Low Temperature Antioxidant Activity QTL Associate with Genomic Regions Involved in Physiological Cold Stress Tolerance Responses in Rice (Oryza sativa L.)
Abstract
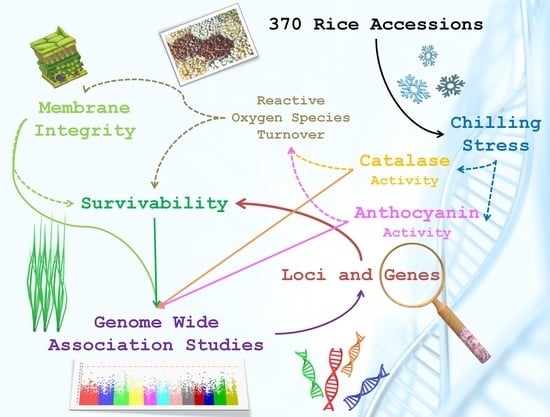
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Growth and Stress Conditions
2.3. Measurement of Electrolyte Leakage (EL)
2.4. Measurement of Catalase Activity (CAT)
2.5. Measurement of Anthocyanin Activity (ANT)
2.6. Measurement of Low Temperature Seedling Survivability (LTSS)
2.7. Genome-Wide Association Study (GWAS)-Based Quantitative Trait Locus (QTL) Mapping
2.8. QTL Mapping
2.9. Gene Identification and Gene Ontology (GO) Term Analysis
2.10. Correlation Analysis and Statistical Analysis
3. Results
3.1. Summary of Low Temperature Seedling Survivability (LTSS) Results
3.2. Summary of Electrolyte Leakage (EL) Results
3.3. Summary of Catalase (CAT) Activity Results
3.4. Summary of Anthocyanin (ANT) Activity Results
3.5. Correlations between LTSS, EL, CAT Activity, and ANT Activity after Low Temperature Treatment
3.5.1. Correlation between EL and LTSS
3.5.2. Correlation between CAT Activity and LTSS
3.5.3. Correlation between ANT Activity and LTSS
3.5.4. Correlation between EL and CAT Activity
3.5.5. Correlation between ANT Activity and EL
3.5.6. Correlation between CAT Activity and ANT Activity
3.6. Genome-Wide Association Study (GWAS)-Based Mapping of Four Phenotypes
3.7. GO Term Analyses of Annotated Genes within Multiple-Phenotype QTL
4. Discussion
4.1. Cold Stress Tolerance Scores of Rice Accessions Cluster with Their Respective Subgroups
4.2. Genome-Wide Association Study (GWAS)-Based Mapping of Four Cold Tolerance Traits
4.3. Gene Ontology (GO) Term Analysis of Genes Found in 20 Multiple-Phenotype QTL
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shi, Y.; Guo, E.; Wang, L.; Li, T.; Jiang, S.; Xiang, H.; Zhang, T.; Cheng, X.; Xhu, X.; Zhou, L. Effects of chilling at the booting and flowering stages on rice phenology and yield: A case study in Northeast China. J. Agron. Crop. Sci. 2021, 1–12. [Google Scholar]
- Arshad, M.S.; Farooq, M.; Asch, F.; Krishna, J.S.V.; Prasad, P.V.V.; Siddique, K.H.M. Thermal stress impacts reproductive development and grain yield in rice. Plant Physiol. Biochem. 2017, 115, 57–72. [Google Scholar] [CrossRef]
- Bodirsky, B.L.; Rolinski, S.; Biewald, A.; Weindl, I.; Popp, A.; Lotze-Campen, H. Global food demand scenarios for the 21st century. PLoS ONE 2015, 10, e0139201. [Google Scholar] [CrossRef] [PubMed]
- Bajzelj, B.; Richards, K.S.; Allwood, J.M.; Smith, P.; Dennis, J.S.; Curmi, E.; Gilligan, C.A. Importance of food-demand management for climate mitigation. Nat. Clim. Chang. 2014, 4, 924–929. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Li, Y.; Zhu, J.-K. Developing naturally stress-resistant crops for a sustainable agriculture. Nat. Plants 2018, 4, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Gambhir, G.; Dass, A.; Tripathi, A.K.; Singh, A.; Jha, A.K.; Yadava, P.; Choudhary, M.; Rakshit, S. Genetically modified crops: Current status and future prospects. Planta 2020, 251, 1–27. [Google Scholar] [CrossRef]
- Wu, X.; Ning, F.; Hu, X.; Wang, W. Genetic modification for improving seed vigor is transitioning from model plants to crop plants. Front. Plant Sci. 2017, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Curr. Genom. 2011, 12, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, L.; DaCosta, M.; Ebdon, J.S.; Watkins, E. Physiological changes during cold acclimation of perennial ryegrass accessions differing in freeze tolerance. Crop. Sci. 2010, 50, 1037–1047. [Google Scholar] [CrossRef]
- Ritonga, F.N.; Su, C. Physiological and molecular mechanism involved in cold stress tolerance in plants. Plants 2020, 9, 560. [Google Scholar] [CrossRef]
- Shi, Y.; Phan, H.; Liu, Y.; Cao, S.; Zhang, Z.; Chu, C.; Schläppi, M.R. Glycosyltransferase OsUGT90A1 helps protect the plasma membrane during chilling stress in rice. J. Exp. Bot. 2020, 71, 2723–2739. [Google Scholar] [CrossRef]
- Takahashi, D.; Li, B.; Nakayama, T.; Kawamura, Y.; Uemura, U. Plant plasma membrane proteomics for improving cold tolerance. Front. Plant Sci. 2013, 4, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrero-Sicilia, C.; Silvestre, S.; Haslam, R.P.; Michaelson, L.V. Lipid remodelling: Unravelling the response to cold stress in Arabidopsis and its extremophile relative Eutrema salsugineum. Plant Sci. 2017, 263, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Aryadeep, R. Abiotic stress, generation of reactive oxygen species, and their consequences: An overview. In Reactive Oxygen Species in Plants: Boon or Bane? Wiley: Hoboken, NJ, USA, 2018; pp. 23–50. [Google Scholar]
- Dreyer, A.; Karl-Josef, D. Reactive oxygen species and the redox-regulatory network in cold stress acclimation. Antioxidants 2018, 7, 169. [Google Scholar] [CrossRef] [Green Version]
- Gupta, D.K.; José, M.P.; Francisco, J.C. (Eds.) Reactive Oxygen Species and Oxidative Damage in Plants under Stress; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Caverzan, A.; Alice, C.; Sandra, P.B. Reactive oxygen species and antioxidant enzymes involved in plant tolerance to stress. SHANKER AK & SHANKER C. Abiotic and Biotic Stress in Plants-Recent Advances and Future Perspectives; InTechOpen Ltd.: London, UK, 2016; pp. 463–480. [Google Scholar]
- Caverzan, A.; Passaia, G.; Barcellos, S.; Carolina, R.; Ribeiro, W.; Lazzarotto, F.; Margis-Pinheiro, M. Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 2012, 35, 1011–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bela, K.; Horváth, E.; Gallé, Á.; Szabados, L.; Tari, I.; Csiszár, J. Plant glutathione peroxidases: Emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 2015, 176, 192–201. [Google Scholar] [CrossRef]
- Mhamdi, A.; Queval, G.; Chaouch, S.; Vanderauwera, S.; Van Breusegem, F.; Noctor, G. Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 2010, 61, 4197–4220. [Google Scholar] [CrossRef] [Green Version]
- Alam, N.B.; Ajit, G. Comprehensive analysis and transcript profiling of Arabidopsis thaliana and Oryza sativa catalase gene family suggests their specific roles in development and stress responses. Plant Physiol. Biochem. 2018, 123, 54–64. [Google Scholar] [CrossRef]
- Vighi, I.L.; Benitez, L.C.; do Amaral, M.N.; Auler, P.A.; Moraes, G.P.; Rodrigues, G.S.; Da Maia, L.C.; Pinto, L.S.; Braga, E.J. Changes in gene expression and catalase activity in Oryza sativa L. under abiotic stress. Genet. Mol. Res. 2016, 15, 1–15. [Google Scholar] [CrossRef]
- Azpilicueta, C.E.; Pena, L.B.; Tomaro, M.L.; Gallego, S.M. Modifications in catalase activity and expression in developing sunflower seedlings under cadmium stress. Redox Rep. 2008, 13, 40–46. [Google Scholar] [CrossRef]
- Moriwaki, T.; Yamamoto, Y.; Aida, T.; Funahashi, T.; Shishido, T.; Asada, M.; Prodhan, S.H.; Komamine, A.; Motohashi, T. Overexpression of the Escherichia coli catalase gene, katE, enhances tolerance to salinity stress in the transgenic indica rice cultivar, BR5. Plant Biotechnol. Rep. 2008, 2, 41–46. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: A review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Landi, M.; Tattini, M.; Kevin, S.G. Multiple functional roles of anthocyanins in plant-environment interactions. Environ. Exp. Bot. 2015, 119, 4–17. [Google Scholar] [CrossRef]
- Goufo, P.; Henrique, T. Rice antioxidants: Phenolic acids, flavonoids, anthocyanins, proanthocyanidins, tocopherols, tocotrienols, γ-oryzanol, and phytic acid. Food Sci. Nutr. 2014, 2, 75–104. [Google Scholar] [CrossRef]
- Kovinich, N.; Kayanja, G.; Chanoca, A.; Riedl, K.; Otegui, M.S.; Grotewold, E. Not all anthocyanins are born equal: Distinct patterns induced by stress in Arabidopsis. Planta 2014, 240, 931–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, W.; Jia, Q.; Wang, Y.; Zhang, Y.; Xia, M. The anthocyanin cyanidin-3-O-β-glucoside, a flavonoid, increases hepatic glutathione synthesis and protects hepatocytes against reactive oxygen species during hyperglycemia: Involvement of a cAMP–PKA-dependent signaling pathway. Free. Radic. Biol. Med. 2012, 52, 314–327. [Google Scholar] [CrossRef]
- Zilic, S. Phenolic compounds of wheat. Their content, antioxidant capacity and bioaccessibility. MOJ Food Process. Technol. 2016, 2, 00037. [Google Scholar] [CrossRef]
- Chutipaijit, S.; Suriyan, C.; Kanokporn, S. High contents of proline and anthocyanin increase protective response to salinity in ‘Oryza sativa’ L. spp. ‘indica’. Aust. J. Crop. Sci. 2011, 5, 1191–1198. [Google Scholar]
- Kovinich, N.; Kayanja, G.; Chanoca, A.; Riedl, K.; Otegui, M.S.; Grotewold, E. Abiotic stresses induce different localizations of anthocyanins in Arabidopsis. Plant Signal. Behav. 2015, 10, e1027850. [Google Scholar] [CrossRef] [Green Version]
- Tisarum, R.; Theerawitaya, C.; Samphumphuang, T.; Cha-um, S. Regulation of anthocyanin accumulation in rice (Oryza sativa L. subsp. indica) using MgSO4 spraying and low temperature. Arch. Agron. Soil Sci. 2018, 64, 1663–1677. [Google Scholar] [CrossRef]
- Zhu, J.-J.; Li, Y.-R.; Liao, J.-X. Involvement of anthocyanins in the resistance to chilling-induced oxidative stress in Saccharum officinarum L. leaves. Plant Physiol. Biochem. 2013, 73, 427–433. [Google Scholar] [CrossRef]
- Zhao, K.; Wright, M.; Kimball, J.; Eizenga, G.; McClung, A.; Kovach, M.; Tyagi, W.; Ali, M.L.; Tung, C.-W.; Reynolds, A.; et al. Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLoS ONE 2010, 5, e10780. [Google Scholar]
- Agrama, H.A.; Eizenga, G.C.; Yan, W.E. Association mapping of yield and its components in rice cultivars. Mol. Breed. 2007, 19, 341–356. [Google Scholar] [CrossRef]
- Schläppi, M.R.; Jackson, A.K.; Eizenga, G.C.; Wang, A.; Chu, C.; Shi, Y.; Shimoyama, S.; Boykin, D.L. Assessment of five chilling tolerance traits and GWAS mapping in rice using the USDA mini-core collection. Front. Plant Sci. 2017, 8, 957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCouch, S.R.; Wright, M.H.; Tung, C.-W.; Maron, L.G.; McNally, K.L.; Fitzgerald, M.; Namrata Singh, N.; DeClerck, G.; Agosto-Perez, F.; Korniliev, P.; et al. Open access resources for genome-wide association mapping in rice. Nat. Commun. 2016, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spindel, J.E.; Begum, H.; Akdemir, D.; Collard, B.; Redoña, E.; Jannink, J.L.; McCouch, S. Genome-wide prediction models that incorporate de novo GWAS are a powerful new tool for tropical rice improvement. Heredity 2016, 116, 395–408. [Google Scholar] [CrossRef] [Green Version]
- Shimoyama, N.; Johnson, M.; Beaumont, A.; Schläppi, M. Multiple cold tolerance trait phenotyping reveals shared quantitative trait loci in Oryza sativa. Rice 2020, 13, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Singh, A.; Mithra, S.V.M.; Krishnamurthy, S.L.; Parida, S.K.; Jain, S.; Tiwari, K.K.; Kumar, P.; Rao, A.R.; Sharma, S.K.; et al. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Res. 2015, 22, 133–145. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Guo, Z.; Lv, Y.; Xiang Cen, X.; Ding, X.; Wu, H.; Li, X.; Huang, J.; Xiong, L. Genetic control of the root system in rice under normal and drought stress conditions by genome-wide association study. PLoS Genet. 2017, 13, e1006889. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Wang, D.; Peng, S.; Chen, Y.; Su, P.; Chen, J.; Zheng, L.; Tan, X.; Liu, J.; Xiao, Y.; et al. Genome-wide association mapping of resistance against rice blast strains in South China and identification of a new Pik allele. Rice 2019, 12, 1–9. [Google Scholar] [CrossRef]
- Eizenga, G.C.; Ali, M.L.; Bryant, R.B.; Yeater, K.M.; McClung, A.M.; McCouch, S.R. Registration of the rice diversity panel 1 for genome wide association studies. J. Plant Regist. 2014, 8, 109–116. [Google Scholar] [CrossRef]
- Chen, H.; Xie, W.; He, H.; Yu, H.; Chen, W.; Li, J.; Yu, R.; Yao, Y.; Zhang, W.; He, Y.; et al. A high-density SNP genotyping array for rice biology and molecular breeding. Mol. Plant 2014, 7, 541–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, Y.; Guo, Z.; Li, X.; Ye, H.; Li, X.; Xiong, L. New insights into the genetic basis of natural chilling and cold shock tolerance in rice by genome-wide association analysis. Plant Cell Environ. 2016, 39, 556–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Liu, J.; Li, C.; Kang, H.; Wang, Y.; Tan, X.; Liu, M.; Deng, Y.; Wang, Z.; Liu, Y.; et al. Genome-wide association mapping of cold tolerance genes at the seedling stage in rice. Rice 2016, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.-I.; Thomas, H.T. Evaluation of seedling cold tolerance in rice cultivars: A comparison of visual ratings and quantitative indicators of physiological changes. Euphytica 2011, 178, 437–447. [Google Scholar] [CrossRef]
- Kazemi-Shahandashti, S.-S.; Reza, M.-A. Global insights of protein responses to cold stress in plants: Signaling, defence, and degradation. J. Plant Physiol. 2018, 226, 123–135. [Google Scholar] [CrossRef]
- Wang, X.; Fang, G.; Li, Y.; Ding, M.; Gong, H.; Li, Y. Differential antioxidant responses to cold stress in cell suspension cultures of two subspecies of rice. Plant Cell Tissue Organ Cult. (PCTOC) 2013, 113, 353–361. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Nayak, D.K.; Behera, L.; Mohapatra, T. Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-Seq analysis. BMC Plant Biol. 2019, 19, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Chawade, A.; Lindlöf, A.; Olsson, B.; Olsson, O. Global expression profiling of low temperature induced genes in the chilling tolerant japonica rice Jumli Marshi. PLoS ONE 2013, 8, e81729. [Google Scholar] [CrossRef]
- Shen, C.; Li, D.; He, R.; Fang, Z.; Xia, Y.; Gao, J.; Shen, H.; Cao, M. Comparative transcriptome analysis of RNA-seq data for cold-tolerant and cold-sensitive rice genotypes under cold stress. J. Plant Biol. 2014, 57, 337–348. [Google Scholar] [CrossRef]
- Zhang, J.; Luo, W.; Zhao, Y.; Xu, Y.; Song, S.; Chong, K. Comparative metabolomic analysis reveals a reactive oxygen species-dominated dynamic model underlying chilling environment adaptation and tolerance in rice. New Phytol. 2016, 211, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, Q.; Wang, S.; Hong, Y.; Wang, Z. Rice and cold stress: Methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice 2014, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, B.; Ma, X.; Cui, D.; Wang, Y.; Geng, L.; Cao, G.; Zhang, H.; Han, L. Comprehensive evaluation and analysis of the mechanism of cold tolerance based on the transcriptome of weedy rice seedlings. Rice 2020, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.; Campbell, M.; Childs, K.; Thibaud-Nissen, F.; Malek, R.L.; Lee, Y.; Zheng, L.; et al. The TIGR Rice Genome Annotation Resource: Improvements and new features. Nucleic Acids Res. 2007, 35, D883–D887. [Google Scholar] [CrossRef] [Green Version]
- Yao, W.; Li, G.; Yu, Y.; Ouyang, Y. funRiceGenes dataset for comprehensive understanding and application of rice functional genes. Gigascience 2018, 7, gix119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Li, J.; Pan, Y.; Li, J.; Shi, H.; Zeng, Y.; Guo, H.; Yang, S.; Zheng, W.; Yu, J.; et al. Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Saito, K.; Hayano-Saito, Y.; Maruyama-Funatsuki, W.; Sato, Y.; Kato, A. Physical mapping and putative candidate gene identification of a quantitative trait locus Ctb1 for cold tolerance at the booting stage of rice. Theor. Appl. Genet. 2004, 109, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Hayano-Saito, Y.; Kuroki, M.; Sato, Y. Map-based cloning of the rice cold tolerance gene Ctb1. Plant Sci. 2010, 179, 97–102. [Google Scholar] [CrossRef]
- Lu, C.-A.; Huang, C.-K.; Huang, W.-S.; Huang, T.-S.; Liu, H.-Y.; Chen, Y.-F. DEAD-box RNA helicase 42 plays a critical role in pre-mRNA splicing under cold stress. Plant Physiol. 2020, 182, 255–271. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.-J.; Fu, S.-F.; Tai, Y.-H.; Chou, W.-C.; Huang, D.-D. Expression of Oryza sativa MAP kinase gene is developmentally regulated and stress-responsive. Physiol. Plant. 2002, 114, 572–580. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, G.K.; Randeep, R.; Hitoshi, I. Isolation of novel rice (Oryza sativa L.) multiple stress responsive MAP kinase gene, OsMSRMK2, whose mRNA accumulates rapidly in response to environmental cues. Biochem. Biophys. Res. Commun. 2002, 294, 1009–1016. [Google Scholar] [CrossRef]
- Xie, G.; Hideki, K.; Ryozo, I. Biochemical identification of the OsMKK6–OsMPK3 signalling pathway for chilling stress tolerance in rice. Biochem. J. 2012, 443, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ni, L.; Liu, Y.; Wang, Y.; Zhang, A.; Tan, M.; Jiang, M. The C2H2-type Zinc Finger Protein ZFP182 is Involved in Abscisic Acid-Induced Antioxidant Defense in Rice, F.J. Integr. Plant Biol. 2012, 54, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Cai, L.; Chen, Z.; Wang, R.; Zhang, L.; Guan, S.; Zhang, S.; Ma, W.; Liu, C.; Pan, G. Identification of candidate genes controlling chilling tolerance of rice in the cold region at the booting stage by BSA-Seq and RNA-Seq. R. Soc. Open Sci. 2020, 7, 201081. [Google Scholar] [CrossRef]
- Ma, Q.; Dai, X.; Xu, Y.; Guo, J.; Liu, Y.; Chen, N.; Xiao, J.; Zhang, D.; Xu, Z.; Zhang, X.; et al. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 2009, 150, 244–256. [Google Scholar] [CrossRef]
- Dai, X.; Xu, Y.; Ma, Q.; Xu, W.; Wang, T.; Xue, Y.; Chong, K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007, 143, 1739–1751. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Li, Q.-F.; Zhang, J.-P.; Zhang, F.; Zhou, Y.-F.; Feng, Y.-Z.; Chen, Y.-Q.; Zhang, Y.-C. Laccase-13 regulates seed setting rate by affecting hydrogen peroxide dynamics and mitochondrial integrity in rice. Front. Plant Sci. 2017, 8, 1324. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, B.; Zhang, Z.; Luo, W.; Tang, Y.; Niu, Y.; Chong, K.; Xu, Y. OsGRF6 interacts with SLR1 to regulate OsGA2ox1 expression for coordinating chilling tolerance and growth in rice. J. Plant Physiol. 2021, 260, 153406. [Google Scholar] [CrossRef]
- Kim, S.-H.; Choi, H.-S.; Cho, Y.-C.; Kim, S.-R. Cold-responsive regulation of a flower-preferential class III peroxidase gene, OsPOX1, in rice (Oryza sativa L.). J. Plant Biol. 2012, 55, 123–131. [Google Scholar] [CrossRef]
- Hashimoto, M.; Setsuko, K. Proteomic analysis of rice seedlings during cold stress. Proteomics 2007, 7, 1293–1302. [Google Scholar] [CrossRef]
- Bonnecarrère, V.; Borsani, O.; Díaz, P.; Capdevielle, F.; Blanco, P.; Monza, J. Response to photoxidative stress induced by cold in japonica rice is genotype dependent. Plant Sci. 2011, 180, 726–732. [Google Scholar] [CrossRef]
- Lin, F.; Manisseri, C.; Fagerström, A.; Peck, M.L.; Vega-Sánchez, M.E.; Williams, B.; Chiniquy, D.M.; Saha, P.; Pattathil, S.; Conlin, B.; et al. Cell wall composition and candidate biosynthesis gene expression during rice development. Plant Cell Physiol. 2016, 57, 2058–2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2015, 5, 771. [Google Scholar] [CrossRef] [Green Version]
- Novakovic, L.; Guo, T.; Bacic, A.; Sampathkumar, A.; Johnson, K.L. Hitting the wall—Sensing and signaling pathways involved in plant cell wall remodeling in response to abiotic stress. Plants 2018, 7, 89. [Google Scholar] [CrossRef] [Green Version]
- Wolf, S.; Steffen, G. Growth control by cell wall pectins. Protoplasma 2012, 249, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Prathi, N.B.; Palit, P.; Madhu, P.; Ramesh, M.; Laha, G.S.; Balachandran, S.M.; Madhav, M.S.; Sundaram, R.M.; Mangrauthia, S.K. Proteomic and transcriptomic approaches to identify resistance and susceptibility related proteins in contrasting rice genotypes infected with fungal pathogen Rhizoctonia solani. Plant Physiol. Biochem. 2018, 130, 258–266. [Google Scholar] [CrossRef]
- Cutter, A.R.; Jeffrey, J.H. A brief review of nucleosome structure. FEBS Lett. 2015, 589, 2914–2922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, A.J.; Karolin, L. Nucleosome structure (s) and stability: Variations on a theme. Annu. Rev. Biophys. 2011, 40, 99–117. [Google Scholar] [CrossRef]
- McGinty, R.K. Nucleosome structure and function. Chem. Rev. 2015, 115, 2255–2273. [Google Scholar] [CrossRef] [Green Version]
- Bhardwaj, D.; Suman, L.; Narendra, T. Can G-proteins be the key proteins for overcoming environmental stresses and increasing crop yield in plants. In Plant Acclimation to Environmental Stress; Springer: New York, NY, USA, 2013; pp. 461–482. [Google Scholar]





| Genetic Background | Number of Accessions | Low Temperature Seedling Survivability (%) | Electrolyte Leakage (%) | Catalase Activity (AU) | Anthocyanin Activity (AU) |
|---|---|---|---|---|---|
| ADMIX | 11 | 80.22 ± 2.05 | 15.74 ± 2.02 | 11.89 ± 1.16 | 15.33 ± 1.14 |
| ARO | 11 | 80.60 ± 3.32 | 19.65 ± 2.41 | 11.96 ± 1.00 | 20.47 ± 1.45 |
| AUS | 53 | 13.11 ± 1.76 | 36.43 ± 1.70 | 6.68 ± 0.26 | 10.30 ± 0.44 |
| IND | 74 | 27.44 ± 1.46 | 32.47 ± 1.22 | 6.77 ± 0.27 | 10.59 ± 0.45 |
| MIX-I | 6 | 14.91 ± 4.26 | 40.38 ± 4.24 | 7.41 ± 0.89 | 8.80 ± 1.26 |
| MIX-J | 30 | 94.39 ± 1.10 | 15.15 ± 0.95 | 13.45 ± 0.68 | 20.54 ± 1.16 |
| TEJ | 92 | 95.03 ± 0.65 | 15.66 ± 0.63 | 14.35 ± 0.47 | 22.28 ± 0.70 |
| TRJ | 93 | 93.89 ± 0.74 | 15.78 ± 0.66 | 13.27 ± 0.39 | 20.57 ± 0.68 |
| TOL (MIX-J+TEJ+TRJ) | 215 | 94.45 ± 0.59 | 15.64 ± 0.39 | 13.76 ± 0.20 | 21.30 ± 0.36 |
| INT (ADMIX+ARO) | 22 | 80.41 ± 5.30 | 17.70 ± 0.36 | 11.93 ± 0.71 | 17.90 ± 1.28 |
| SEN (AUS+IND+MIX-I) | 133 | 21.16 ± 2.11 | 34.40 ± 1.13 | 6.76 ± 0.22 | 10.39 ± 0.29 |
| All | 370 | 67.27 ± 2.01 | 22.51 ± 0.66 | 11.13 ± 0.23 | 17.17 ± 0.36 |
| QTL | Chr | Start | End | Phenotypes | Peak LOD Score | Peak SNP Gene | Published QTL |
|---|---|---|---|---|---|---|---|
| qMP1-1 | 1 | 8815194 | 9418864 | CAT, LTSS | 5.31 | – | qMT1-1 [40] |
| qMP1-2 | 1 | 18792455 | 18916961 | ANT, CAT, LTSS | 4.96 | LOC_Os01g34430 | qMT1-4 [40], qCTS1-2 [47] |
| qMP1-3 | 1 | 22290853 | 22489300 | CAT, EL | 4.27 | LOC_Os01g38740 | – |
| qMP1-4 | 1 | 37834095 | 37862889 | CAT, EL | 5.00 | – | qCTS1-4 [47] |
| qMP3-1 | 3 | 10104198 | 10284651 | ANT, LTSS | 5.46 | – | qLVG3 [46] |
| qMP4-1 | 4 | 2699247 | 2729293 | CAT, EL | 3.44 | LOC_Os04g05420 | qCTS4-1 [47] |
| qMP4-2 | 4 | 4150962 | 4265645 | ANT, EL | 4.35 | – | qCTS4-1 [47] |
| qMP4-3 | 4 | 32076622 | 32401404 | CAT, LTSS | 6.19 | – | qMT4-3 [40], qLTSS4-3 [37], qLTG4 [46] |
| qMP5-1 | 5 | 3512674 | 4377698 | ANT, CAT | 3.45 | – | qLTG5-1 [46], qCTS5-1 [47] |
| qMP5-2 | 5 | 23154623 | 23564490 | CAT, EL | 6.08 | – | qMT5-3 [40], qCTSS-5 [46], qCTS5-4 [47] |
| qMP8-1 | 8 | 3078138 | 3346129 | ANT, CAT | 4.21 | – | qMT8-1 [40], qLTSS8-1 [37] |
| qMP8-2 | 8 | 21298422 | 21317944 | ANT, LTSS | 3.45 | LOC_Os08g34030 | qMT8-3 [40], qLTSS8-2 [37], COLD2 [46], qCTS8-4 [47] |
| qMP8-3 | 8 | 25620369 | 25655572 | ANT, LTSS | 6.22 | – | qMT8-4 [40] |
| qMP10-1 | 10 | 11675173 | 12345517 | ANT, CAT | 3.18 | – | qLTSS10-2 [37], qCTSS-10 [46] |
| qMP10-2 | 10 | 13841307 | 14025821 | ANT, LTSS | 6.80 | LOC_Os10g26550 | qMT10-4 [40], qCTSS-10 [46] |
| qMP11-1 | 11 | 2685973 | 2766157 | CAT, EL, LTSS | 6.43 | – | – |
| qMP11-2 | 11 | 28342262 | 28475216 | ANT, EL | 3.53 | – | qMT11-3 [40] |
| qMP12-1 | 12 | 2192895 | 2390059 | ANT, LTSS | 6.47 | – | qMT12 [40] |
| qMP12-2 | 12 | 8027010 | 8406160 | CAT, EL | 5.26 | LOC_Os12g14680 | qCTSS-12 [46] |
| qMP12-3 | 12 | 9435619 | 9819447 | CAT, EL | 4.14 | – | qCTSS-12 [46] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phan, H.; Schläppi, M. Low Temperature Antioxidant Activity QTL Associate with Genomic Regions Involved in Physiological Cold Stress Tolerance Responses in Rice (Oryza sativa L.). Genes 2021, 12, 1700. https://doi.org/10.3390/genes12111700
Phan H, Schläppi M. Low Temperature Antioxidant Activity QTL Associate with Genomic Regions Involved in Physiological Cold Stress Tolerance Responses in Rice (Oryza sativa L.). Genes. 2021; 12(11):1700. https://doi.org/10.3390/genes12111700
Chicago/Turabian StylePhan, Huy, and Michael Schläppi. 2021. "Low Temperature Antioxidant Activity QTL Associate with Genomic Regions Involved in Physiological Cold Stress Tolerance Responses in Rice (Oryza sativa L.)" Genes 12, no. 11: 1700. https://doi.org/10.3390/genes12111700
APA StylePhan, H., & Schläppi, M. (2021). Low Temperature Antioxidant Activity QTL Associate with Genomic Regions Involved in Physiological Cold Stress Tolerance Responses in Rice (Oryza sativa L.). Genes, 12(11), 1700. https://doi.org/10.3390/genes12111700