PreLnc: An Accurate Tool for Predicting lncRNAs Based on Multiple Features
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
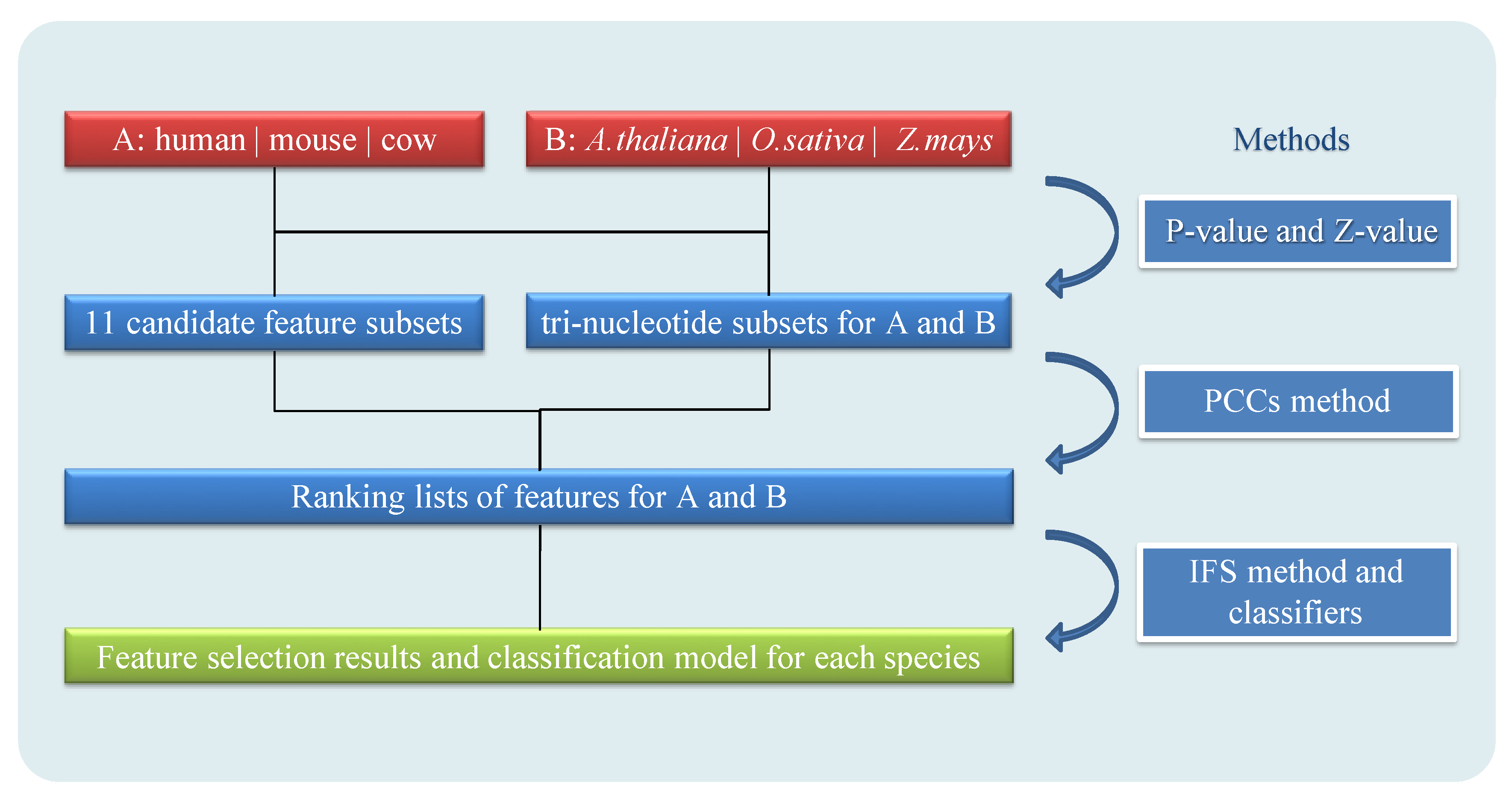
2.2. Feature Selection and Extraction
2.3. Model Foundation and Evaluation
3. Results
3.1. Analysis of Feature Selection and Model Foundation
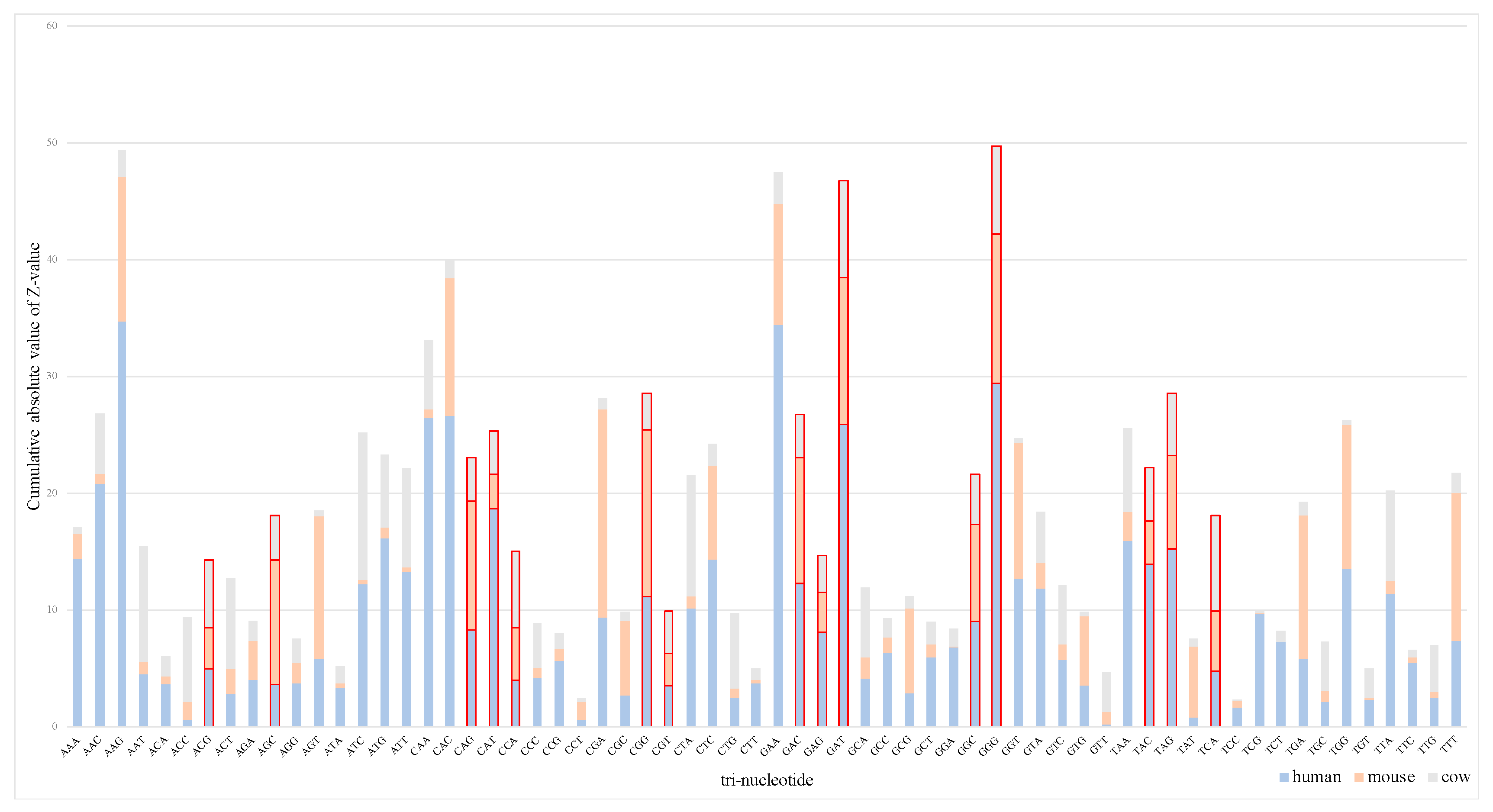
3.1.1. Analysis of Tri-Nucleotide Differences Among Species
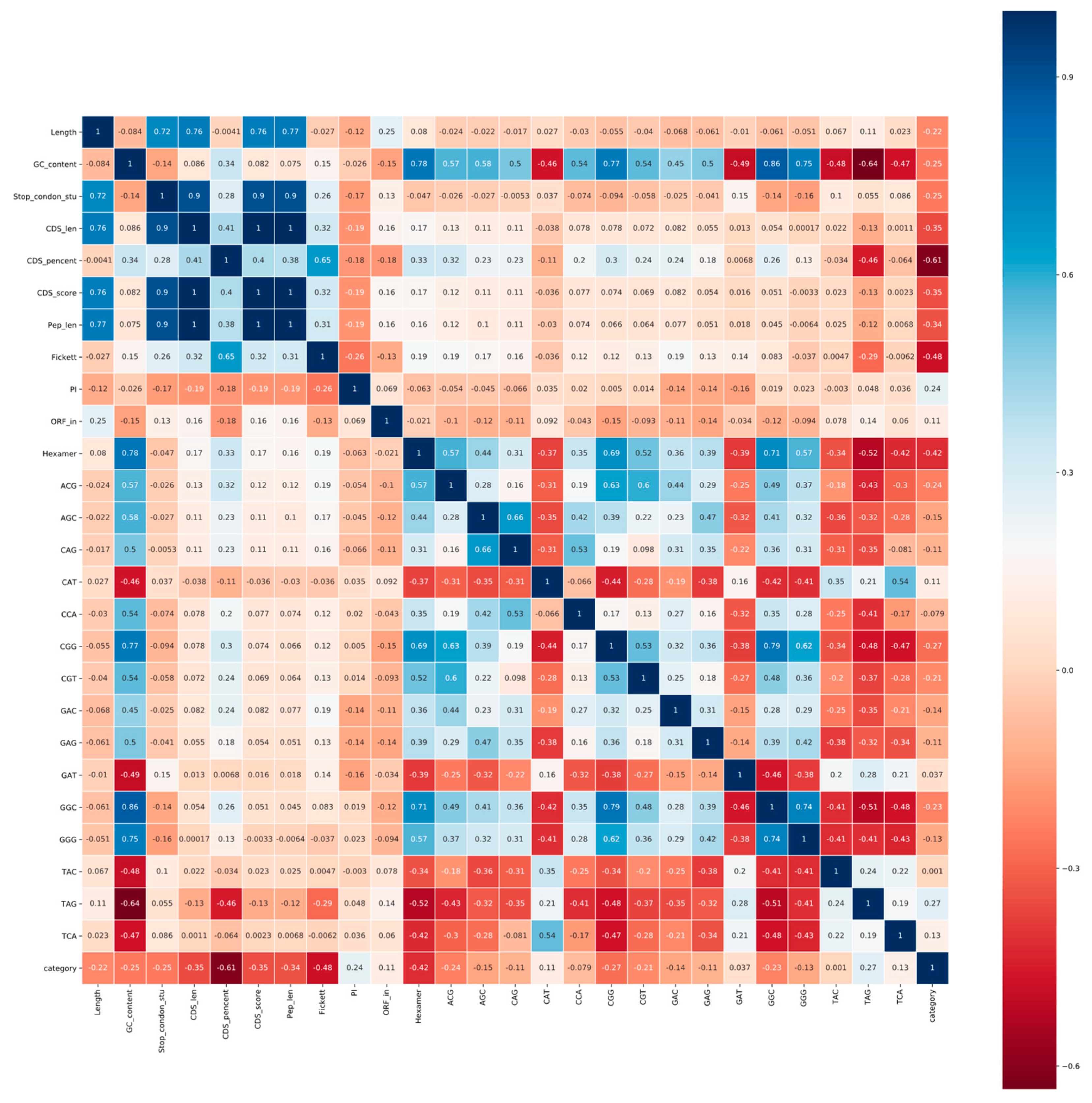
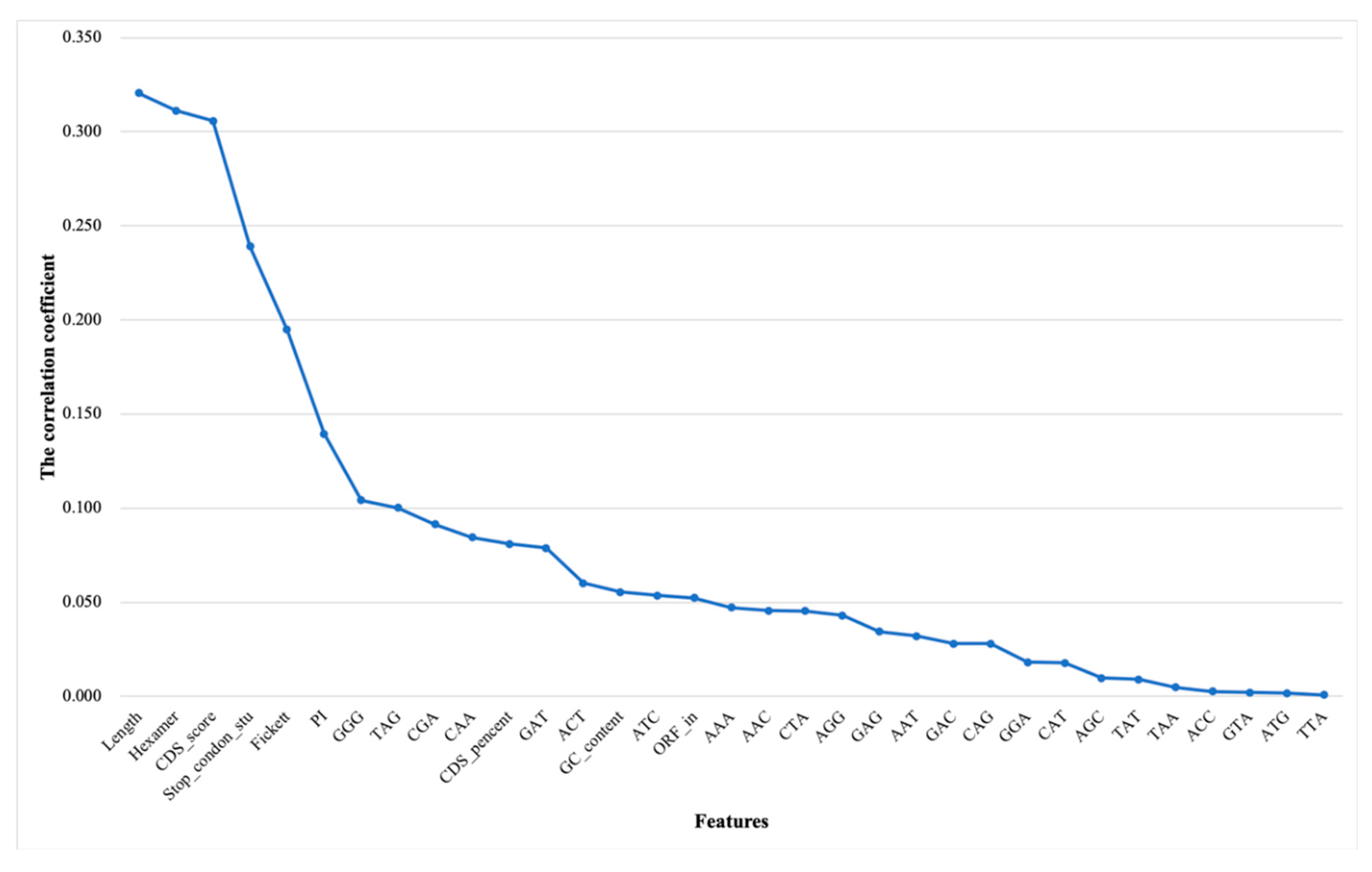
3.1.2. Correlation Analysis and Ranking Lists of Features
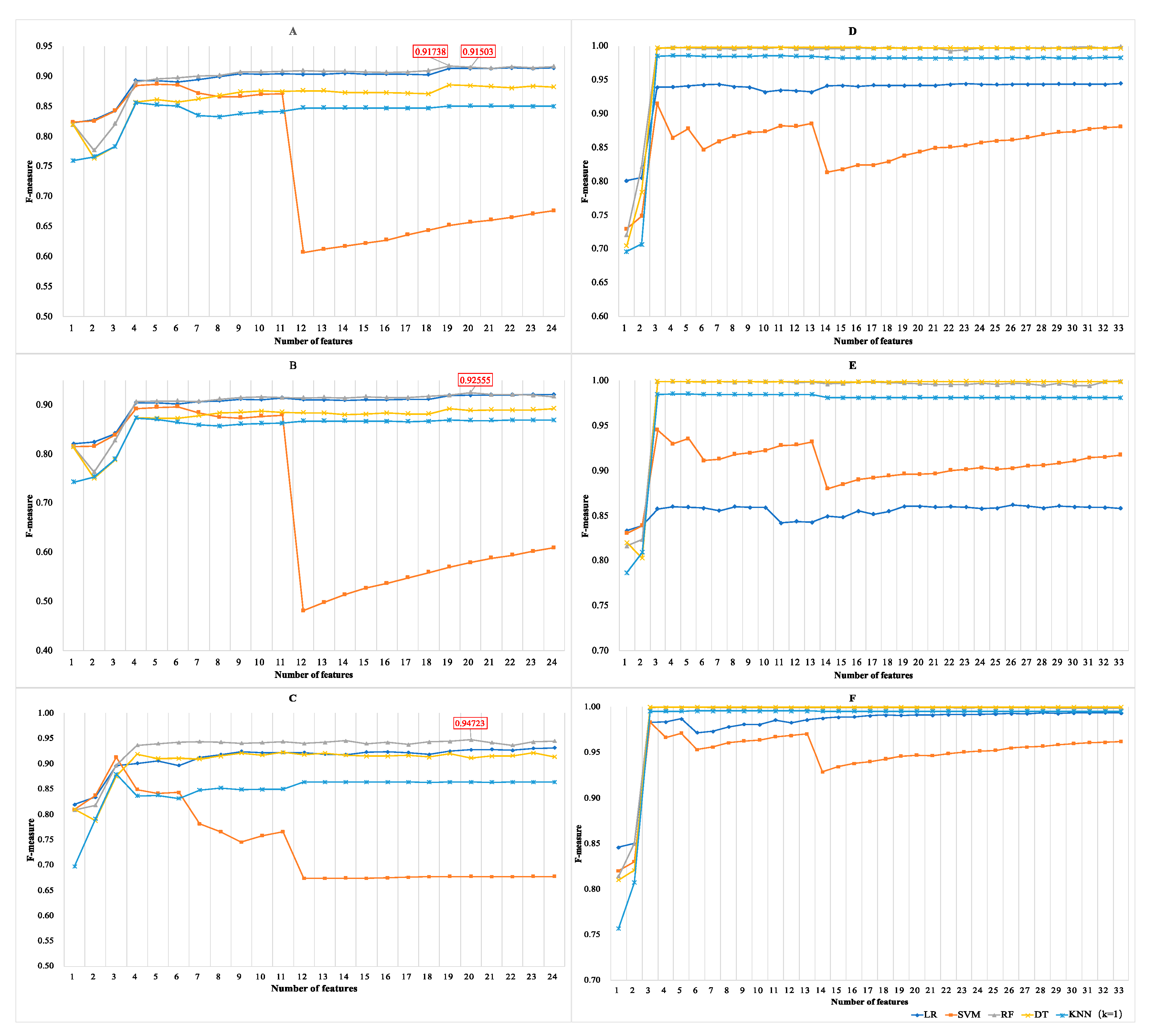
3.1.3. Results of Incremental Feature Selection Method with Multiple Classifiers
3.2. Comparison and Analysis of Prediction Results
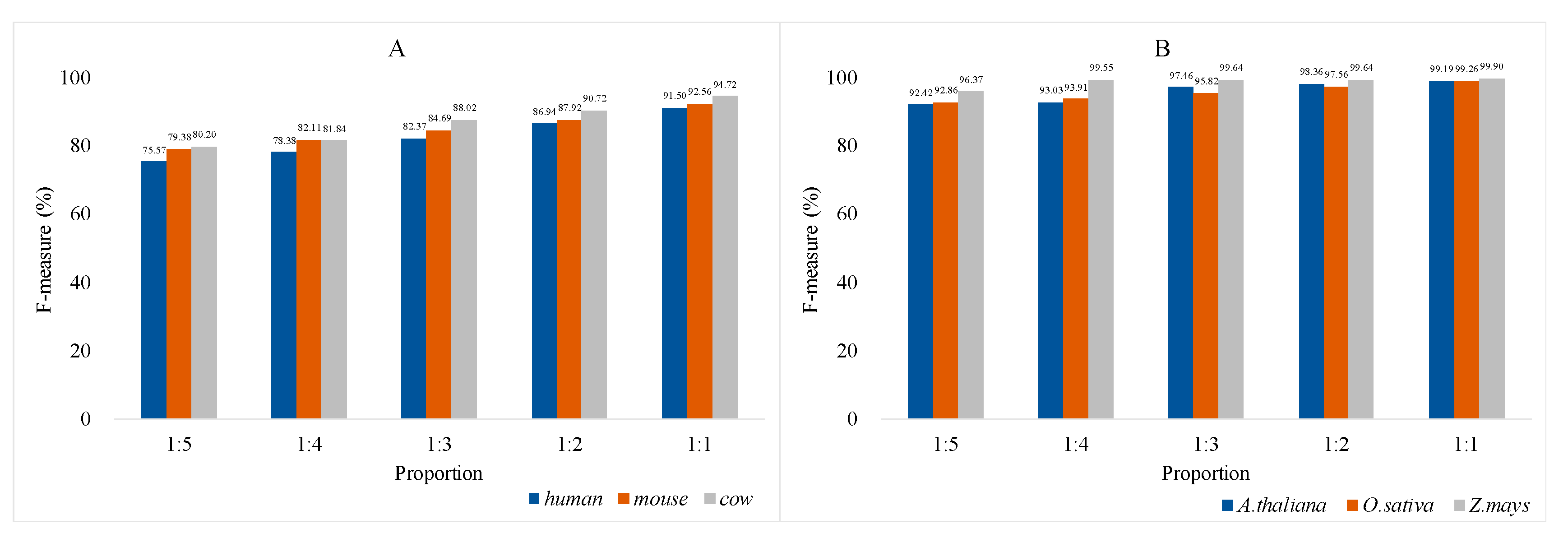
3.3. Prediction on Other Known Datasets
3.4. Analysis of Model Universality and Generalization Ability
3.5. Comparison of Time Consumption
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long Noncoding RNAs: Past, Present, and Future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T. Epigenetic regulation by long noncoding RNAs. Science 2013, 21, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Kraus, W.L. From discovery to function: The expanding roles of long noncoding RNAs in physiology and disease. Endocr. Rev. 2015, 36, 25–64. [Google Scholar] [CrossRef] [PubMed]
- Carl, E.; Morton, C.C. Identification and function of long non-coding RNA. Front. Cell. Neurosci. 2013, 7, 168. [Google Scholar]
- Licatalosi, D.D.; Darnell, R.B. RNA processing and its regulation: Global insights into biological networks. Nat. Rev. Genet. 2009, 11, 75. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Yao, R.; Wang, Y.; Chen, L. Cellular functions of long noncoding RNAs. Nat. Cell. Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef]
- Dinger, M.E.; Amaral, P.P.; Mercer, T.R.; Mattick, J.S. Pervasive transcription of the eukaryotic genome: Functional indices and conceptual implications. Brief. Funct. Genom. 2009, 8, 407–423. [Google Scholar] [CrossRef]
- Song, X.; Sun, L.; Luo, H.; Ma, Q.; Zhao, Y.; Pei, D. Genome-Wide Identification and Characterization of Long Non-Coding RNAs from Mulberry (Morus notabilis) RNA-seq Data. Genes 2016, 7, 11. [Google Scholar] [CrossRef]
- Milligan, M.J.; Lipovich, L. Pseudogene-derived lncRNAs: Emerging regulators of gene expression. Front. Genet. 2014, 5, 476. [Google Scholar] [CrossRef]
- Alcid, E.A.; Tsukiyama, T. Systematic approaches to identify functional lncRNAs. Curr. Opin. Genet. Dev. 2016, 37, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, P.; Lipovich, L.; Grandér, D.; Morris, K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 2014, 1840, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Liu, X.; Liu, L.; Deng, H.; Zhang, J.; Xu, Q.; Cen, B.; Ji, A. Regulation of lncRNA expression. Cell. Mol. Biol. Lett. 2014, 19, 561–575. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, 74. [Google Scholar] [CrossRef] [PubMed]
- Aimin, L.; Junying, Z.; Zhongyin, Z. PLEK: A tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinform. 2014, 15, 311. [Google Scholar]
- Kang, Y.; Yang, D.; Kong, L.; Hou, M.; Meng, Y.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, 12–16. [Google Scholar] [CrossRef]
- Sun, L.; Liu, H.; Zhang, L.; Meng, J. lncRScan-SVM: A Tool for Predicting Long Non-Coding RNAs Using Support Vector Machine. PLoS ONE 2015, 10, 0139654. [Google Scholar] [CrossRef]
- Han, S.; Liang, Y.; Ma, Q.; Xu, Y.; Zhang, Y.; Du, W.; Wang, C.; Li, Y. LncFinder: An integrated platform for long non-coding RNA identification utilizing sequence intrinsic composition, structural information and physicochemical property. Brief. Bioinform. 2019, 20, 2009–2027. [Google Scholar] [CrossRef]
- Singh, U.; Khemka, N.; Rajkumar, M.S.; Garg, R.; Jain, M. PLncPRO for prediction of long non-coding RNAs (lncRNAs) in plants and its application for discovery of abiotic stress-responsive lncRNAs in rice and chickpea. Nucleic Acids Res. 2017, 45, 183. [Google Scholar] [CrossRef]
- Costa, N.T.D.; Luz, A.W.A.; Henrique, B.P.; Maeda, P.T.S.; Silva, D.D.; Rossi, A.P. Pattern recognition analysis on long noncoding RNAs: A tool for prediction in plants. Brief. Bioinform. 2019, 20, 682–689. [Google Scholar]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Girón, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2017, 46, 754–761. [Google Scholar] [CrossRef] [PubMed]
- Paytuví Gallart, A.; Hermoso Pulido, A.; Anzar Martínez De Lagrán, I.; Sanseverino, W.; Aiese Cigliano, R. GREENC: A Wiki-based database of plant lncRNAs. Nucleic Acids Res. 2015, 44, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Bolser, D.M.; Staines, D.M.; Perry, E.; Kersey, P.J. Ensembl Plants: Integrating Tools for Visualizing, Mining, and Analyzing Plant Genomic Data. In Plant Genom. Databases: Methods Protocols; Van Dijk, A.D.J., Ed.; Springer: New York, NY, USA, 2017; pp. 1–31. [Google Scholar]
- Weizhong, L.; Lukasz, J.; Adam, G. Tolerating some redundancy significantly speeds up clustering, of large protein databases. Bioinformatics 2002, 18, 77–82. [Google Scholar]
- Sun, K.; Chen, X.; Jiang, P.; Song, X.; Wang, H.; Sun, H. iSeeRNA: Identification of long intergenic non-coding RNA transcripts from transcriptome sequencing data. BMC Genom. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef] [PubMed]
- Fickett, J.W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982, 10, 5303–5318. [Google Scholar] [CrossRef]
- Yáñez, J.; Argüello, M.; Osuna, J.; Soberón, X.; Gaytán, P. Combinatorial codon-based amino acid substitutions. Nucleic Acids Res. 2004, 32, 158. [Google Scholar] [CrossRef]
- Panwar, B.; Arora, A.; Raghava, G.P. Prediction and classification of ncRNAs using structural information. BMC Genom. 2014, 15, 1–13. [Google Scholar] [CrossRef]
- Meng, J.; Chang, Z.; Zhang, P.; Shi, W.; Luan, Y. lncRNA-LSTM: Prediction of Plant Long Non-coding RNAs Using Long Short-Term Memory Based on p-nts Encoding. In Intelligent Computing Methodologies; Huang, D.S., Huang, Z.K., Hussain, A., Eds.; Springer: Cham, Switzerland, 2019; Volume 11645, pp. 347–357. [Google Scholar]
- Hertz, G.Z.; Stormo, G.D. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 1999, 15, 563–577. [Google Scholar] [CrossRef]
- Storey, J.D. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann. Stat. 2003, 31, 2013–2035. [Google Scholar] [CrossRef]
- Kirk, J.M.; Kim, S.O.; Inoue, K.; Smola, M.J.; Lee, D.M.; Schertzer, M.D.; Wooten, J.S.; Baker, A.R.; Sprague, D.; Collins, D.W.; et al. Functional classification of long non-coding RNAs by k-mer content. Nat. Genet. 2018, 50, 1474–1482. [Google Scholar] [CrossRef] [PubMed]
- Bastien, O.; Aude, J.; Roy, S.; Maréchal, E. Fundamentals of massive automatic pairwise alignments of protein sequences: Theoretical significance of Z-value statistics. Bioinformatics 2004, 20, 534–537. [Google Scholar] [CrossRef] [PubMed]
- Artusi, R.; Verderio, P.; Marubini, E. Bravais-Pearson and Spearman correlation coefficients: Meaning, test of hypothesis and confidence interval. Int. J. Biol. Markers 2002, 17, 148. [Google Scholar] [CrossRef]
- Liu, H.; Setiono, R. Incremental Feature Selection. Appl. Intell. 1998, 9, 217–230. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.H.; Pan, X.; Liu, M.; Wang, S.; Huang, T.; Cai, Y.D. Tissue Expression Difference between mRNAs and lncRNAs. Int. J. Mol. Sci. 2018, 19, 3416. [Google Scholar] [CrossRef]
- Le Cessie, S.; Van Houwelingen, J.C. Ridge estimators in logistic regression. J. R. Stat. Soc. Ser. C (Appl. Stat.) 1992, 41, 191–201. [Google Scholar] [CrossRef]
- Mavroforakis, M.; Theodoridis, S. A geometric approach to Support Vector Machine (SVM) classification. Ieee Trans. Neural Netw. 2006, 17, 671–682. [Google Scholar] [CrossRef]
- Galligan, D.T.; Ramberg, C.; Curtis, C.; Ferguson, J.R.; Fetrow, J. Application of portfolio theory in decision tree analysis. J. Dairy Sci. 1991, 747, 2138–2144. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Peterson, L.E. K-nearest neighbor. Scholarpedia 2009, 4, 1883. [Google Scholar] [CrossRef]
- Khoshgoftaar, T.M.; Golawala, M.; Hulse, J.V. An Empirical Study of Learning from Imbalanced Data Using Random Forest. In Proceedings of the IEEE International Conference on Tools with Artificial Intelligence, Patras, Greece, 29–31 October 2007. [Google Scholar]
- Anaissi, A.; Kennedy, P.J.; Goyal, M.; Catchpoole, D. A balanced iterative random forest for gene selection from microarray data. BMC Bioinform. 2013, 14, 261. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2017, 46, D308–D314. [Google Scholar] [CrossRef] [PubMed]
- Azlan, A.; Obeidat, S.M.; Yunus, M.A.; Azzam, G. Systematic identification and characterization of Aedes aegypti long noncoding RNAs (lncRNAs). Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Diamond, J. Evolution, consequences and future of plant and animal domestication. Nature 2002, 418, 700–707. [Google Scholar] [CrossRef]
- Knoll, A.H.; Nowak, M.A. The timetable of evolution. Sci. Adv. 2017, 3, e1603076. [Google Scholar] [CrossRef]
- Volkova, O.A.; Kondrakhin, Y.V.; Kashapov, T.A.; Sharipov, R.N. Comparative analysis of protein-coding and long non-coding transcripts based on RNA sequence features. J. Bioinform. Comput. Biol. 2018, 16, 1840013. [Google Scholar] [CrossRef]
- Xu, J.; Bai, J.; Zhang, X.; Lv, Y.; Gong, Y.; Liu, L.; Zhao, H.; Yu, F.; Ping, Y.; Zhang, G.; et al. A comprehensive overview of lncRNA annotation resources. Brief. Bioinform. 2017, 18, 236–249. [Google Scholar] [CrossRef]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef]
- Wang, J.; Meng, X.; Dobrovolskaya, O.B.; Orlov, Y.L.; Chen, M. Non-coding RNAs and Their Roles in Stress Response in Plants. Genom. Proteom. Bioinform. 2017, 15, 301–312. [Google Scholar] [CrossRef]
- Angrand, P.-O.; Vennin, C.; Le Bourhis, X.; Adriaenssens, E. The role of long non-coding RNAs in genome formatting and expression. Front. Genet. 2015, 6, 165. [Google Scholar] [CrossRef] [PubMed]
- Moses, A. Statistical tests for natural selection on regulatory regions based on the strength of transcription factor binding sites. BMC Evol. Boil. 2009, 9, 286. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Du, S.Y.; Armenia, J.; Qu, F.; Fan, J.; Wang, X.; Fei, T.; Komura, K.; Liu, S.X.; Lee, G.M.; et al. Expression of lncRNA MIR222HG co-transcribed from the miR-221/222 gene promoter facilitates the development of castration-resistant prostate cancer. Oncogenesis 2018, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, C.; Robertson, M.; Finnegan, E.J.; Buzas, D.M.; Dennis, E.S. Vernalization-Repression of Arabidopsis FLC Requires Promoter Sequences but Not Antisense Transcripts. PloS ONE 2011, 6, e21513. [Google Scholar] [CrossRef] [PubMed]





| Dataset | Species | Training | Testing | |
|---|---|---|---|---|
| 1 | Humans | positive | 12,000 | 24,323 |
| negative | 12,000 | 80,324 | ||
| 2 | Mice | positive | 6500 | 6534 |
| negative | 6500 | 53,239 | ||
| 3 | Cows | positive | 1000 | 976 |
| negative | 1000 | 36,346 | ||
| 4 | Arabidopsis thaliana | positive | 1400 | 1339 |
| negative | 1400 | 33,345 | ||
| 5 | Oryza sativa | positive | 2500 | 2487 |
| negative | 2500 | 33,970 | ||
| 6 | Zea mays | positive | 7500 | 7843 |
| negative | 7500 | 68,460 | ||
| No. | Feature | Introduction |
|---|---|---|
| 1 | Length | sequence length |
| 2 | GC_content | GC content |
| 3 | Stop_condon_std | standard deviation of stop codon counts (TAA, TAG, TGA) |
| 4 | ORF_in | open reading frame integrity |
| 5 | CDS_len | CDS length |
| 6 | CDS_score | CDS score of txCdsPredict prediction |
| 7 | CDS_percent | CDS percentage |
| 8 | Pep_len | peptide length |
| 9 | PI | isoelectric point |
| 10 | Hexamer | hexamer score |
| 11 | Fickett | fickett score |
| Species | Methods | SEN% | SPE% | ACC% | MCC% | AUC% |
|---|---|---|---|---|---|---|
| Humans | CPAT | 95.321 | 81.759 | 84.911 | 67.763 | 94.857 |
| CPC2 | 94.742 | 69.398 | 75.288 | 54.402 | 90.122 | |
| PreLnc | 96.633 | 85.000 | 87.703 | 72.801 | 96.199 | |
| PLEK | 87.761 | 81.219 | 82.739 | 61.160 | 86.971 | |
| LncFinder | 95.663 | 84.005 | 86.714 | 70.781 | / | |
| Mice | CPAT | 95.883 | 86.842 | 87.831 | 62.111 | 96.414 |
| CPC2 | 94.781 | 76.235 | 78.263 | 47.693 | 92.155 | |
| PreLnc | 94.154 | 89.927 | 90.389 | 66.525 | 97.087 | |
| PLEK | 91.185 | 74.534 | 76.354 | 43.730 | 85.574 | |
| LncFinder | 95.638 | 89.448 | 90.219 | 66.557 | / | |
| Cows | CPAT | 94.570 | 91.391 | 91.474 | 44.095 | 97.657 |
| CPC2 | 87.602 | 94.759 | 94.572 | 50.225 | 97.172 | |
| PreLnc | 95.389 | 93.743 | 93.787 | 50.768 | 97.881 | |
| PLEK | 90.164 | 83.228 | 83.409 | 30.043 | 86.190 | |
| LncFinder | 93.955 | 95.251 | 95.217 | 55.497 | / | |
| A. thaliana | CPAT | 99.851 | 90.097 | 90.474 | 50.910 | 96.663 |
| CPC2 | 82.450 | 92.473 | 92.086 | 47.245 | 95.976 | |
| PreLnc | 99.925 | 93.138 | 93.400 | 58.598 | 97.483 | |
| PLEK | 88.200 | 82.876 | 83.082 | 34.318 | 85.566 | |
| LncFinder | 99.477 | 90.610 | 90.953 | 51.833 | / | |
| PLncPRO | 73.786 | 88.925 | 88.340 | 35.359 | / | |
| RNAplonc | 99.776 | 90.814 | 91.160 | 52.444 | / | |
| O. sativa | CPAT | 94.692 | 82.437 | 83.273 | 46.333 | 92.944 |
| CPC2 | 78.166 | 77.265 | 77.327 | 31.660 | 83.629 | |
| PreLnc | 96.220 | 82.058 | 83.024 | 46.697 | 96.085 | |
| PLEK | 89.867 | 85.514 | 85.811 | 47.849 | 89.763 | |
| LncFinder | 94.094 | 87.861 | 88.241 | 53.995 | / | |
| PLncPRO | 30.197 | 76.334 | 73.186 | 3.849 | / | |
| RNAplonc | 99.517 | 77.489 | 78.991 | 43.352 | / | |
| Z. mays | CPAT | 98.126 | 91.788 | 92.439 | 71.936 | 97.826 |
| CPC2 | 88.920 | 89.239 | 89.206 | 60.756 | 95.351 | |
| PreLnc | 99.796 | 97.547 | 97.779 | 89.514 | 99.892 | |
| PLEK | 90.960 | 90.879 | 90.888 | 65.360 | 95.757 | |
| LncFinder | 98.546 | 92.331 | 92.970 | 73.453 | / | |
| PLncPRO | 66.888 | 77.615 | 76.512 | 30.455 | / | |
| RNAplonc | 99.308 | 85.465 | 86.883 | 60.881 | / |
| Species | Methods | SEN% | SPE% | ACC% | MCC% |
|---|---|---|---|---|---|
| Humans | CPAT | 90.247 | 95.409 | 92.574 | 85.285 |
| CPC2 | 92.692 | 95.995 | 94.181 | 88.386 | |
| PreLnc | 96.072 | 98.893 | 97.344 | 94.705 | |
| PLEK | 92.866 | 89.086 | 91.163 | 82.132 | |
| LncFinder | 93.106 | 96.239 | 94.518 | 89.054 | |
| Mice | CPAT | 97.879 | 91.003 | 93.601 | 87.154 |
| CPC2 | 94.892 | 93.871 | 94.257 | 87.972 | |
| PreLnc | 92.167 | 92.254 | 92.221 | 83.677 | |
| PLEK | 96.409 | 85.888 | 89.986 | 80.498 | |
| LncFinder | 96.703 | 95.412 | 95.912 | 91.428 | |
| A. thaliana | CPAT | 99.961 | 93.371 | 94.391 | 82.777 |
| CPC2 | 99.649 | 95.310 | 95.981 | 95.981 | |
| PreLnc | 98.205 | 100.000 | 99.722 | 98.936 | |
| PLEK | 98.712 | 86.004 | 87.972 | 68.947 | |
| LncFinder | 96.721 | 93.443 | 93.951 | 80.768 | |
| RNAplonc | 98.205 | 94.101 | 94.737 | 83.181 | |
| PLncPRO | 99.141 | 94.881 | 95.540 | 85.552 |
| Methods | NONCODEv5_Humans (172,216 Total) | NONCODEv5_Mice (131,697 Total) |
|---|---|---|
| PreLnc | 95.319% | 96.315% |
| CPAT | 93.188% | 97.594% |
| CPC2 | 93.946% | 95.928% |
| PLEK | 89.321% | 95.195% |
| LncFinder | 94.256% | 96.913% |
| Methods | Humans (13,627 Total) | Mice (17,098 Total) | A. thaliana (16,542 Total) |
|---|---|---|---|
| CPAT | 37 s | 46 s | 39 s |
| CPC2 | 26 s | 34 s | 25 s |
| PreLnc | 269 s | 346 s | 279 s |
| PLEK | 400 s | 307 s | 291 s |
| LncFinder | 198 s | 212 s | 164 s |
| RNAplonc | / | / | 34 s |
| PLncPRO | / | / | 56 s |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, L.; Wang, Y.; Bi, C.; Ye, Q.; Yin, T.; Ye, N. PreLnc: An Accurate Tool for Predicting lncRNAs Based on Multiple Features. Genes 2020, 11, 981. https://doi.org/10.3390/genes11090981
Cao L, Wang Y, Bi C, Ye Q, Yin T, Ye N. PreLnc: An Accurate Tool for Predicting lncRNAs Based on Multiple Features. Genes. 2020; 11(9):981. https://doi.org/10.3390/genes11090981
Chicago/Turabian StyleCao, Lei, Yupeng Wang, Changwei Bi, Qiaolin Ye, Tongming Yin, and Ning Ye. 2020. "PreLnc: An Accurate Tool for Predicting lncRNAs Based on Multiple Features" Genes 11, no. 9: 981. https://doi.org/10.3390/genes11090981
APA StyleCao, L., Wang, Y., Bi, C., Ye, Q., Yin, T., & Ye, N. (2020). PreLnc: An Accurate Tool for Predicting lncRNAs Based on Multiple Features. Genes, 11(9), 981. https://doi.org/10.3390/genes11090981







