Polycomb Repressive Complex(es) and Their Role in Adult Stem Cells
Abstract
:1. Introduction
2. Classification of Mammalian Polycomb Complexes
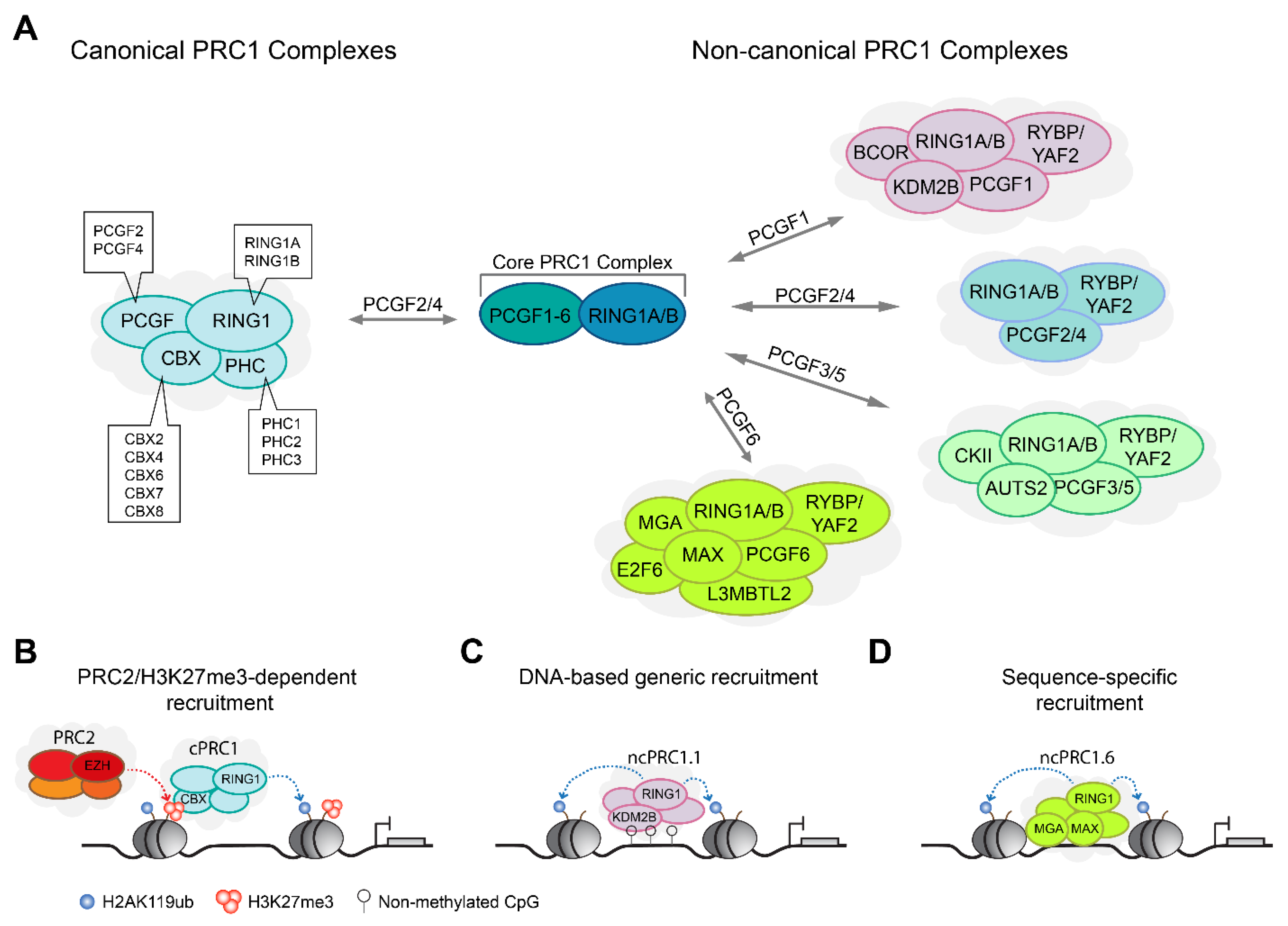
2.1. PRC1 Complexes Form Canonical and Non-Canonical Complex Assemblies
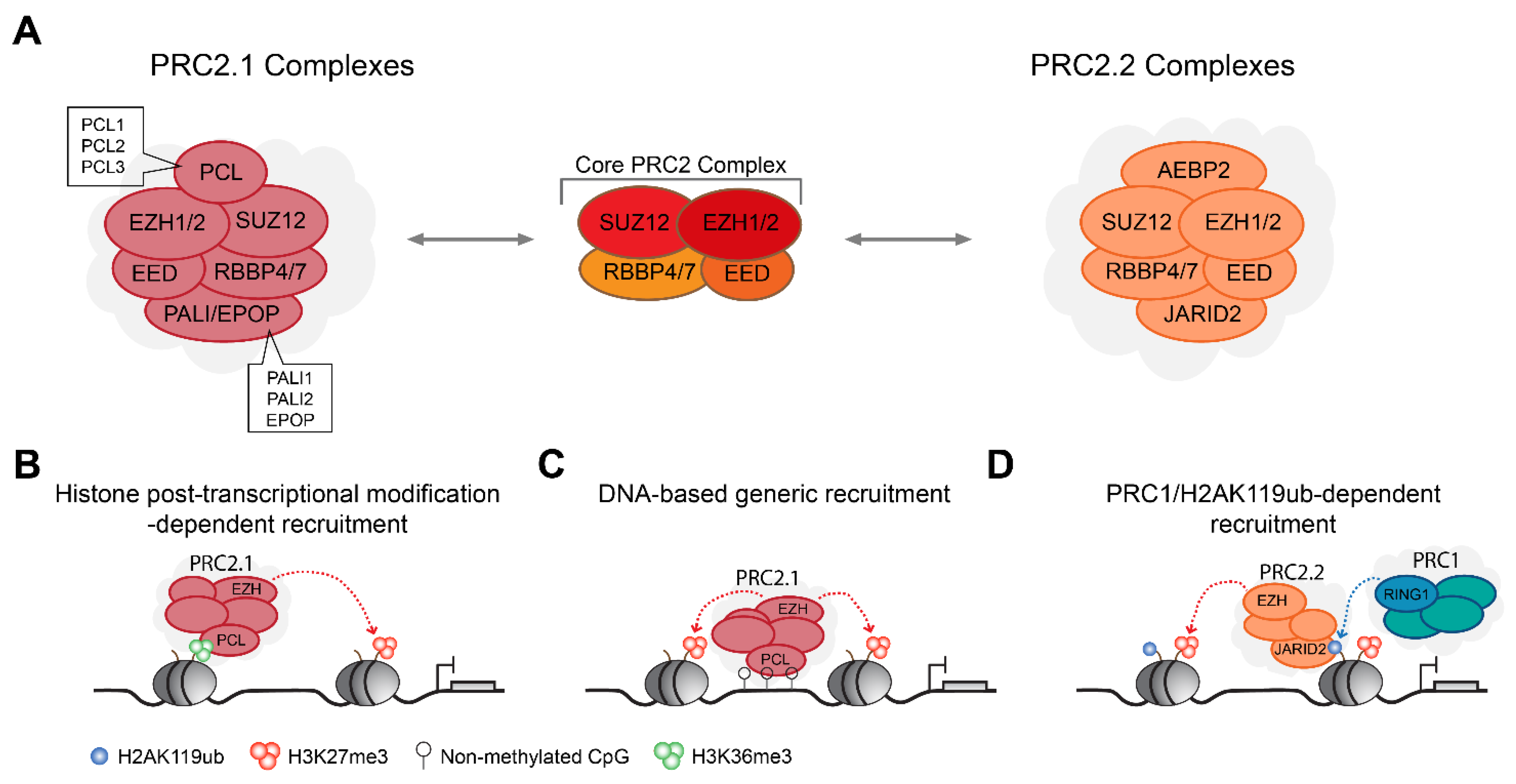
2.2. PRC2 Complexes Form Type 2.1 and Type 2.2 Complex Assemblies
3. Transcriptional Regulation by Polycomb Complexes
3.1. Role of PRC1 Catalysis and H2AK119ub
3.2. Role of PRC2-Mediated H3K27me3
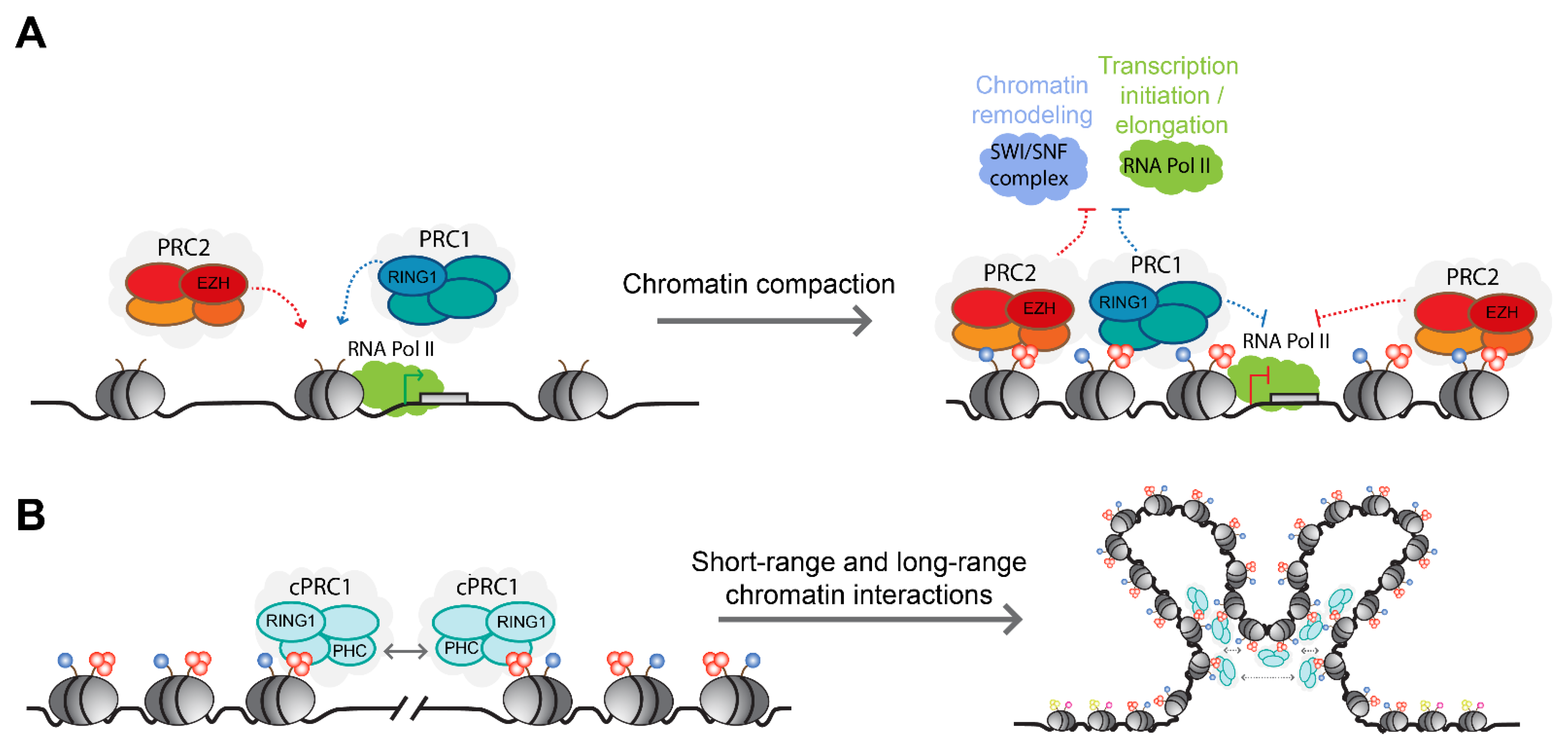
3.3. Non-Catalytic Activities and Transcriptional Regulation by Polycomb Complexes
4. The Role of PRC1 Complexes at Active Genes
5. Role of Polycomb Complexes in Adult Stem Cells
5.1. Polycomb Complexes Are Required for Adult Intestinal Regeneration
5.2. Polycomb Complexes Play a Critical Role in Adult Hematopoiesis
5.3. Role of Polycomb Complexes in Adult Epidermal Stem Cells
5.4. Polycomb Group Proteins Are Expressed in the Adult Olfactory Epithelium
5.5. The Role of Polycomb Repressive Complex 1 in Other Adult Stem Cell Systems
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clevers, H.; Watt, F.M. Defining Adult Stem Cells by Function, not by Phenotype. Ann. Rev. Biochem. 2018, 87, 1015–1027. [Google Scholar] [CrossRef]
- Avgustinova, A.; Benitah, S.A. Epigenetic control of adult stem cell function. Nat. Rev. Mol. Cell Biol. 2016, 17, 643–658. [Google Scholar] [CrossRef]
- Schuettengruber, B.; Bourbon, H.M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jürgens, G. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 1985, 316, 153–155. [Google Scholar] [CrossRef]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.; Chiang, A.; Bender, W. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 1992, 114, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Struhl, G. A gene product required for correct initiation of segmental determination in Drosophila. Nature 1981, 293, 36–41. [Google Scholar] [CrossRef]
- Leeb, M.; Pasini, D.; Novatchkova, M.; Jaritz, M.; Helin, K.; Wutz, A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010, 24, 265–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.; Liu, Y.; Hsu, Y.J.; Fujiwara, Y.; Kim, J.; Mao, X.; Yuan, G.C.; Orkin, S.H. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 2008, 32, 491–502. [Google Scholar] [CrossRef] [Green Version]
- Endoh, M.; Endo, T.A.; Endoh, T.; Fujimura, Y.; Ohara, O.; Toyoda, T.; Otte, A.P.; Okano, M.; Brockdorff, N.; Vidal, M.; et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 2008, 135, 1513–1524. [Google Scholar] [CrossRef] [Green Version]
- Bracken, A.P.; Dietrich, N.; Pasini, D.; Hansen, K.H.; Helin, K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006, 20, 1123–1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyer, L.A.; Plath, K.; Zeitlinger, J.; Brambrink, T.; Medeiros, L.A.; Lee, T.I.; Levine, S.S.; Wernig, M.; Tajonar, A.; Ray, M.K.; et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006, 441, 349–353. [Google Scholar] [CrossRef]
- Pasini, D.; Bracken, A.P.; Jensen, M.R.; Lazzerini Denchi, E.; Helin, K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004, 23, 4061–4071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voncken, J.W.; Roelen, B.A.; Roefs, M.; de Vries, S.; Verhoeven, E.; Marino, S.; Deschamps, J.; van Lohuizen, M. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl. Acad. Sci. USA 2003, 100, 2468–2473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Carroll, D.; Erhardt, S.; Pagani, M.; Barton, S.C.; Surani, M.A.; Jenuwein, T. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell Biol. 2001, 21, 4330–4336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faust, C.; Schumacher, A.; Holdener, B.; Magnuson, T. The eed mutation disrupts anterior mesoderm production in mice. Development 1995, 121, 273–285. [Google Scholar] [CrossRef]
- Bar, C.; Cohen, I.; Zhao, D.; Pothula, V.; Litskevitch, A.; Koseki, H.; Zheng, D.; Ezhkova, E. Polycomb Repressive Complex 1 Controls Maintenance of Fungiform Papillae by Repressing Sonic Hedgehog Expression. Cell Rep. 2019, 28, 257–266.e255. [Google Scholar] [CrossRef] [Green Version]
- Vidal, M.; Starowicz, K. Polycomb complexes PRC1 and their function in hematopoiesis. Exp. Hematol. 2017, 48, 12–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- von Schimmelmann, M.; Feinberg, P.A.; Sullivan, J.M.; Ku, S.M.; Badimon, A.; Duff, M.K.; Wang, Z.; Lachmann, A.; Dewell, S.; Ma’ayan, A.; et al. Polycomb repressive complex 2 (PRC2) silences genes responsible for neurodegeneration. Nat. Neurosci. 2016, 19, 1321–1330. [Google Scholar] [CrossRef]
- Koppens, M.A.; Bounova, G.; Gargiulo, G.; Tanger, E.; Janssen, H.; Cornelissen-Steijger, P.; Blom, M.; Song, J.Y.; Wessels, L.F.; van Lohuizen, M. Deletion of Polycomb Repressive Complex 2 From Mouse Intestine Causes Loss of Stem Cells. Gastroenterology 2016, 151, 684–697.e612. [Google Scholar] [CrossRef] [Green Version]
- Ikawa, T.; Masuda, K.; Endo, T.A.; Endo, M.; Isono, K.; Koseki, Y.; Nakagawa, R.; Kometani, K.; Takano, J.; Agata, Y.; et al. Conversion of T cells to B cells by inactivation of polycomb-mediated epigenetic suppression of the B-lineage program. Genes Dev. 2016, 30, 2475–2485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiacchiera, F.; Rossi, A.; Jammula, S.; Piunti, A.; Scelfo, A.; Ordonez-Moran, P.; Huelsken, J.; Koseki, H.; Pasini, D. Polycomb Complex PRC1 Preserves Intestinal Stem Cell Identity by Sustaining Wnt/beta-Catenin Transcriptional Activity. Cell Stem Cell 2016, 18, 91–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, H.; Xu, J.; Hsu, J.H.; Nguyen, M.; Fujiwara, Y.; Peng, C.; Orkin, S.H. Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell 2014, 14, 68–80. [Google Scholar] [CrossRef] [Green Version]
- Mu, W.; Starmer, J.; Fedoriw, A.M.; Yee, D.; Magnuson, T. Repression of the soma-specific transcriptome by Polycomb-repressive complex 2 promotes male germ cell development. Genes Dev. 2014, 28, 2056–2069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bantignies, F.; Roure, V.; Comet, I.; Leblanc, B.; Schuettengruber, B.; Bonnet, J.; Tixier, V.; Mas, A.; Cavalli, G. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 2011, 144, 214–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akasaka, T.; van Lohuizen, M.; van der Lugt, N.; Mizutani-Koseki, Y.; Kanno, M.; Taniguchi, M.; Vidal, M.; Alkema, M.; Berns, A.; Koseki, H. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development 2001, 128, 1587–1597. [Google Scholar] [CrossRef]
- Simon, J.A.; Kingston, R.E. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 2013, 49, 808–824. [Google Scholar] [CrossRef] [Green Version]
- Cohen, I.; Bar, C.; Ezhkova, E. Activity of PRC1 and Histone H2AK119 Monoubiquitination: Revising Popular Misconceptions. Bioessays 2020, 42, e1900192. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef]
- de Napoles, M.; Mermoud, J.E.; Wakao, R.; Tang, Y.A.; Endoh, M.; Appanah, R.; Nesterova, T.B.; Silva, J.; Otte, A.P.; Vidal, M.; et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 2004, 7, 663–676. [Google Scholar] [CrossRef]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzmichev, A.; Nishioka, K.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002, 16, 2893–2905. [Google Scholar] [CrossRef] [Green Version]
- Czermin, B.; Melfi, R.; McCabe, D.; Seitz, V.; Imhof, A.; Pirrotta, V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 2002, 111, 185–196. [Google Scholar] [CrossRef] [Green Version]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [Green Version]
- Muller, J.; Hart, C.M.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.B.; Kingston, R.E.; Simon, J.A. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 2002, 111, 197–208. [Google Scholar] [CrossRef] [Green Version]
- Di Croce, L.; Helin, K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 2013, 20, 1147–1155. [Google Scholar] [CrossRef]
- Ku, M.; Koche, R.P.; Rheinbay, E.; Mendenhall, E.M.; Endoh, M.; Mikkelsen, T.S.; Presser, A.; Nusbaum, C.; Xie, X.; Chi, A.S.; et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008, 4, e1000242. [Google Scholar] [CrossRef]
- Tolhuis, B.; de Wit, E.; Muijrers, I.; Teunissen, H.; Talhout, W.; van Steensel, B.; van Lohuizen, M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 2006, 38, 694–699. [Google Scholar] [CrossRef]
- Cohen, I.; Bar, C.; Liu, H.; Valdes, V.J.; Zhao, D.; Galbo, P.M., Jr.; Silva, J.M.; Koseki, H.; Zheng, D.; Ezhkova, E. Polycomb complexes redundantly maintain epidermal stem cell identity during development. Genes Dev. 2021, 35, 354–366. [Google Scholar] [CrossRef]
- Wang, L.; Brown, J.L.; Cao, R.; Zhang, Y.; Kassis, J.A.; Jones, R.S. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 2004, 14, 637–646. [Google Scholar] [CrossRef]
- Min, J.; Zhang, Y.; Xu, R.M. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003, 17, 1823–1828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidal, M. Polycomb assemblies multitask to regulate transcription. Epigenomes 2019, 3, 12. [Google Scholar] [CrossRef] [Green Version]
- Aranda, S.; Mas, G.; Di Croce, L. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 2015, 1, e1500737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef] [Green Version]
- Piunti, A.; Shilatifard, A. The roles of Polycombpolycomb repressive complexes in mammalian development and cancer. Nat. Rev. Mol. Cell Biol. 2021, 22, 326–345. [Google Scholar] [CrossRef] [PubMed]
- Conway, E.; Jerman, E.; Healy, E.; Ito, S.; Holoch, D.; Oliviero, G.; Deevy, O.; Glancy, E.; Fitzpatrick, D.J.; Mucha, M.; et al. A Family of Vertebrate-Specific Polycombs Encoded by the LCOR/LCORL Genes Balance PRC2 Subtype Activities. Mol. Cell 2018, 70, 408–421.e408. [Google Scholar] [CrossRef] [Green Version]
- Cohen, I.; Zhao, D.; Bar, C.; Valdes, V.J.; Dauber-Decker, K.L.; Nguyen, M.B.; Nakayama, M.; Rendl, M.; Bickmore, W.A.; Koseki, H.; et al. PRC1 Fine-tunes Gene Repression and Activation to Safeguard Skin Development and Stem Cell Specification. Cell Stem Cell 2018, 22, 726–739.e727. [Google Scholar] [CrossRef] [Green Version]
- Kloet, S.L.; Makowski, M.M.; Baymaz, H.I.; van Voorthuijsen, L.; Karemaker, I.D.; Santanach, A.; Jansen, P.; Di Croce, L.; Vermeulen, M. The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat. Struct. Mol. Biol. 2016, 23, 682–690. [Google Scholar] [CrossRef] [Green Version]
- Frangini, A.; Sjoberg, M.; Roman-Trufero, M.; Dharmalingam, G.; Haberle, V.; Bartke, T.; Lenhard, B.; Malumbres, M.; Vidal, M.; Dillon, N. The aurora B kinase and the polycomb protein ring1B combine to regulate active promoters in quiescent lymphocytes. Mol. Cell 2013, 51, 647–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mousavi, K.; Zare, H.; Wang, A.H.; Sartorelli, V. Protein Ezh1 promotes RNA polymerase II elongation. Mol. Cell 2012, 45, 255–262. [Google Scholar] [CrossRef] [Green Version]
- Taherbhoy, A.M.; Huang, O.W.; Cochran, A.G. BMI1-RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat. Commun. 2015, 6, 7621. [Google Scholar] [CrossRef] [PubMed]
- Metzger, M.B.; Pruneda, J.N.; Klevit, R.E.; Weissman, A.M. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 2014, 1843, 47–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borden, K.L. RING domains: Master builders of molecular scaffolds? J. Mol. Biol. 2000, 295, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- Chittock, E.C.; Latwiel, S.; Miller, T.C.; Muller, C.W. Molecular architecture of polycomb repressive complexes. Biochem. Soc. Trans. 2017, 45, 193–205. [Google Scholar] [CrossRef] [Green Version]
- Bezsonova, I.; Walker, J.R.; Bacik, J.P.; Duan, S.; Dhe-Paganon, S.; Arrowsmith, C.H. Ring1B contains a ubiquitin-like docking module for interaction with Cbx proteins. Biochemistry 2009, 48, 10542–10548. [Google Scholar] [CrossRef]
- Sanchez-Pulido, L.; Devos, D.; Sung, Z.R.; Calonje, M. RAWUL: A new ubiquitin-like domain in PRC1 ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genom. 2008, 9, 308. [Google Scholar] [CrossRef] [Green Version]
- Levine, S.S.; Weiss, A.; Erdjument-Bromage, H.; Shao, Z.; Tempst, P.; Kingston, R.E. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell Biol. 2002, 22, 6070–6078. [Google Scholar] [CrossRef] [Green Version]
- Francis, N.J.; Saurin, A.J.; Shao, Z.; Kingston, R.E. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 2001, 8, 545–556. [Google Scholar] [CrossRef]
- Shao, Z.; Raible, F.; Mollaaghababa, R.; Guyon, J.R.; Wu, C.T.; Bender, W.; Kingston, R.E. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 1999, 98, 37–46. [Google Scholar] [CrossRef] [Green Version]
- Fischle, W.; Wang, Y.; Jacobs, S.A.; Kim, Y.; Allis, C.D.; Khorasanizadeh, S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003, 17, 1870–1881. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.G.; Villa, R.; Trojer, P.; Norman, J.; Yan, K.P.; Reinberg, D.; Di Croce, L.; Shiekhattar, R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 2007, 318, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, N.P.; Farcas, A.M.; Kondo, T.; King, H.W.; McGouran, J.F.; Hanssen, L.L.P.; Ito, S.; Cooper, S.; Kondo, K.; Koseki, Y.; et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 2014, 157, 1445–1459. [Google Scholar] [CrossRef] [Green Version]
- Tavares, L.; Dimitrova, E.; Oxley, D.; Webster, J.; Poot, R.; Demmers, J.; Bezstarosti, K.; Taylor, S.; Ura, H.; Koide, H.; et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 2012, 148, 664–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagarou, A.; Mohd-Sarip, A.; Moshkin, Y.M.; Chalkley, G.E.; Bezstarosti, K.; Demmers, J.A.; Verrijzer, C.P. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev. 2008, 22, 2799–2810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez, C.; Sanchez, I.; Demmers, J.A.; Rodriguez, P.; Strouboulis, J.; Vidal, M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol. Cell Proteom. 2007, 6, 820–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gearhart, M.D.; Corcoran, C.M.; Wamstad, J.A.; Bardwell, V.J. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell Biol. 2006, 26, 6880–6889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Johansen, J.V.; Helin, K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol. Cell 2013, 49, 1134–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Shen, L.; Wan, M.; Taranova, O.; Wu, H.; Zhang, Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat. Cell Biol. 2013, 15, 373–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farcas, A.M.; Blackledge, N.P.; Sudbery, I.; Long, H.K.; McGouran, J.F.; Rose, N.R.; Lee, S.; Sims, D.; Cerase, A.; Sheahan, T.W.; et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. eLife 2012, 1, e00205. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Zhou, J.C.; Tolstorukov, M.Y.; Farcas, A.M.; Park, P.J.; Klose, R.J. CpG islands recruit a histone H3 lysine 36 demethylase. Mol. Cell 2010, 38, 179–190. [Google Scholar] [CrossRef]
- He, J.; Kallin, E.M.; Tsukada, Y.; Zhang, Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat. Struct. Mol. Biol. 2008, 15, 1169–1175. [Google Scholar] [CrossRef]
- Dahlet, T.; Truss, M.; Frede, U.; Al Adhami, H.; Bardet, A.F.; Dumas, M.; Vallet, J.; Chicher, J.; Hammann, P.; Kottnik, S.; et al. E2F6 initiates stable epigenetic silencing of germline genes during embryonic development. Nat. Commun. 2021, 12, 3582. [Google Scholar] [CrossRef]
- Scelfo, A.; Fernandez-Perez, D.; Tamburri, S.; Zanotti, M.; Lavarone, E.; Soldi, M.; Bonaldi, T.; Ferrari, K.J.; Pasini, D. Functional Landscape of PCGF Proteins Reveals Both RING1A/B-Dependent-and RING1A/B-Independent-Specific Activities. Mol. Cell 2019, 74, 1037–1052.e1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stielow, B.; Finkernagel, F.; Stiewe, T.; Nist, A.; Suske, G. MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6. PLoS Genet. 2018, 14, e1007193. [Google Scholar] [CrossRef]
- Endoh, M.; Endo, T.A.; Shinga, J.; Hayashi, K.; Farcas, A.; Ma, K.W.; Ito, S.; Sharif, J.; Endoh, T.; Onaga, N.; et al. PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. eLife 2017, 6, e21064. [Google Scholar] [CrossRef] [PubMed]
- Fursova, N.A.; Blackledge, N.P.; Nakayama, M.; Ito, S.; Koseki, Y.; Farcas, A.M.; King, H.W.; Koseki, H.; Klose, R.J. Synergy between Variant PRC1 Complexes Defines Polycomb-Mediated Gene Repression. Mol. Cell 2019, 74, 1020–1036.e1028. [Google Scholar] [CrossRef] [Green Version]
- Hauri, S.; Comoglio, F.; Seimiya, M.; Gerstung, M.; Glatter, T.; Hansen, K.; Aebersold, R.; Paro, R.; Gstaiger, M.; Beisel, C. A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep. 2016, 17, 583–595. [Google Scholar] [CrossRef] [Green Version]
- Margueron, R.; Li, G.; Sarma, K.; Blais, A.; Zavadil, J.; Woodcock, C.L.; Dynlacht, B.D.; Reinberg, D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 2008, 32, 503–518. [Google Scholar] [CrossRef] [Green Version]
- Margueron, R.; Justin, N.; Ohno, K.; Sharpe, M.L.; Son, J.; Drury, W.J., 3rd; Voigt, P.; Martin, S.R.; Taylor, W.R.; De Marco, V.; et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 2009, 461, 762–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tie, F.; Stratton, C.A.; Kurzhals, R.L.; Harte, P.J. The N terminus of Drosophila ESC binds directly to histone H3 and is required for E(Z)-dependent trimethylation of H3 lysine 27. Mol. Cell Biol. 2007, 27, 2014–2026. [Google Scholar] [CrossRef] [Green Version]
- Laugesen, A.; Hojfeldt, J.W.; Helin, K. Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol. Cell 2019, 74, 8–18. [Google Scholar] [CrossRef] [Green Version]
- Song, J.J.; Garlick, J.D.; Kingston, R.E. Structural basis of histone H4 recognition by p55. Genes Dev. 2008, 22, 1313–1318. [Google Scholar] [CrossRef] [Green Version]
- Murzina, N.V.; Pei, X.Y.; Zhang, W.; Sparkes, M.; Vicente-Garcia, J.; Pratap, J.V.; McLaughlin, S.H.; Ben-Shahar, T.R.; Verreault, A.; Luisi, B.F.; et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure 2008, 16, 1077–1085. [Google Scholar] [CrossRef]
- Glancy, E.; Ciferri, C.; Bracken, A.P. Structural basis for PRC2 engagement with chromatin. Curr. Opin. Struct. Biol. 2021, 67, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Rothbart, S.B.; Lu, R.; Xu, B.; Chen, W.Y.; Tripathy, A.; Rockowitz, S.; Zheng, D.; Patel, D.J.; Allis, C.D.; et al. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol. Cell 2013, 49, 571–582. [Google Scholar] [CrossRef] [Green Version]
- Musselman, C.A.; Avvakumov, N.; Watanabe, R.; Abraham, C.G.; Lalonde, M.E.; Hong, Z.; Allen, C.; Roy, S.; Nunez, J.K.; Nickoloff, J.; et al. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat. Struct. Mol. Biol. 2012, 19, 1266–1272. [Google Scholar] [CrossRef] [PubMed]
- Brien, G.L.; Gambero, G.; O’Connell, D.J.; Jerman, E.; Turner, S.A.; Egan, C.M.; Dunne, E.J.; Jurgens, M.C.; Wynne, K.; Piao, L.; et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat. Struct. Mol. Biol. 2012, 19, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Ballare, C.; Lange, M.; Lapinaite, A.; Martin, G.M.; Morey, L.; Pascual, G.; Liefke, R.; Simon, B.; Shi, Y.; Gozani, O.; et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat. Struct. Mol. Biol. 2012, 19, 1257–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Healy, E.; Mucha, M.; Glancy, E.; Fitzpatrick, D.J.; Conway, E.; Neikes, H.K.; Monger, C.; Van Mierlo, G.; Baltissen, M.P.; Koseki, Y.; et al. PRC2.1 and PRC2.2 Synergize to Coordinate H3K27 Trimethylation. Mol. Cell 2019, 76, 437–452.e436. [Google Scholar] [CrossRef] [PubMed]
- Liefke, R.; Karwacki-Neisius, V.; Shi, Y. EPOP Interacts with Elongin BC and USP7 to Modulate the Chromatin Landscape. Mol. Cell 2016, 64, 659–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beringer, M.; Pisano, P.; Di Carlo, V.; Blanco, E.; Chammas, P.; Vizan, P.; Gutierrez, A.; Aranda, S.; Payer, B.; Wierer, M.; et al. EPOP Functionally Links Elongin and Polycomb in Pluripotent Stem Cells. Mol. Cell 2016, 64, 645–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Jones, A.; Sun, C.W.; Li, C.; Chang, C.W.; Joo, H.Y.; Dai, Q.; Mysliwiec, M.R.; Wu, L.C.; Guo, Y.; et al. PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells 2011, 29, 229–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perino, M.; van Mierlo, G.; Karemaker, I.D.; van Genesen, S.; Vermeulen, M.; Marks, H.; van Heeringen, S.J.; Veenstra, G.J.C. MTF2 recruits Repressive Complex 2 by helical-shape-selective DNA binding. Nat. Genet. 2018, 50, 1002–1010. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Jiao, L.; Liu, X.; Yang, X.; Liu, X. A Dimeric Structural Scaffold for PRC2-PCL Targeting to CpG Island Chromatin. Mol. Cell 2020, 77, 1265–1278.e1267. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liefke, R.; Jiang, J.; Kurland, J.V.; Tian, W.; Deng, P.; Zhang, W.; He, Q.; Patel, D.J.; Bulyk, M.L.; et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 2017, 549, 287–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, J.C.; Valouev, A.; Swigut, T.; Zhang, J.; Zhao, Y.; Sidow, A.; Wysocka, J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 2009, 139, 1290–1302. [Google Scholar] [CrossRef] [Green Version]
- Klose, R.J.; Kallin, E.M.; Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006, 7, 715–727. [Google Scholar] [CrossRef]
- Cooper, S.; Grijzenhout, A.; Underwood, E.; Ancelin, K.; Zhang, T.; Nesterova, T.B.; Anil-Kirmizitas, B.; Bassett, A.; Kooistra, S.M.; Agger, K.; et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 2016, 7, 13661. [Google Scholar] [CrossRef] [Green Version]
- Son, J.; Shen, S.S.; Margueron, R.; Reinberg, D. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 2013, 27, 2663–2677. [Google Scholar] [CrossRef] [Green Version]
- Pasini, D.; Cloos, P.A.; Walfridsson, J.; Olsson, L.; Bukowski, J.P.; Johansen, J.V.; Bak, M.; Tommerup, N.; Rappsilber, J.; Helin, K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 2010, 464, 306–310. [Google Scholar] [CrossRef]
- Sanulli, S.; Justin, N.; Teissandier, A.; Ancelin, K.; Portoso, M.; Caron, M.; Michaud, A.; Lombard, B.; da Rocha, S.T.; Offer, J.; et al. Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation. Mol. Cell 2015, 57, 769–783. [Google Scholar] [CrossRef] [Green Version]
- Kasinath, V.; Faini, M.; Poepsel, S.; Reif, D.; Feng, X.A.; Stjepanovic, G.; Aebersold, R.; Nogales, E. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 2018, 359, 940–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Kang, K.; Kim, J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res. 2009, 37, 2940–2950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loubiere, V.; Martinez, A.M.; Cavalli, G. Cell Fate and Developmental Regulation Dynamics by Polycomb Proteins and 3D Genome Architecture. Bioessays 2019, 41, e1800222. [Google Scholar] [CrossRef] [Green Version]
- Rose, N.R.; King, H.W.; Blackledge, N.P.; Fursova, N.A.; Ember, K.J.; Fischer, R.; Kessler, B.M.; Klose, R.J. RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes. eLife 2016, 5, e18591. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Huang, Y.; Zhang, J.; Liu, M.; Ji, H.; Wang, C.; Cao, N.; Li, C.; Xia, Y.; Jiang, Q.; et al. Polycomb group RING finger proteins 3/5 activate transcription via an interaction with the pluripotency factor Tex10 in embryonic stem cells. J. Biol. Chem. 2017, 292, 21527–21537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perdigoto, C.N.; Dauber, K.L.; Bar, C.; Tsai, P.C.; Valdes, V.J.; Cohen, I.; Santoriello, F.J.; Zhao, D.; Zheng, D.; Hsu, Y.C.; et al. Polycomb-Mediated Repression and Sonic Hedgehog Signaling Interact to Regulate Merkel Cell Specification during Skin Development. PLoS Genet. 2016, 12, e1006151. [Google Scholar] [CrossRef] [PubMed]
- Dauber, K.L.; Perdigoto, C.N.; Valdes, V.J.; Santoriello, F.J.; Cohen, I.; Ezhkova, E. Dissecting the Roles of Polycomb Repressive Complex 2 Subunits in the Control of Skin Development. J. Investig. Derm. 2016, 136, 1647–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blackledge, N.P.; Fursova, N.A.; Kelley, J.R.; Huseyin, M.K.; Feldmann, A.; Klose, R.J. PRC1 Catalytic Activity Is Central to Polycomb System Function. Mol. Cell 2020, 77, 857–874.e859. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, L.; Ferrari, R.; Vashisht, A.A.; Wohlschlegel, J.A.; Kurdistani, S.K.; Carey, M. Polycomb repressive complex 1 (PRC1) disassembles RNA polymerase II preinitiation complexes. J. Biol. Chem. 2012, 287, 35784–35794. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Zhu, P.; Wang, J.; Pascual, G.; Ohgi, K.A.; Lozach, J.; Glass, C.K.; Rosenfeld, M.G. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol. Cell 2008, 29, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Stock, J.K.; Giadrossi, S.; Casanova, M.; Brookes, E.; Vidal, M.; Koseki, H.; Brockdorff, N.; Fisher, A.G.; Pombo, A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 2007, 9, 1428–1435. [Google Scholar] [CrossRef] [Green Version]
- Dellino, G.I.; Schwartz, Y.B.; Farkas, G.; McCabe, D.; Elgin, S.C.; Pirrotta, V. Polycomb silencing blocks transcription initiation. Mol. Cell 2004, 13, 887–893. [Google Scholar] [CrossRef] [Green Version]
- Kalb, R.; Latwiel, S.; Baymaz, H.I.; Jansen, P.W.; Muller, C.W.; Vermeulen, M.; Muller, J. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 2014, 21, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Tamburri, S.; Lavarone, E.; Fernandez-Perez, D.; Conway, E.; Zanotti, M.; Manganaro, D.; Pasini, D. Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Mol. Cell 2020, 77, 840–856.e845. [Google Scholar] [CrossRef] [Green Version]
- Ezhkova, E.; Pasolli, H.A.; Parker, J.S.; Stokes, N.; Su, I.H.; Hannon, G.; Tarakhovsky, A.; Fuchs, E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 2009, 136, 1122–1135. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Shao, Z.; Li, D.; Xie, H.; Kim, W.; Huang, J.; Taylor, J.E.; Pinello, L.; Glass, K.; Jaffe, J.D.; et al. Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol. Cell 2015, 57, 304–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.H.; Holder, M.; Grau, D.; Saldana-Meyer, R.; Yu, J.R.; Ganai, R.A.; Zhang, J.; Wang, M.; LeRoy, G.; Dobenecker, M.W.; et al. Distinct Stimulatory Mechanisms Regulate the Catalytic Activity of Polycomb Repressive Complex 2. Mol. Cell 2018, 70, 435–448.e435. [Google Scholar] [CrossRef] [Green Version]
- Cao, R.; Zhang, Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 2004, 15, 57–67. [Google Scholar] [CrossRef]
- Ragazzini, R.; Perez-Palacios, R.; Baymaz, I.H.; Diop, S.; Ancelin, K.; Zielinski, D.; Michaud, A.; Givelet, M.; Borsos, M.; Aflaki, S.; et al. EZHIP constrains Polycomb Repressive Complex 2 activity in germ cells. Nat. Commun. 2019, 10, 3858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piunti, A.; Smith, E.R.; Morgan, M.A.J.; Ugarenko, M.; Khaltyan, N.; Helmin, K.A.; Ryan, C.A.; Murray, D.C.; Rickels, R.A.; Yilmaz, B.D.; et al. CATACOMB: An endogenous inducible gene that antagonizes H3K27 methylation activity of Polycomb repressive complex 2 via an H3K27M-like mechanism. Sci. Adv. 2019, 5, eaax2887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, S.U.; Do, T.J.; Lund, P.J.; Rashoff, A.Q.; Diehl, K.L.; Cieslik, M.; Bajic, A.; Juretic, N.; Deshmukh, S.; Venneti, S.; et al. PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat. Commun. 2019, 10, 2146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pengelly, A.R.; Copur, O.; Jackle, H.; Herzig, A.; Muller, J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 2013, 339, 698–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chopra, V.S.; Hendrix, D.A.; Core, L.J.; Tsui, C.; Lis, J.T.; Levine, M. The polycomb group mutant esc leads to augmented levels of paused Pol II in the Drosophila embryo. Mol. Cell 2011, 42, 837–844. [Google Scholar] [CrossRef] [Green Version]
- Landeira, D.; Sauer, S.; Poot, R.; Dvorkina, M.; Mazzarella, L.; Jorgensen, H.F.; Pereira, C.F.; Leleu, M.; Piccolo, F.M.; Spivakov, M.; et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat. Cell Biol. 2010, 12, 618–624. [Google Scholar] [CrossRef] [Green Version]
- Wani, A.H.; Boettiger, A.N.; Schorderet, P.; Ergun, A.; Munger, C.; Sadreyev, R.I.; Zhuang, X.; Kingston, R.E.; Francis, N.J. Chromatin topology is coupled to Polycomb group protein subnuclear organization. Nat. Commun. 2016, 7, 10291. [Google Scholar] [CrossRef]
- Isono, K.; Endo, T.A.; Ku, M.; Yamada, D.; Suzuki, R.; Sharif, J.; Ishikura, T.; Toyoda, T.; Bernstein, B.E.; Koseki, H. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev. Cell 2013, 26, 565–577. [Google Scholar] [CrossRef] [Green Version]
- Cohen, I.; Zhao, D.; Menon, G.; Nakayama, M.; Koseki, H.; Zheng, D.; Ezhkova, E. PRC1 preserves epidermal tissue integrity independently of PRC2. Genes Dev. 2019, 33, 55–60. [Google Scholar] [CrossRef] [Green Version]
- King, H.W.; Fursova, N.A.; Blackledge, N.P.; Klose, R.J. Polycomb repressive complex 1 shapes the nucleosome landscape but not accessibility at target genes. Genome Res. 2018, 28, 1494–1507. [Google Scholar] [CrossRef] [Green Version]
- Poepsel, S.; Kasinath, V.; Nogales, E. Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Biol. 2018, 25, 154–162. [Google Scholar] [CrossRef]
- Reveron-Gomez, N.; Gonzalez-Aguilera, C.; Stewart-Morgan, K.R.; Petryk, N.; Flury, V.; Graziano, S.; Johansen, J.V.; Jakobsen, J.S.; Alabert, C.; Groth, A. Accurate Recycling of Parental Histones Reproduces the Histone Modification Landscape during DNA Replication. Mol. Cell 2018, 72, 239–249.e235. [Google Scholar] [CrossRef] [Green Version]
- Laprell, F.; Finkl, K.; Muller, J. Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science 2017, 356, 85–88. [Google Scholar] [CrossRef]
- Alabert, C.; Barth, T.K.; Reveron-Gomez, N.; Sidoli, S.; Schmidt, A.; Jensen, O.N.; Imhof, A.; Groth, A. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 2015, 29, 585–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coleman, R.T.; Struhl, G. Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science 2017, 356, eaai8236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saurin, A.J.; Shiels, C.; Williamson, J.; Satijn, D.P.; Otte, A.P.; Sheer, D.; Freemont, P.S. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 1998, 142, 887–898. [Google Scholar] [CrossRef] [Green Version]
- Satijn, D.P.; Gunster, M.J.; van der Vlag, J.; Hamer, K.M.; Schul, W.; Alkema, M.J.; Saurin, A.J.; Freemont, P.S.; van Driel, R.; Otte, A.P. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol. Cell Biol. 1997, 17, 4105–4113. [Google Scholar] [CrossRef] [Green Version]
- Kundu, S.; Ji, F.; Sunwoo, H.; Jain, G.; Lee, J.T.; Sadreyev, R.I.; Dekker, J.; Kingston, R.E. Polycomb Repressive Complex 1 Generates Discrete Compacted Domains that Change during Differentiation. Mol. Cell 2017, 65, 432–446.e435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boettiger, A.N.; Bintu, B.; Moffitt, J.R.; Wang, S.; Beliveau, B.J.; Fudenberg, G.; Imakaev, M.; Mirny, L.A.; Wu, C.T.; Zhuang, X. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 2016, 529, 418–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, A.K.; Leal, B.Z.; Chadwell, L.V.; Wang, R.; Ilangovan, U.; Kaur, Y.; Junco, S.E.; Schirf, V.; Osmulski, P.A.; Gaczynska, M.; et al. The growth-suppressive function of the polycomb group protein polyhomeotic is mediated by polymerization of its sterile alpha motif (SAM) domain. J. Biol. Chem. 2012, 287, 8702–8713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.A.; Gingery, M.; Pilpa, R.M.; Bowie, J.U. The SAM domain of polyhomeotic forms a helical polymer. Nat. Struct. Biol. 2002, 9, 453–457. [Google Scholar] [CrossRef]
- Schoenfelder, S.; Sugar, R.; Dimond, A.; Javierre, B.M.; Armstrong, H.; Mifsud, B.; Dimitrova, E.; Matheson, L.; Tavares-Cadete, F.; Furlan-Magaril, M.; et al. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat. Genet. 2015, 47, 1179–1186. [Google Scholar] [CrossRef]
- Boyle, S.; Flyamer, I.M.; Williamson, I.; Sengupta, D.; Bickmore, W.A.; Illingworth, R.S. A central role for canonical PRC1 in shaping the 3D nuclear landscape. Genes Dev. 2020, 34, 931–949. [Google Scholar] [CrossRef]
- Eskeland, R.; Leeb, M.; Grimes, G.R.; Kress, C.; Boyle, S.; Sproul, D.; Gilbert, N.; Fan, Y.; Skoultchi, A.I.; Wutz, A.; et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell 2010, 38, 452–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, H.L.; Beckedorff, F.; Zhang, Y.; Garcia-Huidobro, J.; Jiang, H.; Colaprico, A.; Bilbao, D.; Figueroa, M.E.; LaCava, J.; Shiekhattar, R.; et al. Polycomb complexes associate with enhancers and promote oncogenic transcriptional programs in cancer through multiple mechanisms. Nat. Commun. 2018, 9, 3377. [Google Scholar] [CrossRef] [Green Version]
- van den Boom, V.; Maat, H.; Geugien, M.; Rodriguez Lopez, A.; Sotoca, A.M.; Jaques, J.; Brouwers-Vos, A.Z.; Fusetti, F.; Groen, R.W.; Yuan, H.; et al. Non-canonical PRC1.1 Targets Active Genes Independent of H3K27me3 and Is Essential for Leukemogenesis. Cell Rep. 2016, 14, 332–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loubiere, V.; Delest, A.; Thomas, A.; Bonev, B.; Schuettengruber, B.; Sati, S.; Martinez, A.M.; Cavalli, G. Coordinate redeployment of PRC1 proteins suppresses tumor formation during Drosophila development. Nat. Genet. 2016, 48, 1436–1442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morey, L.; Santanach, A.; Blanco, E.; Aloia, L.; Nora, E.P.; Bruneau, B.G.; Di Croce, L. Polycomb Regulates Mesoderm Cell Fate-Specification in Embryonic Stem Cells through Activation and Repression Mechanisms. Cell Stem Cell 2015, 17, 300–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.; Lee, P.; Stafford, J.M.; von Schimmelmann, M.; Schaefer, A.; Reinberg, D. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature 2014, 516, 349–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maezawa, S.; Hasegawa, K.; Yukawa, M.; Sakashita, A.; Alavattam, K.G.; Andreassen, P.R.; Vidal, M.; Koseki, H.; Barski, A.; Namekawa, S.H. Polycomb directs timely activation of germline genes in spermatogenesis. Genes Dev. 2017, 31, 1693–1703. [Google Scholar] [CrossRef] [Green Version]
- Buczacki, S.J.; Zecchini, H.I.; Nicholson, A.M.; Russell, R.; Vermeulen, L.; Kemp, R.; Winton, D.J. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 2013, 495, 65–69. [Google Scholar] [CrossRef]
- Takeda, N.; Jain, R.; LeBoeuf, M.R.; Wang, Q.; Lu, M.M.; Epstein, J.A. Interconversion between intestinal stem cell populations in distinct niches. Science 2011, 334, 1420–1424. [Google Scholar] [CrossRef] [Green Version]
- van der Flier, L.G.; Clevers, H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Ann. Rev. Physiol. 2009, 71, 241–260. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; Bartfeld, S.; Clevers, H. Tissue-resident adult stem cell populations of rapidly self-renewing organs. Cell Stem Cell 2010, 7, 656–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pourebrahim, R.; Houtmeyers, R.; Ghogomu, S.; Janssens, S.; Thelie, A.; Tran, H.T.; Langenberg, T.; Vleminckx, K.; Bellefroid, E.; Cassiman, J.J.; et al. Transcription factor Zic2 inhibits Wnt/beta-catenin protein signaling. J. Biol. Chem. 2011, 286, 37732–37740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujimi, T.J.; Hatayama, M.; Aruga, J. Xenopus Zic3 controls notochord and organizer development through suppression of the Wnt/beta-catenin signaling pathway. Dev. Biol. 2012, 361, 220–231. [Google Scholar] [CrossRef] [Green Version]
- Krausova, M.; Korinek, V. Wnt signaling in adult intestinal stem cells and cancer. Cell Signal. 2014, 26, 570–579. [Google Scholar] [CrossRef] [Green Version]
- Chiacchiera, F.; Rossi, A.; Jammula, S.; Zanotti, M.; Pasini, D. PRC2 preserves intestinal progenitors and restricts secretory lineage commitment. EMBO J. 2016, 35, 2301–2314. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Kitano, A.; Luu, V.; Dawson, B.; Hoegenauer, K.A.; Lee, B.H.; Nakada, D. Bmi1 Suppresses Adipogenesis in the Hematopoietic Stem Cell Niche. Stem Cell Rep. 2019, 13, 545–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nitta, E.; Itokawa, N.; Yabata, S.; Koide, S.; Hou, L.B.; Oshima, M.; Aoyama, K.; Saraya, A.; Iwama, A. Bmi1 counteracts hematopoietic stem cell aging by repressing target genes and enforcing the stem cell gene signature. Biochem. Biophys. Res. Commun. 2020, 521, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Ueda, T.; Yamasaki, N.; Nakata, Y.; Sera, Y.; Nagamachi, A.; Miyama, T.; Kobayashi, H.; Takubo, K.; Kanai, A.; et al. Maintenance of the functional integrity of mouse hematopoiesis by EED and promotion of leukemogenesis by EED haploinsufficiency. Sci. Rep. 2016, 6, 29454. [Google Scholar] [CrossRef] [PubMed]
- Vizan, P.; Gutierrez, A.; Espejo, I.; Garcia-Montolio, M.; Lange, M.; Carretero, A.; Lafzi, A.; de Andres-Aguayo, L.; Blanco, E.; Thambyrajah, R.; et al. The Polycomb-associated factor PHF19 controls hematopoietic stem cell state and differentiation. Sci. Adv. 2020, 6, eabb2745. [Google Scholar] [CrossRef]
- Li, M.Y.; Flora, P.; Pu, H.; Bar, C.; Silva, J.; Cohen, I.; Galbo, P.M., Jr.; Liu, H.; Yu, X.; Jin, J.; et al. UV-induced reduction in Polycomb repression promotes epidermal pigmentation. Dev. Cell 2021. [Google Scholar] [CrossRef] [PubMed]
- Mejetta, S.; Morey, L.; Pascual, G.; Kuebler, B.; Mysliwiec, M.R.; Lee, Y.; Shiekhattar, R.; Di Croce, L.; Benitah, S.A. Jarid2 regulates mouse epidermal stem cell activation and differentiation. EMBO J. 2011, 30, 3635–3646. [Google Scholar] [CrossRef]
- Pivetti, S.; Fernandez-Perez, D.; D’Ambrosio, A.; Barbieri, C.M.; Manganaro, D.; Rossi, A.; Barnabei, L.; Zanotti, M.; Scelfo, A.; Chiacchiera, F.; et al. Loss of PRC1 activity in different stem cell compartments activates a common transcriptional program with cell type-dependent outcomes. Sci. Adv. 2019, 5, eaav1594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapthanasupkul, P.; Feng, J.; Mantesso, A.; Takada-Horisawa, Y.; Vidal, M.; Koseki, H.; Wang, L.; An, Z.; Miletich, I.; Sharpe, P.T. Ring1a/b polycomb proteins regulate the mesenchymal stem cell niche in continuously growing incisors. Dev. Biol. 2012, 367, 140–153. [Google Scholar] [CrossRef] [Green Version]
- An, Z.; Akily, B.; Sabalic, M.; Zong, G.; Chai, Y.; Sharpe, P.T. Regulation of Mesenchymal Stem to Transit-Amplifying Cell Transition in the Continuously Growing Mouse Incisor. Cell Rep. 2018, 23, 3102–3111. [Google Scholar] [CrossRef]
- Biehs, B.; Hu, J.K.; Strauli, N.B.; Sangiorgi, E.; Jung, H.; Heber, R.P.; Ho, S.; Goodwin, A.F.; Dasen, J.S.; Capecchi, M.R.; et al. BMI1 represses Ink4a/Arf and Hox genes to regulate stem cells in the rodent incisor. Nat. Cell Biol. 2013, 15, 846–852. [Google Scholar] [CrossRef]
- Yan, K.S.; Chia, L.A.; Li, X.; Ootani, A.; Su, J.; Lee, J.Y.; Su, N.; Luo, Y.; Heilshorn, S.C.; Amieva, M.R.; et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. USA 2012, 109, 466–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dun, Y.; Chen, J.; Liu, J.; Guo, Y.; Zhang, C.; Yuan, D. Changes of Wnt/beta-catenin signalling, BMP2, and BMP4 in the jejunum during ageing in rats. Arab. J. Gastroenterol. 2020, 21, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Pinto, D.; Gregorieff, A.; Begthel, H.; Clevers, H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003, 17, 1709–1713. [Google Scholar] [CrossRef] [Green Version]
- Kuhnert, F.; Davis, C.R.; Wang, H.T.; Chu, P.; Lee, M.; Yuan, J.; Nusse, R.; Kuo, C.J. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc. Natl. Acad. Sci. USA 2004, 101, 266–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, J.H.; Dimri, M.; Dimri, G.P. A positive feedback loop regulates the expression of polycomb group protein BMI1 via WNT signaling pathway. J. Biol. Chem. 2013, 288, 3406–3418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jadhav, U.; Manieri, E.; Nalapareddy, K.; Madha, S.; Chakrabarti, S.; Wucherpfennig, K.; Barefoot, M.; Shivdasani, R.A. Replicational Dilution of H3K27me3 in Mammalian Cells and the Role of Poised Promoters. Mol. Cell 2020, 78, 141–151.e145. [Google Scholar] [CrossRef] [PubMed]
- Eaves, C.J. Hematopoietic stem cells: Concepts, definitions, and the new reality. Blood 2015, 125, 2605–2613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, K.; Sedello, A.K.; Todd, G.P.; Paszkowski-Rogacz, M.; Bird, A.W.; Ding, L.; Grinenko, T.; Behrens, K.; Hubner, N.; Mann, M.; et al. Polycomb group ring finger 1 cooperates with Runx1 in regulating differentiation and self-renewal of hematopoietic cells. Blood 2012, 119, 4152–4161. [Google Scholar] [CrossRef]
- Mochizuki-Kashio, M.; Mishima, Y.; Miyagi, S.; Negishi, M.; Saraya, A.; Konuma, T.; Shinga, J.; Koseki, H.; Iwama, A. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood 2011, 118, 6553–6561. [Google Scholar] [CrossRef]
- Hidalgo, I.; Herrera-Merchan, A.; Ligos, J.M.; Carramolino, L.; Nunez, J.; Martinez, F.; Dominguez, O.; Torres, M.; Gonzalez, S. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell 2012, 11, 649–662. [Google Scholar] [CrossRef] [Green Version]
- Cales, C.; Roman-Trufero, M.; Pavon, L.; Serrano, I.; Melgar, T.; Endoh, M.; Perez, C.; Koseki, H.; Vidal, M. Inactivation of the polycomb group protein Ring1B unveils an antiproliferative role in hematopoietic cell expansion and cooperation with tumorigenesis associated with Ink4a deletion. Mol. Cell Biol. 2008, 28, 1018–1028. [Google Scholar] [CrossRef] [Green Version]
- Di Carlo, V.; Mocavini, I.; Di Croce, L. Polycomb complexes in normal and malignant hematopoiesis. J. Cell Biol. 2019, 218, 55–69. [Google Scholar] [CrossRef] [Green Version]
- Oguro, H.; Iwama, A.; Morita, Y.; Kamijo, T.; van Lohuizen, M.; Nakauchi, H. Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice. J. Exp. Med. 2006, 203, 2247–2253. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, J.J.; Kieboom, K.; Marino, S.; DePinho, R.A.; van Lohuizen, M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 1999, 397, 164–168. [Google Scholar] [CrossRef]
- Nakamura, S.; Oshima, M.; Yuan, J.; Saraya, A.; Miyagi, S.; Konuma, T.; Yamazaki, S.; Osawa, M.; Nakauchi, H.; Koseki, H.; et al. Bmi1 confers resistance to oxidative stress on hematopoietic stem cells. PLoS ONE 2012, 7, e36209. [Google Scholar] [CrossRef] [Green Version]
- Sacchetti, B.; Funari, A.; Michienzi, S.; Di Cesare, S.; Piersanti, S.; Saggio, I.; Tagliafico, E.; Ferrari, S.; Robey, P.G.; Riminucci, M.; et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell 2007, 131, 324–336. [Google Scholar] [CrossRef] [Green Version]
- Frisch, B.J.; Porter, R.L.; Calvi, L.M. Hematopoietic niche and bone meet. Curr. Opin. Support. Palliat. Care 2008, 2, 211–217. [Google Scholar] [CrossRef] [Green Version]
- Iwama, A. polycomb repressive complexes in hematological malignancies. Blood 2017, 130, 23–29. [Google Scholar] [CrossRef] [Green Version]
- San Roman, A.K.; Tovaglieri, A.; Breault, D.T.; Shivdasani, R.A. Distinct Processes and Transcriptional Targets Underlie CDX2 Requirements in Intestinal Stem Cells and Differentiated Villus Cells. Stem Cell Rep. 2015, 5, 673–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doynova, M.D.; Markworth, J.F.; Cameron-Smith, D.; Vickers, M.H.; O’Sullivan, J.M. Linkages between changes in the 3D organization of the genome and transcription during myotube differentiation in vitro. Skelet. Muscle 2017, 7, 5. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; He, F.; Xiong, W.; Gu, S.; Liu, H.; Zhang, T.; Yu, X.; Chen, Y. Polycomb-like-2-deficient mice exhibit normal left-right asymmetry. Dev. Dyn. 2007, 236, 853–861. [Google Scholar] [CrossRef]
- Fuchs, E. Epithelial Skin Biology: Three Decades of Developmental Biology, a Hundred Questions Answered and a Thousand New Ones to Address. Curr. Top. Dev. Biol. 2016, 116, 357–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miroshnikova, Y.A.; Cohen, I.; Ezhkova, E.; Wickstrom, S.A. Epigenetic gene regulation, chromatin structure, and force-induced chromatin remodelling in epidermal development and homeostasis. Curr. Opin. Genet. Dev. 2019, 55, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Chovatiya, G.; Tumbar, T. Epigenetic control in skin development, homeostasis and injury repair. Exp. Derm. 2019, 28, 453–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ezhkova, E.; Lien, W.H.; Stokes, N.; Pasolli, H.A.; Silva, J.M.; Fuchs, E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011, 25, 485–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mardaryev, A.N.; Liu, B.; Rapisarda, V.; Poterlowicz, K.; Malashchuk, I.; Rudolf, J.; Sharov, A.A.; Jahoda, C.A.; Fessing, M.Y.; Benitah, S.A.; et al. Cbx4 maintains the epithelial lineage identity and cell proliferation in the developing stratified epithelium. J. Cell Biol. 2016, 212, 77–89. [Google Scholar] [CrossRef] [Green Version]
- Rompolas, P.; Greco, V. Stem cell dynamics in the hair follicle niche. Semin. Cell Dev. Biol. 2014, 25-26, 34–42. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.A.; Li, K.N.; Tumbar, T. Stem cell-intrinsic mechanisms regulating adult hair follicle homeostasis. Exp. Derm. 2021, 30, 430–447. [Google Scholar] [CrossRef]
- Fuchs, E. Skin Stem Cells in Silence, Action, and Cancer. Stem Cell Rep. 2018, 10, 1432–1438. [Google Scholar] [CrossRef] [PubMed]
- Jaks, V.; Barker, N.; Kasper, M.; van Es, J.H.; Snippert, H.J.; Clevers, H.; Toftgard, R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 2008, 40, 1291–1299. [Google Scholar] [CrossRef]
- Hoeck, J.D.; Biehs, B.; Kurtova, A.V.; Kljavin, N.M.; de Sousa, E.M.F.; Alicke, B.; Koeppen, H.; Modrusan, Z.; Piskol, R.; de Sauvage, F.J. Stem cell plasticity enables hair regeneration following Lgr5+ cell loss. Nat. Cell Biol. 2017, 19, 666–676. [Google Scholar] [CrossRef]
- Haegebarth, A.; Clevers, H. Wnt signaling, lgr5, and stem cells in the intestine and skin. Am. J. Pathol. 2009, 174, 715–721. [Google Scholar] [CrossRef] [Green Version]
- Lien, W.H.; Guo, X.; Polak, L.; Lawton, L.N.; Young, R.A.; Zheng, D.; Fuchs, E. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell 2011, 9, 219–232. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Kang, S.; Lilja, K.C.; Colletier, K.J.; Scheitz, C.J.; Zhang, Y.V.; Tumbar, T. Signalling couples hair follicle stem cell quiescence with reduced histone H3 K4/K9/K27me3 for proper tissue homeostasis. Nat. Commun. 2016, 7, 11278. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.; Long, K.; Wang, S.; Sada, A.; Tumbar, T. Histone H3 K4/9/27 Trimethylation Levels Affect Wound Healing and Stem Cell Dynamics in Adult Skin. Stem Cell Rep. 2020, 14, 34–48. [Google Scholar] [CrossRef] [Green Version]
- Graziadei, G.A.; Graziadei, P.P. Neurogenesis and neuron regeneration in the olfactory system of mammals. II. Degeneration and reconstitution of the olfactory sensory neurons after axotomy. J. Neurocytol. 1979, 8, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Sokpor, G.; Abbas, E.; Rosenbusch, J.; Staiger, J.F.; Tuoc, T. Transcriptional and Epigenetic Control of Mammalian Olfactory Epithelium Development. Mol. Neurobiol. 2018, 55, 8306–8327. [Google Scholar] [CrossRef]
- Schwob, J.E. Neural regeneration and the peripheral olfactory system. Anat. Rec. 2002, 269, 33–49. [Google Scholar] [CrossRef]
- Morrison, E.E.; Costanzo, R.M. Morphology of olfactory epithelium in humans and other vertebrates. Microsc. Res. Tech. 1992, 23, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Choi, R.; Goldstein, B.J. Olfactory epithelium: Cells, clinical disorders, and insights from an adult stem cell niche. Laryngoscope Investig. Otolaryngol. 2018, 3, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Calof, A.L.; Bonnin, A.; Crocker, C.; Kawauchi, S.; Murray, R.C.; Shou, J.; Wu, H.H. Progenitor cells of the olfactory receptor neuron lineage. Microsc. Res. Tech. 2002, 58, 176–188. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, B.J.; Goss, G.M.; Choi, R.; Saur, D.; Seidler, B.; Hare, J.M.; Chaudhari, N. Contribution of Polycomb group proteins to olfactory basal stem cell self-renewal in a novel c-KIT+ culture model and in vivo. Development 2016, 143, 4394–4404. [Google Scholar] [CrossRef] [Green Version]
- Krolewski, R.C.; Packard, A.; Jang, W.; Wildner, H.; Schwob, J.E. Ascl1 (Mash1) knockout perturbs differentiation of nonneuronal cells in olfactory epithelium. PLoS ONE 2012, 7, e51737. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, B.J.; Goss, G.M.; Hatzistergos, K.E.; Rangel, E.B.; Seidler, B.; Saur, D.; Hare, J.M. Adult c-Kit+ progenitor cells are necessary for maintenance and regeneration of olfactory neurons. J. Comp. Neurol. 2015, 523, 15–31. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, B.J.; Choi, R.; Goss, G.M. Multiple polycomb epigenetic regulatory proteins are active in normal and regenerating adult olfactory epithelium. Laryngoscope Investig. Otolaryngol. 2018, 3, 337–344. [Google Scholar] [CrossRef]
- Laflamme, M.A.; Murry, C.E. Heart regeneration. Nature 2011, 473, 326–335. [Google Scholar] [CrossRef] [Green Version]
- Herrero, D.; Canon, S.; Pelacho, B.; Salvador-Bernaldez, M.; Aguilar, S.; Pogontke, C.; Carmona, R.M.; Salvador, J.M.; Perez-Pomares, J.M.; Klein, O.D.; et al. Bmi1-Progenitor Cell Ablation Impairs the Angiogenic Response to Myocardial Infarction. Arter. Thromb. Vasc. Biol. 2018, 38, 2160–2173. [Google Scholar] [CrossRef] [Green Version]
- Beltrami, A.P.; Barlucchi, L.; Torella, D.; Baker, M.; Limana, F.; Chimenti, S.; Kasahara, H.; Rota, M.; Musso, E.; Urbanek, K.; et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 2003, 114, 763–776. [Google Scholar] [CrossRef] [Green Version]
- Senyo, S.E.; Steinhauser, M.L.; Pizzimenti, C.L.; Yang, V.K.; Cai, L.; Wang, M.; Wu, T.D.; Guerquin-Kern, J.L.; Lechene, C.P.; Lee, R.T. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 2013, 493, 433–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valiente-Alandi, I.; Albo-Castellanos, C.; Herrero, D.; Arza, E.; Garcia-Gomez, M.; Segovia, J.C.; Capecchi, M.; Bernad, A. Cardiac Bmi1+ cells contribute to myocardial renewal in the murine adult heart. Stem Cell Res. Ther. 2015, 6, 205. [Google Scholar] [CrossRef] [Green Version]
- Valiente-Alandi, I.; Albo-Castellanos, C.; Herrero, D.; Sanchez, I.; Bernad, A. Bmi1+ cardiac progenitor cells contribute to myocardial repair following acute injury. Stem Cell Res. Ther. 2016, 7, 100. [Google Scholar] [CrossRef] [Green Version]
- Sharpe, P.T. Dental mesenchymal stem cells. Development 2016, 143, 2273–2280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, D.C.; Shyu, W.C.; Lin, S.Z. Mesenchymal stem cells. Cell Transpl. 2011, 20, 5–14. [Google Scholar] [CrossRef] [Green Version]
- Yu, T.; Volponi, A.A.; Babb, R.; An, Z.; Sharpe, P.T. Stem Cells in Tooth Development, Growth, Repair, and Regeneration. Curr. Top. Dev. Biol. 2015, 115, 187–212. [Google Scholar] [CrossRef] [PubMed]
- Sedgley, C.M.; Botero, T.M. Dental stem cells and their sources. Dent. Clin. N. Am. 2012, 56, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Pecina-Slaus, N. Wnt signal transduction pathway and apoptosis: A review. Cancer Cell Int. 2010, 10, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vizan, P.; Beringer, M.; Di Croce, L. Polycomb-dependent control of cell fate in adult tissue. EMBO J. 2016, 35, 2268–2269. [Google Scholar] [CrossRef] [Green Version]
- Brand, M.; Nakka, K.; Zhu, J.; Dilworth, F.J. Polycomb/Trithorax Antagonism: Cellular Memory in Stem Cell Fate and Function. Cell Stem Cell 2019, 24, 518–533. [Google Scholar] [CrossRef] [Green Version]
- Aloia, L.; Di Stefano, B.; Di Croce, L. Polycomb complexes in stem cells and embryonic development. Development 2013, 140, 2525–2534. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Komai, Y.; Tokuyama, Y.; Yanai, H.; Ohe, S.; Okazaki, K.; Ueno, H. Identification of stem cells that maintain and regenerate lingual keratinized epithelial cells. Nat. Cell Biol. 2013, 15, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Hugues, A.; Jacobs, C.S.; Roudier, F. Mitotic Inheritance of PRC2-Mediated Silencing: Mechanistic Insights and Developmental Perspectives. Front. Plant. Sci. 2020, 11, 262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciabrelli, F.; Comoglio, F.; Fellous, S.; Bonev, B.; Ninova, M.; Szabo, Q.; Xuereb, A.; Klopp, C.; Aravin, A.; Paro, R.; et al. Stable Polycomb-dependent transgenerational inheritance of chromatin states in Drosophila. Nat. Genet. 2017, 49, 876–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cales, C.; Pavon, L.; Starowicz, K.; Perez, C.; Bravo, M.; Ikawa, T.; Koseki, H.; Vidal, M. Role of Polycomb RYBP in Maintaining the B-1-to-B-2 B-Cell Lineage Switch in Adult Hematopoiesis. Mol. Cell Biol. 2015, 36, 900–912. [Google Scholar] [CrossRef] [Green Version]
- Sauvageau, M.; Sauvageau, G. Polycombpolycomb group proteins: Multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 2010, 7, 299–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koppens, M.; van Lohuizen, M. Context-dependent actions of Polycomb repressors in cancer. Oncogene 2016, 35, 1341–1352. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.L.; Morey, L. Emerging Roles for Polycomb-Group Proteins in Stem Cells and Cancer. Trends Biochem. Sci. 2019, 44, 688–700. [Google Scholar] [CrossRef] [PubMed]
| Tissue Studied | Targeted Gene | Mouse Model Used | Observed Phenotypes | Biological Process Affected | References |
|---|---|---|---|---|---|
| Intestine | Ring1a/b | Ring1a−/−;Ring1bfl/fl; LGR5-eGFP-IRES-CreERT2 | lower body weight, thinner intestine, defect in crypt architecture | reduced self-renewal and proliferation of intestinal stem cells, expression of non-lineage transcription factors | Chiacchiera et al., 2016 [22] |
| Eed | Eedfl/fl; AhCre | alteration in crypt-villus architecture, expansion of goblet cell and enteroendocrine cells | reduced proliferation of transit amplifying cells, lowered secretory lineage commitment | Chiacchiera et al., 2016 [157] | |
| Eed | Eedfl/fl; AhCre | lower body weight, extensive crypt and microcystic degeneration, necrosis, hypertrophy, and hyperplasia of crypts | reduced proliferation of intestinal crypt cells | Koppens et al., 2016 [20] | |
| Bone marrow stromal cells | Bmi1 | Bmi1fl/fl; Prx1-Cre | accumulation of bone marrow adipocytes, reduction in bone marrow cellularity | increased senescence, depletion of HSCs and progenitor cells, increased adipogenic differentiation | Hu et al., 2019 [158] |
| Hematopoietic cells | Bmi1 | Vav-Cre;Rosa26StopFLBmi1 (over-expression of Bmi1) | reduction in age-related anemia | attenuated age-related HSC function, maintenance of HSC signature gene expression | Nitta et al., 2020 [159] |
| Eed | Eedfl/fl; CreERT2 | thymic and splenic atrophy, pale bone marrow, hematopoietic dysplasia | reduction in all hematopoietic cellular lineages, abnormal cell cycle, upregulation of adhesion molecule genes | Ikeda et al., 2016 [160] | |
| Phf19 | Phf19−/− | high penetrance of splenomegaly | upregulation of retinoic acid pathway, downregulation of Myc network and genes related to biosynthesis and energy production | Vizán et al., 2020 [161] | |
| Interfollicular epidermis | Eed and Ring1a/b | Eedfl/fl; K14-CreERT2 and Ring1a−/−;Ring1bfl/fl; K14-CreERT2 | epidermal pigmentation | upregulation of UV-responsive genes in EpSCs, induction of COL2A1 expression which promotes epidermal pigmentation | Li et al., 2021 [162] |
| Interfollicular epidermis and hair follicle | Jarid2 | Jarid2fl/fl; K14CreYFP::Rosa26 | defective epidermal thickness and delayed hair cycle | reduced proliferation of basal cells of the epidermis and hair germ cells of the hair follicle, enhanced differentiation of basal cells | Mejetta et al., 2011 [163] |
| Hair follicle | Ring1a/b | Ring1a−/−;Ring1bfl/fl; LGR5-eGFP-IRES-CreERT2 | arrested hair follicle growth | defective Lgr5+ HFSC differentiation due to upregulation of non-lineage genes | Pivetti et al., 2019 [164] |
| Incisors | Ring1a/b | Ring1a−/−;Ring1bfl/fl; Rosa26::CreERT2 | Defective enamel and dentin formation | Reduced proliferation of mesenchymal transit amplifying cells | Lapthanasupkul et al., 2012 [165], An et al., 2018 [166] |
| Bmi1 | Bmi1GFP/GFP (Bmi1-null mice) | defective enamel production | fewer stem cells due to upregulation of Ink4a/Arf, upregulation of Hox genes leading to premature differentiation and loss of stem cell population | Biehs et al., 2013 [167] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flora, P.; Dalal, G.; Cohen, I.; Ezhkova, E. Polycomb Repressive Complex(es) and Their Role in Adult Stem Cells. Genes 2021, 12, 1485. https://doi.org/10.3390/genes12101485
Flora P, Dalal G, Cohen I, Ezhkova E. Polycomb Repressive Complex(es) and Their Role in Adult Stem Cells. Genes. 2021; 12(10):1485. https://doi.org/10.3390/genes12101485
Chicago/Turabian StyleFlora, Pooja, Gil Dalal, Idan Cohen, and Elena Ezhkova. 2021. "Polycomb Repressive Complex(es) and Their Role in Adult Stem Cells" Genes 12, no. 10: 1485. https://doi.org/10.3390/genes12101485





