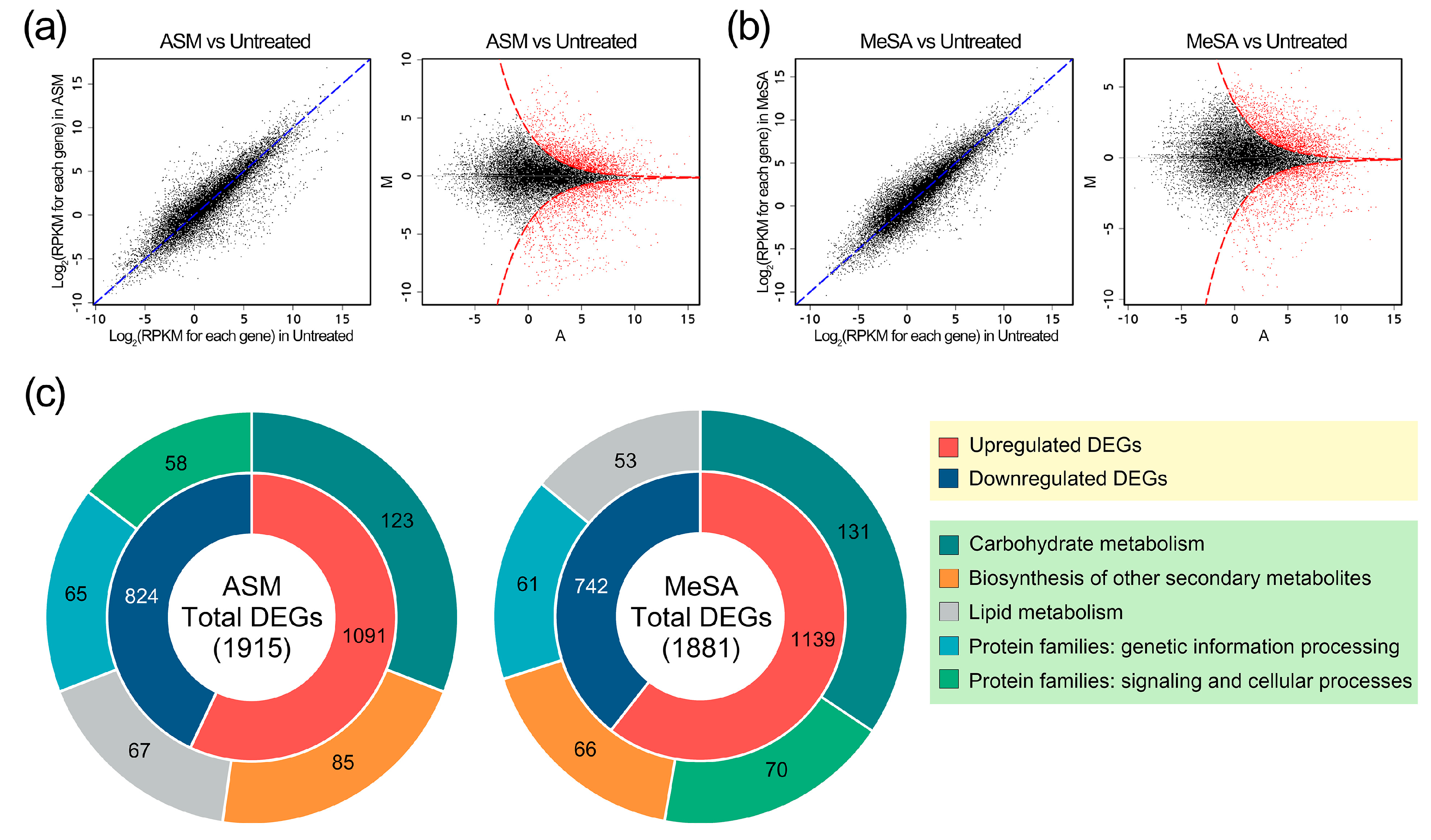
3.2. Overview of the In Vivo Transcriptomes in PWN-Infected Pine Trees
3.3. Genes Induced by PWN Infection under SAR Induction
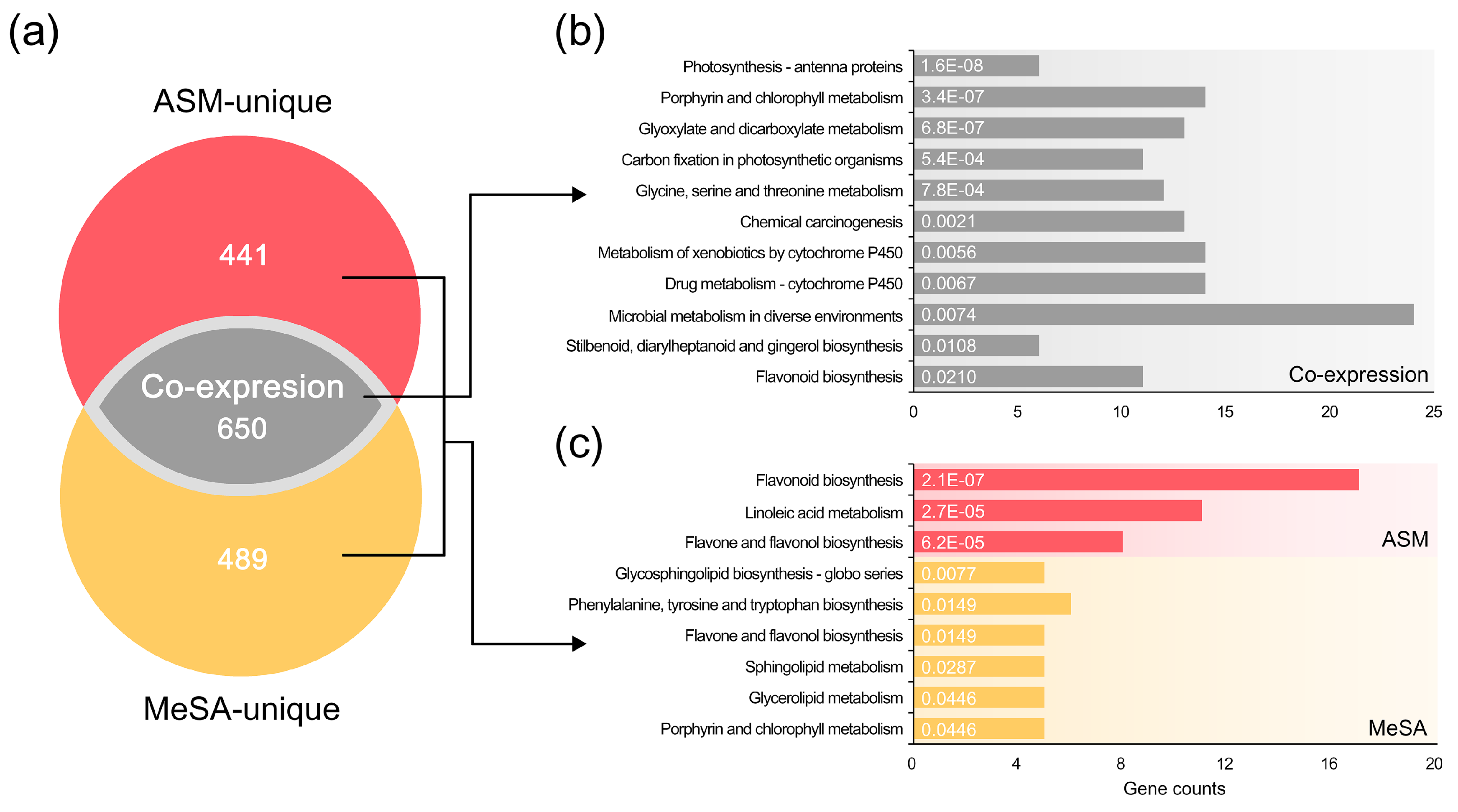
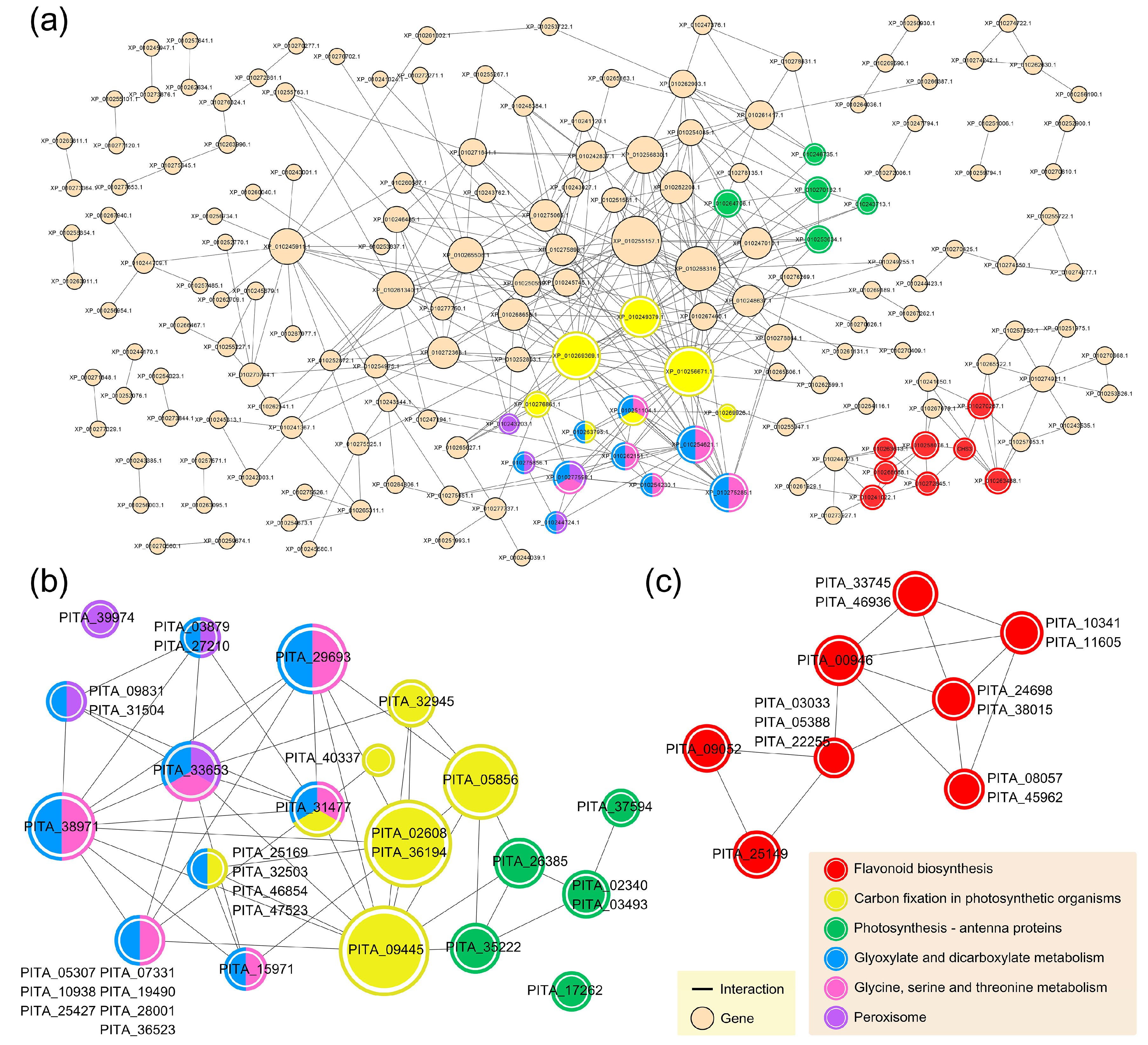
3.4. Common Systems Activated by the RISs for Response to PWN Infection
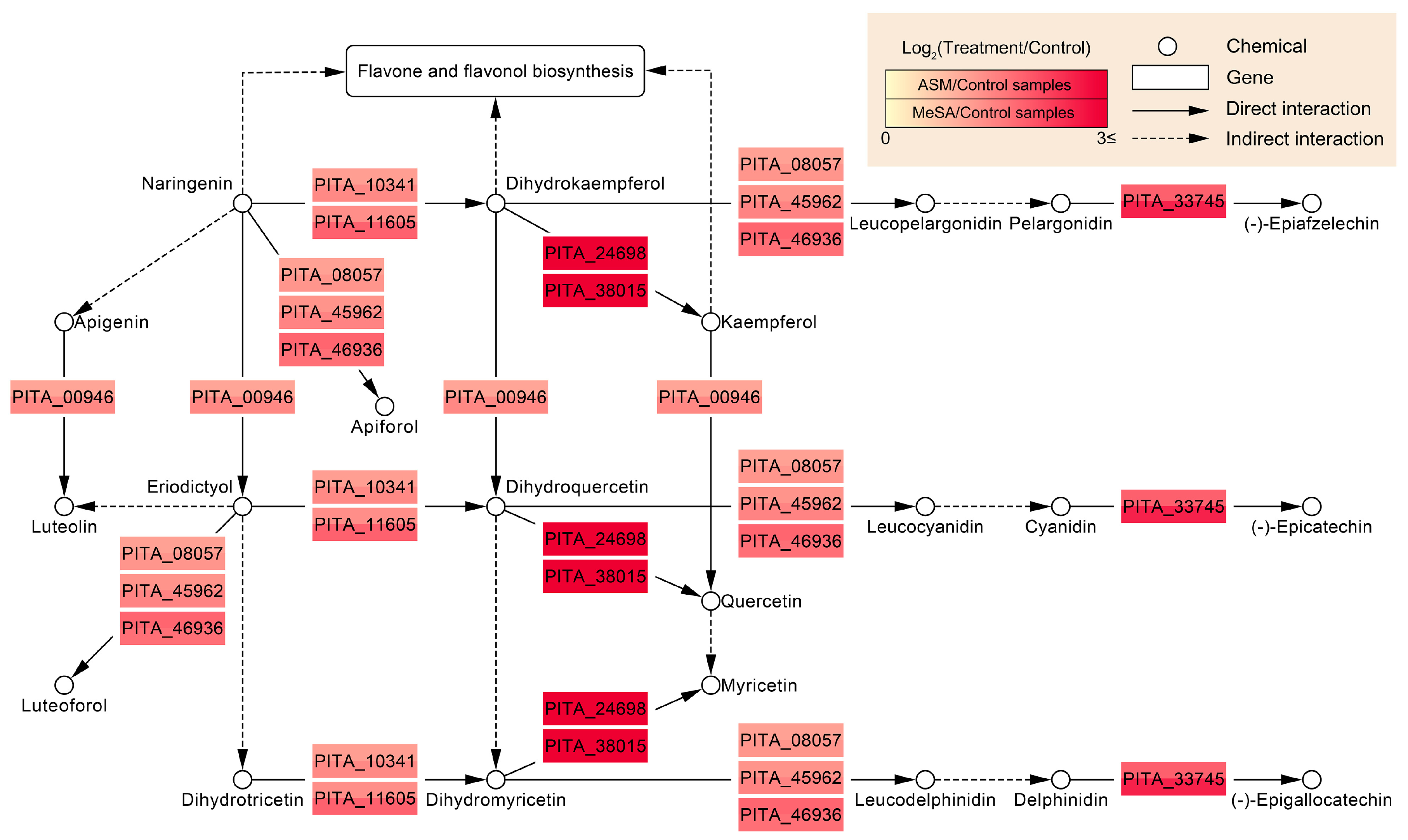
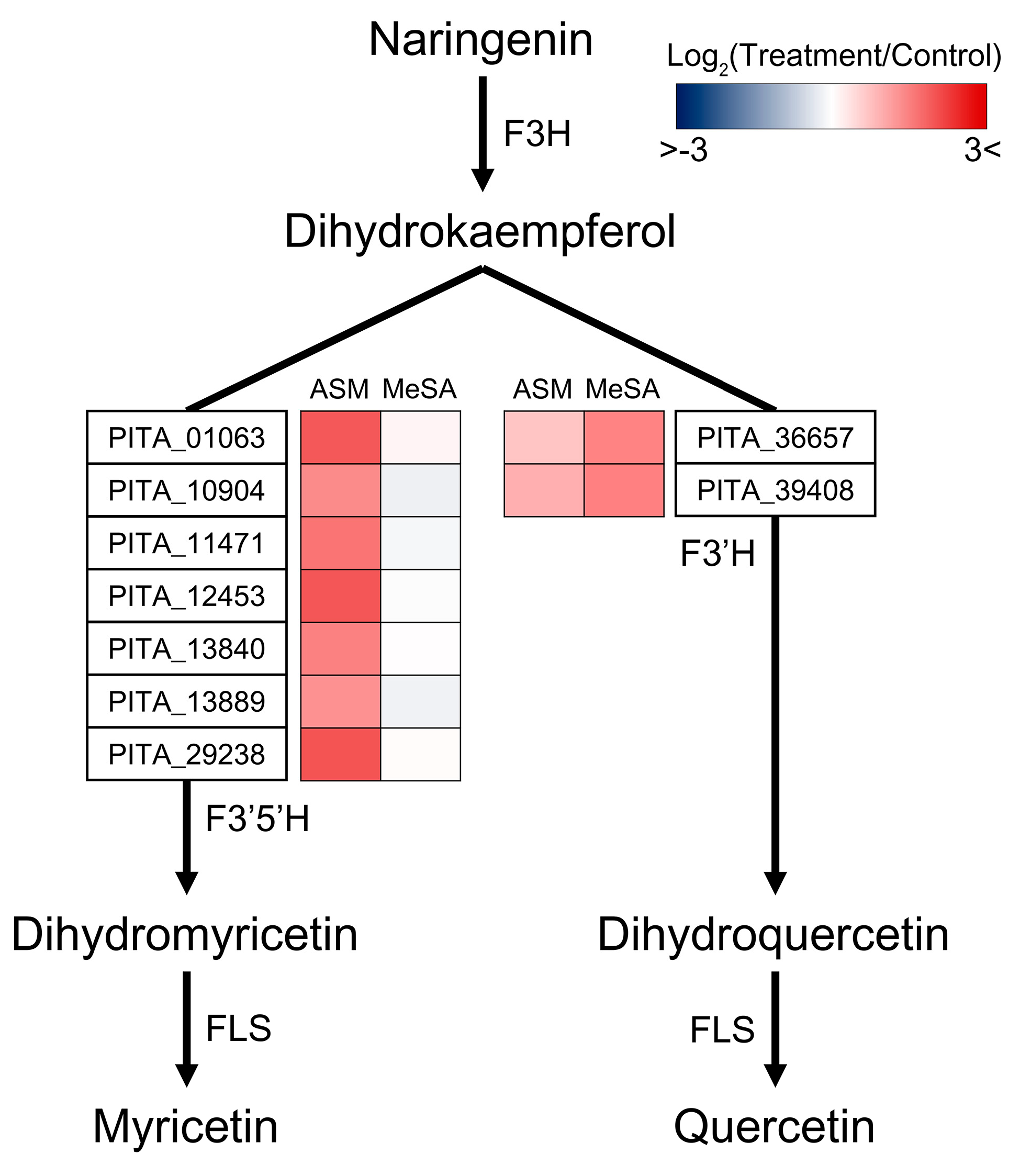
3.5. Functional Systems of ASM- and MeSA-Specific Patterns
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ghildiyal, S.K.; Gairola, S.; Sharma, C.M. Variation in morphological characters of mycorrhizal seedlings of various provenances of Pinus roxburghii Sargent. N. Y. Sci. J. 2010, 3, 1–8. [Google Scholar]
- Semerci, A.; Semerci, H.; Çalişkan, B.; Çiçek, N.; Ekmekçi, Y.; Mencuccini, M. Morphological and physiological responses to drought stress of European provenances of Scots pine. Eur. J. For. Res. 2017, 136, 91–104. [Google Scholar] [CrossRef]
- Montwé, D.; Isaac-Renton, M.; Hamann, A.; Spiecker, H. Cold adaptation recorded in tree rings highlights risks associated with climate change and assisted migration. Nat. Commun. 2018, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cañas, R.A.; Canales, J.; Muñoz-Hernández, C.; Granados, J.M.; Ávila, C.; García-Martín, M.L.; Cánovas, F.M. Understanding developmental and adaptive cues in pine through metabolite profiling and co-expression network analysis. J. Exp. Bot. 2015, 66, 3113–3127. [Google Scholar] [CrossRef] [PubMed]
- Kwak, C.S.; Moon, S.C.; Lee, M.S. Antioxidant, antimutagenic, and antitumor effects of pine needles (Pinus densiflora). Nutr. Cancer 2006, 56, 162–171. [Google Scholar] [CrossRef]
- Kim, Y.B.; Kim, S.M.; Kang, M.K.; Kuzuyama, T.; Lee, J.K.; Park, S.C.; Shin, S.C.; Kim, S.U. Regulation of resin acid synthesis in Pinus densiflora by differential transcription of genes encoding multiple 1-deoxy-d-xylulose 5-phosphate synthase and 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase genes. Tree Physiol. 2009, 29, 737–749. [Google Scholar] [CrossRef]
- Toyohara, G.; Fujihara, M. Succession of secondary forests affected by pine wilt disease in western Japan followed on vegetation maps. Appl. Veg. Sci. 1998, 1, 259–266. [Google Scholar] [CrossRef]
- Kosaka, H.; Aikawa, T.; Ogura, N.; Tabata, K.; Kiyohara, T. Pine wilt disease caused by the pine wood nematode: The induced resistance of pine trees by the avirulent isolates of nematode. Eur. J. Plant Pathol. 2001, 107, 667–675. [Google Scholar] [CrossRef]
- Kiyohara, T.; Tokushige, Y. Inoculation experiments of a nematode, Bursaphelenchus sp., onto pine trees. J. Jpn. For. Soc. 1971, 53, 210–218. [Google Scholar]
- Cheng, H.; Lin, M.; Qian, R. A study on morphological diagnosis and pathogenicity of the pine wood nematode. J. Nanjing Agric. Univ. 1986, 2, 55–59. [Google Scholar]
- Yi, C.K.; Byun, B.H.; Park, J.D.; Yang, S.I.; Chang, K.H. First finding of the pine wood nematode, Bursaphelenchus xylophilus (Steiner et Buhrer) Nickle and its insect vector in Korea. Res. Rep. For. Res. Inst. 1989, 38, 141–149. [Google Scholar]
- Abelleira, A.; Picoaga, A.; Mansilla, J.P.; Aguin, O. Detection of Bursaphelenchus xylophilus, causal agent of pine wilt disease on Pinus pinaster in Northwestern Spain. Plant Dis. 2011, 95, 776. [Google Scholar] [CrossRef] [PubMed]
- Mota, M.M.; Braasch, H.; Bravo, M.A.; Penas, A.C.; Burgermeister, W.; Metge, K.; Sousa, E. First report of Bursaphelenchus xylophilus in Portugal and in Europe. Nematology 1999, 1, 727–734. [Google Scholar] [CrossRef]
- MAMiyA, Y. History of pine wilt disease in Japan. J. Nematol. 1988, 20, 219–226. [Google Scholar] [PubMed]
- Zhao, L.; Mota, M.; Vieira, P.; Butcher, R.A.; Sun, J. Interspecific communication between pinewood nematode, its insect vector, and associated microbes. Trends Parasitol. 2014, 30, 299–308. [Google Scholar] [CrossRef]
- Kulinich, O.; Mota, M.; Vieira, P.; Ryss, A. A synopsis of the genus Bursaphelenchus Fuchs, 1937 (Aphelenchida: Parasitaphelenchidae) with keys to species. Nematology 2005, 7, 393–458. [Google Scholar] [CrossRef]
- Ye, W.; Giblin-Davis, R.M.; Braasch, H.; Morris, K.; Thomas, W.K. Phylogenetic relationships among Bursaphelenchus species (Nematoda: Parasitaphelenchidae) inferred from nuclear ribosomal and mitochondrial DNA sequence data. Mol. Phylogenet. Evol. 2007, 43, 1185–1197. [Google Scholar] [CrossRef]
- Linit, M.J. Nematode-vector relationships in the pine wilt disease system. J. Nematol. 1988, 20, 227–235. [Google Scholar]
- Fukuda, K. Physiological process of the symptom development and resistance mechanism in pine wilt disease. J. For. Res. 1997, 2, 171–181. [Google Scholar] [CrossRef]
- MAMiyA, Y. Initial pathological changes and disease development in pine trees induced by the pine wood nematode, Bursaphelenchus xylophilus. Jpn. J. Phytopathol. 1985, 51, 546–555. [Google Scholar] [CrossRef]
- Ichihara, Y.; Fukuda, K.; Suzuki, K. Early symptom development and histological changes associated with migration of Bursaphelenchus xylophilus in seedling tissues of Pinus thunbergii. Plant Dis. 2000, 84, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Hogetsu, T.; Fukuda, K.; Suzuki, K. Cortical responses in Japanese black pine to attack by the pine wood nematode. Can. J. Bot. 1993, 71, 1399–1405. [Google Scholar] [CrossRef]
- Lee, S.; Chung, Y.; Lee, S.; Lee, D.; Choo, H.; Park, C. Toxic effects of some insecticides on the Japanese pine sawyer, Monochamus alternatus. J. Korean For. Soc. 2003. [Google Scholar]
- Vicente, C.; Espada, M.; Vieira, P.; Mota, M. Pine wilt disease: A threat to European forestry. Eur. J. Plant Pathol. 2012, 133, 89–99. [Google Scholar] [CrossRef]
- Kurinobu, S. Current status of resistance breeding of Japanese pine species to pine wilt disease. For. Sci. Technol. 2008, 4, 51–57. [Google Scholar] [CrossRef]
- Park, I.K.; Kim, J.; Lee, S.G.; Shin, S.C. Nematicidal activity of plant essential oils and components from ajowan (Trachyspermum ammi), allspice (Pimenta dioica) and litsea (Litsea cubeba) essential oils against pine wood nematode (Bursaphelenchus xylophilus). J. Nematol. 2007, 39, 275–279. [Google Scholar]
- Cao, H.; Li, X.; Dong, X. Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA 1998, 95, 6531–6536. [Google Scholar] [CrossRef]
- Romero, A.M.; Ritchie, D.F. Systemic acquired resistance delays race shifts to major resistance genes in bell pepper. Phytopathology 2004, 94, 1376–1382. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef]
- Alexandersson, E.; Mulugeta, T.; Lankinen, Å.; Liljeroth, E.; Andreasson, E. Plant resistance inducers against pathogens in Solanaceae species—From molecular mechanisms to field application. Int. J. Mol. Sci. 2016, 17, 1673. [Google Scholar] [CrossRef]
- Mannaa, M.; Han, G.; Jeon, H.W.; Kim, J.; Kim, N.; Park, A.R.; Kim, J.C.; Seo, Y.S. Influence of resistance-inducing chemical elicitors against pine wilt disease on the rhizosphere microbiome. Microorganisms 2020, 8, 884. [Google Scholar] [CrossRef] [PubMed]
- Vallad, G.E.; Goodman, R.M. Systemic acquired resistance and induced systemic resistance in conventional agriculture. Crop Sci. 2004, 44, 1920–1934. [Google Scholar] [CrossRef]
- Tripathi, D.; Raikhy, G.; Kumar, D. Chemical elicitors of systemic acquired resistance—Salicylic acid and its functional analogs. Curr. Plant Biol. 2019, 17, 48–59. [Google Scholar] [CrossRef]
- Klessig, D.F.; Choi, H.W.; Dempsey, D.A. Systemic acquired resistance and salicylic acid: Past, present, and future. Mol. Plant-Microbe Interact. 2018, 31, 871–888. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.R.; Fatma, M.; Per, T.S.; Anjum, N.A.; Khan, N.A. Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 2015, 6, 462. [Google Scholar] [CrossRef]
- Mandlik, A.; Livny, J.; Robins, W.P.; Ritchie, J.M.; Mekalanos, J.J.; Waldor, M.K. RNA-seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 2011, 10, 165–174. [Google Scholar] [CrossRef]
- Shukla, N.; Yadav, R.; Kaur, P.; Rasmussen, S.; Goel, S.; Agarwal, M.; Jagannath, A.; Gupta, R.; Kumar, A. Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-infected tomato (Solanum lycopersicum) roots reveals complex gene expression profiles and metabolic networks of both host and nematode during susceptible and resistance responses. Mol. Plant Pathol. 2018, 19, 615–633. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhao, D.; Shuang, L.; Xiao, D.; Xuan, Y.; Duan, Y.; Chen, L.; Wang, Y.; Liu, X.; Fan, H.; et al. Transcriptome analysis of rice roots in response to root-knot nematode infection. Int. J. Mol. Sci. 2020, 21, 848. [Google Scholar] [CrossRef]
- Capoen, W.; Den Herder, J.; Rombauts, S.; De Gussem, J.; De Keyser, A.; Holsters, M.; Goormachtig, S. Comparative transcriptome analysis reveals common and specific tags for root hair and crack-entry invasion in Sesbania rostrata. Plant Physiol. 2007, 144, 1878–1889. [Google Scholar] [CrossRef][Green Version]
- Du, H.; Wang, Y.; Yang, J.; Yang, W. Comparative transcriptome analysis of resistant and susceptible tomato lines in response to infection by Xanthomonas perforans race T3. Front. Plant Sci. 2015, 6, 1173. [Google Scholar] [CrossRef]
- Wei, Z.; Zeng, X.; Qin, C.; Wang, Y.; Bai, L.; Xu, Q.; Yuan, H.; Tang, Y.; Nyima, T. Comparative transcriptome analysis revealed genes commonly responsive to varied nitrate stress in leaves of Tibetan hulless barley. Front. Plant Sci. 2016, 7, 1067. [Google Scholar] [CrossRef] [PubMed]
- Myers, R.F. Pathogenesis in pine wilt caused by pinewood nematode, Bursaphelenchus xylophilus. J. Nematol. 1988, 20, 244. [Google Scholar]
- Xue, Q.; Xiang, Y.; Wu, X.Q.; Li, M.J. Bacterial communities and virulence associated with pine wood nematode Bursaphelenchus xylophilus from different Pinus spp. Int. J. Mol. Sci. 2019, 20, 3342. [Google Scholar] [CrossRef] [PubMed]
- Viglierchio, D.R.; Schmitt, R.V. On the methodology of nematode extraction from field samples: Baermann funnel modifications. J. Nematol. 1983, 15, 438–444. [Google Scholar]
- Azevedo, H.; Lino-Neto, T.; Tavares, R.M. An improved method for high-quality RNA isolation from needles of adult maritime pine trees. Plant Mol. Biol. Rep. 2003, 21, 333–338. [Google Scholar] [CrossRef]
- FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 26 February 2019).
- PineRefSeq Database. Available online: https://treegenesdb.org/FTP/Genomes/Pita/ (accessed on 23 April 2019).
- Zimin, A.V.; Stevens, K.A.; Crepeau, M.W.; Puiu, D.; Wegrzyn, J.L.; Yorke, J.A.; Langley, C.H.; Neale, D.B.; Salzberg, S.L. An improved assembly of the loblolly pine mega-genome using long-read single-molecule sequencing. Gigascience 2017, 6, giw016. [Google Scholar]
- FASTX-Toolkit. Available online: http://hannonlab.cshl.edu/fastx_toolkit/ (accessed on 26 February 2019).
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map (SAM) format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.H.; Dudoit, S.; Luu, P.; Lin, D.M.; Peng, V.; Ngai, J.; Speed, T.P. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002, 30, e15. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Qureshi, R.; Sacan, A. Weighted set enrichment of gene expression data. BMC Syst. Biol. 2013, 7, S10. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Jeong, S.T.; Goto-Yamamoto, N.; Hashizume, K.; Esaka, M. Expression of the flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase genes and flavonoid composition in grape (Vitis vinifera). Plant Sci. 2006, 170, 61–69. [Google Scholar] [CrossRef]
- Lim, W.; Li, J. Synergetic effect of the Onion CHI gene on the PAP1 regulatory gene for enhancing the flavonoid profile of tomato skin. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Jeewani, D.C.; Hua, W.Z. Recent advances in anthocyanin biosynthesis in colored wheat. Res. J. Biotechnol. 2017, 12, 6. [Google Scholar]
- Pancaldi, V.; Schubert, F.; Bähler, J. Meta-analysis of genome regulation and expression variability across hundreds of environmental and genetic perturbations in fission yeast. Mol. Biosyst. 2010, 6, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.W.; Wright, A.E.; Mank, J.E. The evolution of gene expression and the transcriptome-phenotype relationship. Semin. Cell Dev. Biol. 2012, 23, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Xia, W.; Cao, P.; Xiao, Z.; Zhang, Y.; Liu, M.; Zhan, C.; Wang, N. Integrated transcriptome analysis reveals plant hormones jasmonic acid and salicylic acid coordinate growth and defense responses upon fungal infection in poplar. Biomolecules 2019, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhi, J.; Zhang, Z.; Wang, L.; Suo, Y.; Xie, C.; Li, M.; Zhang, B.; Du, J.; Gu, L.; et al. Transcriptome analysis of salicylic acid treatment in Rehmannia glutinosa hairy roots using RNA-seq technique for identification of genes involved in acteoside biosynthesis. Front. Plant Sci. 2017, 8, 787. [Google Scholar] [CrossRef]
- Gaspar, D.; Trindade, C.; Usié, A.; Meireles, B.; Fortes, A.M.; Guimarães, J.B.; Simões, F.; Costa, R.L.; Ramos, A.M. Comparative transcriptomic response of two Pinus species to infection with the pine wood nematode Bursaphelenchus xylophilus. Forests 2020, 11, 204. [Google Scholar] [CrossRef]
- Bektas, Y.; Eulgem, T. Synthetic plant defense elicitors. Front. Plant Sci. 2015, 5, 1–9. [Google Scholar] [CrossRef]
- Kunz, W.; Schurter, R.; Maetzke, T. The chemistry of benzothiadiazole plant activators. Pestic. Sci. 1997, 50, 275–282. [Google Scholar] [CrossRef]
- Park, S.W.; Kaimoyo, E.; Kumar, D.; Mosher, S.; Klessig, D.F. Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science (80-) 2007, 318, 113–116. [Google Scholar] [CrossRef]
- Shulaev, V.; Silverman, P.; Raskin, I. Airborne signalling by methyl salicylate in plant pathogen resistance. Nature 1997, 385, 718–721. [Google Scholar] [CrossRef]
- Kunwar, S.; Paret, M.L.; Freeman, J.H.; Ritchie, L.; Olson, S.M.; Colee, J.; Jones, J.B. Foliar applications of acibenzolar-S-methyl negatively affect the yield of grafted tomatoes in fields infested with Ralstonia solanacearum. Plant Dis. 2017, 101, 890–894. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.M.; Zhu, S.; Kachroo, P.; Kachroo, A. Signal regulators of systemic acquired resistance. Front. Plant Sci. 2015, 6, 228. [Google Scholar] [CrossRef]
- Gerstein, M.B.; Rozowsky, J.; Yan, K.K.; Wang, D.; Cheng, C.; Brown, J.B.; Davis, C.A.; Hillier, L.; Sisu, C.; Li, J.J.; et al. Comparative analysis of the transcriptome across distant species. Nature 2014, 512, 445–448. [Google Scholar] [CrossRef]
- Chan, E.T.; Quon, G.T.; Chua, G.; Babak, T.; Trochesset, M.; Zirngibl, R.A.; Aubin, J.; Ratcliffe, M.J.; Wilde, A.; Brudno, M.; et al. Conservation of core gene expression in vertebrate tissues. J. Biol. 2009, 8, 33. [Google Scholar] [CrossRef] [PubMed]
- Bernsdorff, F.; Döring, A.C.; Gruner, K.; Schuck, S.; Bräutigam, A.; Zeier, J. Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and -independent pathways. Plant Cell 2016, 28, 102–129. [Google Scholar] [CrossRef] [PubMed]
- Traw, M.B.; Kniskern, J.M.; Bergelson, J. SAR increases fitness of Arabidopsis thaliana in the presence of natural bacterial pathogens. Evol. Int. J. Org. Evol. 2007, 61, 2444–2449. [Google Scholar] [CrossRef]
- Rivas-San Vicente, M.; Plasencia, J. Salicylic acid beyond defence: Its role in plant growth and development. J. Exp. Bot. 2011, 62, 3321–3338. [Google Scholar] [CrossRef]
- Fukuda, K.; Utsuzawa, S.; Sakaue, D. Correlation between acoustic emission, water status and xylem embolism in pine wilt disease. Tree Physiol. 2007, 27, 969–976. [Google Scholar] [CrossRef]
- Melakeberhan, H.; Toivonen, P.; Vidaver, W.; Webster, J.; Dube, S. Effect of Bursaphelenchus xylophilus on the water potential and water-splitting complex of photosystem II of Pinus sylvestris seedlings. Physiol. Mol. Plant Pathol. 1991, 38, 83–91. [Google Scholar] [CrossRef]
- Melakeberhan, H.; Brooke, R.; Webster, J.; D’Auria, J. The influence of Meloidogyne incognita on the growth, physiology and nutrient content of Phaseolus vulgaris. Physiol. Plant Pathol. 1985, 26, 259–268. [Google Scholar] [CrossRef]
- Wingler, A.; Lea, P.J.; Quick, W.P.; Leegood, R.C. Photorespiration: Metabolic pathways and their role in stress protection. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2000, 355, 1517–1529. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Bloom, A.J.; Queval, G.; Noctor, G. Photorespiratory metabolism: Genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 2009, 60, 455–484. [Google Scholar] [CrossRef] [PubMed]
- Loggini, B.; Scartazza, A.; Brugnoli, E.; Navari-Izzo, F. Antioxidative defense system, pigment composition, and photosynthetic efficiency in two wheat cultivars subjected to drought. Plant Physiol. 1999, 119, 1100. [Google Scholar] [CrossRef] [PubMed]
- Cruz De Carvalho, M.H. Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant Signal. Behav. 2008, 3, 156–165. [Google Scholar] [CrossRef]
- Labudda, M.; Różańska, E.; Czarnocka, W.; Sobczak, M.; Dzik, J.M. Systemic changes in photosynthesis and reactive oxygen species homeostasis in shoots of Arabidopsis thaliana infected with the beet cyst nematode Heterodera schachtii. Mol. Plant Pathol. 2018, 19, 1690–1704. [Google Scholar] [CrossRef]
- Sicher, R.C.; Barnaby, J.Y. Impact of carbon dioxide enrichment on the responses of maize leaf transcripts and metabolites to water stress. Physiol. Plant. 2012, 144, 238–253. [Google Scholar] [CrossRef]
- Noctor, G.; Arisi, A.C.M.; Jouanin, L.; Foyer, C.H. Photorespiratory glycine enhances glutathione accumulation in both the chloroplastic and cytosolic compartments. J. Exp. Bot. 1999, 50, 1157–1167. [Google Scholar] [CrossRef]
- Koda, Y.; Takahashi, K.; Kikuta, Y.; Greulich, F.; Toshima, H.; Ichihara, A. Similarities of the biological activities of coronatine and coronafacic acid to those of jasmonic acid. Phytochemistry 1996, 41, 93–96. [Google Scholar] [CrossRef]
- Maekawa, T.; Kufer, T.A.; Schulze-Lefert, P. NLR functions in plant and animal immune systems: So far and yet so close. Nat. Immunol. 2011, 12, 817. [Google Scholar] [CrossRef]
- Navarro, L.; Zipfel, C.; Rowland, O.; Keller, I.; Robatzek, S.; Boller, T.; Jones, J.D.G. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 2004, 135, 1113–1128. [Google Scholar] [CrossRef]
- Tao, Y.; Xie, Z.; Chen, W.; Glazebrook, J.; Chang, H.S.; Han, B.; Zhu, T.; Zou, G.; Katagiri, F. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 2003, 15, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Preciado-Rangel, P.; Reyes-Pérez, J.; Ramírez-Rodríguez, S.; Salas-Pérez, L.; Fortis-Hernández, M.; Murillo-Amador, B.; Troyo-Diéguez, E. Foliar aspersion of salicylic acid improves phenolic and flavonoid compounds, and also the fruit yield in cucumber (Cucumis sativus L.). Plants 2019, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, L.P.; Zhang, L.; Yan, P.; Ahammed, G.; Han, W.Y. Methyl salicylate enhances flavonoid biosynthesis in tea leaves by stimulating the phenylpropanoid pathway. Molecules 2019, 24, 362. [Google Scholar] [CrossRef] [PubMed]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Chin, S.; Behm, C.A.; Mathesius, U. Functions of flavonoids in plant–nematode interactions. Plants 2018, 7, 85. [Google Scholar] [CrossRef]
- Wuyts, N.; Swennen, R.; De Waele, D. Effects of plant phenylpropanoid pathway products and selected terpenoids and alkaloids on the behaviour of the plant-parasitic nematodes Radopholus similis, Pratylenchus penetrans and Meloidogyne incognita. Nematology 2006, 8, 89–101. [Google Scholar] [CrossRef]
- Faizi, S.; Fayyaz, S.; Bano, S.; Yawar Iqbal, E.; Lubna, L.; Siddiqi, H.; Naz, A. Isolation of nematicidal compounds from Tagetes patula L. yellow flowers: Structure-activity relationship studies against cyst nematode Heterodera zeae infective stage larvae. J. Agric. Food Chem. 2011, 59, 9080–9093. [Google Scholar] [CrossRef]
- Bano, S.; Iqbal, E.Y.; Lubna; Zik-ur-Rehman, S.; Fayyaz, S.; Faizi, S. Nematicidal activity of flavonoids with structure activity relationship (SAR) studies against root knot nematode Meloidogyne incognita. Eur. J. Plant Pathol. 2020, 157, 299–309. [Google Scholar] [CrossRef]
- Grundler, F.; Betka, M.; Wyss, U. Influence of changes in the nurse cell system (syncytium) on sex determination and development of the cyst nematode Heterodera schachtii: Total amounts of proteins and amino acids. Phytopathology 1991, 81, 70–74. [Google Scholar] [CrossRef]
- Jones, J.T.; Furlanetto, C.; Phillips, M.S. The role of flavonoids produced in response to cyst nematode infection of Arabidopsis thaliana. Nematology 2007, 9, 671–677. [Google Scholar] [CrossRef]
- Uzal, E.N.; Gómez-Ros, L.V.; Hernández, J.A.; Pedreño, M.A.; Cuello, J.; Ros Barceló, A. Analysis of the soluble cell wall proteome of gymnosperms. J. Plant Physiol. 2009, 166, 831–843. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, K.A.; Othman, A.; Meon, S.; Abdullah, F.; Ho, C.L. Sequence analysis and gene expression of putative exo- and endo-glucanases from oil palm (Elaeis guineensis) during fungal infection. J. Plant Physiol. 2012, 169, 1565–1570. [Google Scholar] [CrossRef] [PubMed]
- Aryal, S.K.; Davis, R.F.; Stevenson, K.L.; Timper, P.; Ji, P. Influence of infection of cotton by Rotylenchulus reniformis and Meloidogyne incognita on the production of enzymes involved in systemic acquired resistance. J. Nematol. 2011, 43, 152–159. [Google Scholar] [PubMed]

| Sample | Read Type | GC (%) | Quality 1 | Total Reads | Total Length (bp) |
|---|---|---|---|---|---|
| ASM_1 | Paired end | 47 | 40 | 27,814,772 | 2,805,933,290 |
| ASM_2 | Paired end | 45 | 40 | 35,228,444 | 3,554,276,515 |
| ASM_3 | Paired end | 46 | 40 | 33,425,670 | 3,372,510,853 |
| MeSA_1 | Paired end | 45 | 40 | 31,973,654 | 3,226,375,119 |
| MeSA_2 | Paired end | 45 | 40 | 33,377,584 | 3,367,938,501 |
| MeSA_3 | Paired end | 46 | 40 | 33,589,898 | 3,389,002,001 |
| Untreated_1 | Paired end | 45 | 40 | 31,763,374 | 3,205,080,196 |
| Untreated_2 | Paired end | 46 | 40 | 33,120,212 | 3,341,300,502 |
| Untreated_3 | Paired end | 45 | 40 | 34,069,162 | 3,437,840,259 |
| GO ID | Terms | Count | FDR 1 |
|---|---|---|---|
| GO:0048585 | Negative regulation of response to stimulus | 34 | 0 |
| GO:0006855 | Drug transmembrane transport | 26 | 0 |
| GO:0009817 | Defence response to fungus, incompatible interaction | 26 | 0 |
| GO:0042344 | Indole glucosinolate catabolic process | 26 | 0 |
| GO:0052544 | Defence response by callose deposition in cell wall | 26 | 0 |
| GO:0070574 | Cadmium ion transmembrane transport | 26 | 0 |
| GO:0071366 | Cellular response to indolebutyric acid stimulus | 26 | 0 |
| GO:1901140 | p-Coumaryl alcohol transport | 17 | 0 |
| GO:1901141 | Regulation of lignin biosynthetic process | 17 | 0 |
| GO:0009737 | Response to abscisic acid | 44 | 6.44 × 10−11 |
| GO ID | Terms | Count | FDR 1 |
|---|---|---|---|
| GO:0002215 | Defence response to nematode | 9 | 1.27 × 10−7 |
| GO:0009624 | Response to nematode | 14 | 1.16× 10−5 |
| GO:0006833 | Water transport | 10 | 2.37 × 10−5 |
| GO:0009094 | l-Phenylalanine biosynthetic process | 5 | 2.37 × 10−5 |
| GO:0003333 | Amino acid transmembrane transport | 7 | 0.0056 |
| GO:0009817 | Defence response to fungus, incompatible interaction | 8 | 0.0096 |
| GO:0015804 | Neutral amino acid transport | 5 | 0.0096 |
| GO:0051704 | Multi-organism process | 40 | 0.0407 |
| GO:0097305 | Response to alcohol | 25 | 0.0443 |
| GO:0009409 | Response to cold | 14 | 0.0443 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.; Jeon, H.W.; Jung, H.; Lee, H.-H.; Kim, J.; Park, A.R.; Kim, N.; Han, G.; Kim, J.-C.; Seo, Y.-S. Comparative Transcriptome Analysis of Pine Trees Treated with Resistance-Inducing Substances against the Nematode Bursaphelenchus xylophilus. Genes 2020, 11, 1000. https://doi.org/10.3390/genes11091000
Park J, Jeon HW, Jung H, Lee H-H, Kim J, Park AR, Kim N, Han G, Kim J-C, Seo Y-S. Comparative Transcriptome Analysis of Pine Trees Treated with Resistance-Inducing Substances against the Nematode Bursaphelenchus xylophilus. Genes. 2020; 11(9):1000. https://doi.org/10.3390/genes11091000
Chicago/Turabian StylePark, Jungwook, Hee Won Jeon, Hyejung Jung, Hyun-Hee Lee, Junheon Kim, Ae Ran Park, Namgyu Kim, Gil Han, Jin-Cheol Kim, and Young-Su Seo. 2020. "Comparative Transcriptome Analysis of Pine Trees Treated with Resistance-Inducing Substances against the Nematode Bursaphelenchus xylophilus" Genes 11, no. 9: 1000. https://doi.org/10.3390/genes11091000
APA StylePark, J., Jeon, H. W., Jung, H., Lee, H.-H., Kim, J., Park, A. R., Kim, N., Han, G., Kim, J.-C., & Seo, Y.-S. (2020). Comparative Transcriptome Analysis of Pine Trees Treated with Resistance-Inducing Substances against the Nematode Bursaphelenchus xylophilus. Genes, 11(9), 1000. https://doi.org/10.3390/genes11091000







