Metabolite Genome-Wide Association Study for Indoleamine 2,3-Dioxygenase Activity Associated with Chronic Kidney Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Study Participants
2.3. General Characteristics
2.4. Metabolite Measurements
2.5. Genotyping and Imputation
2.6. Statistical Analysis
3. Results
3.1. Participant Characteristics
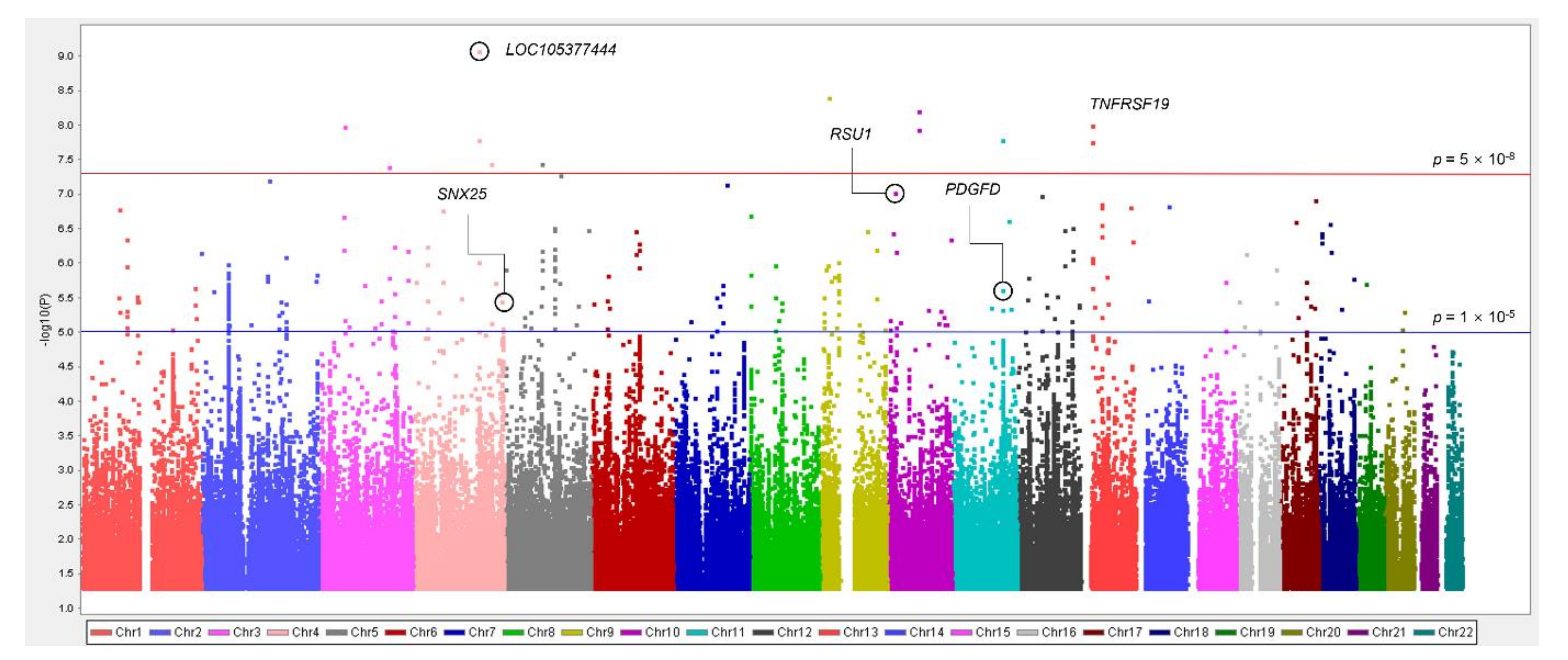
3.2. Associations between Common Variants and IDO Activity Related to CKD
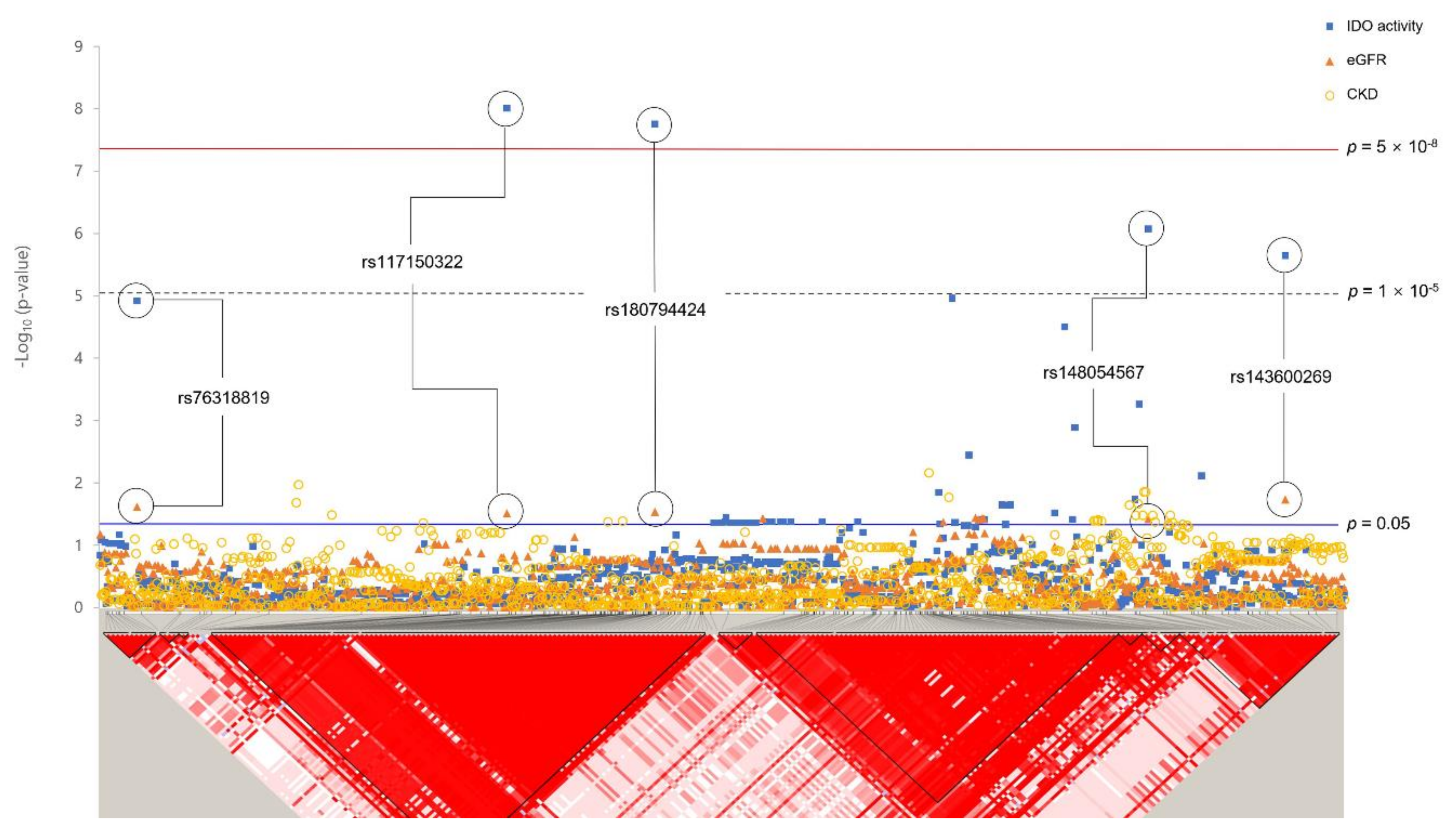
3.3. Associations between Rare Variants and IDO Activity Related to CKD
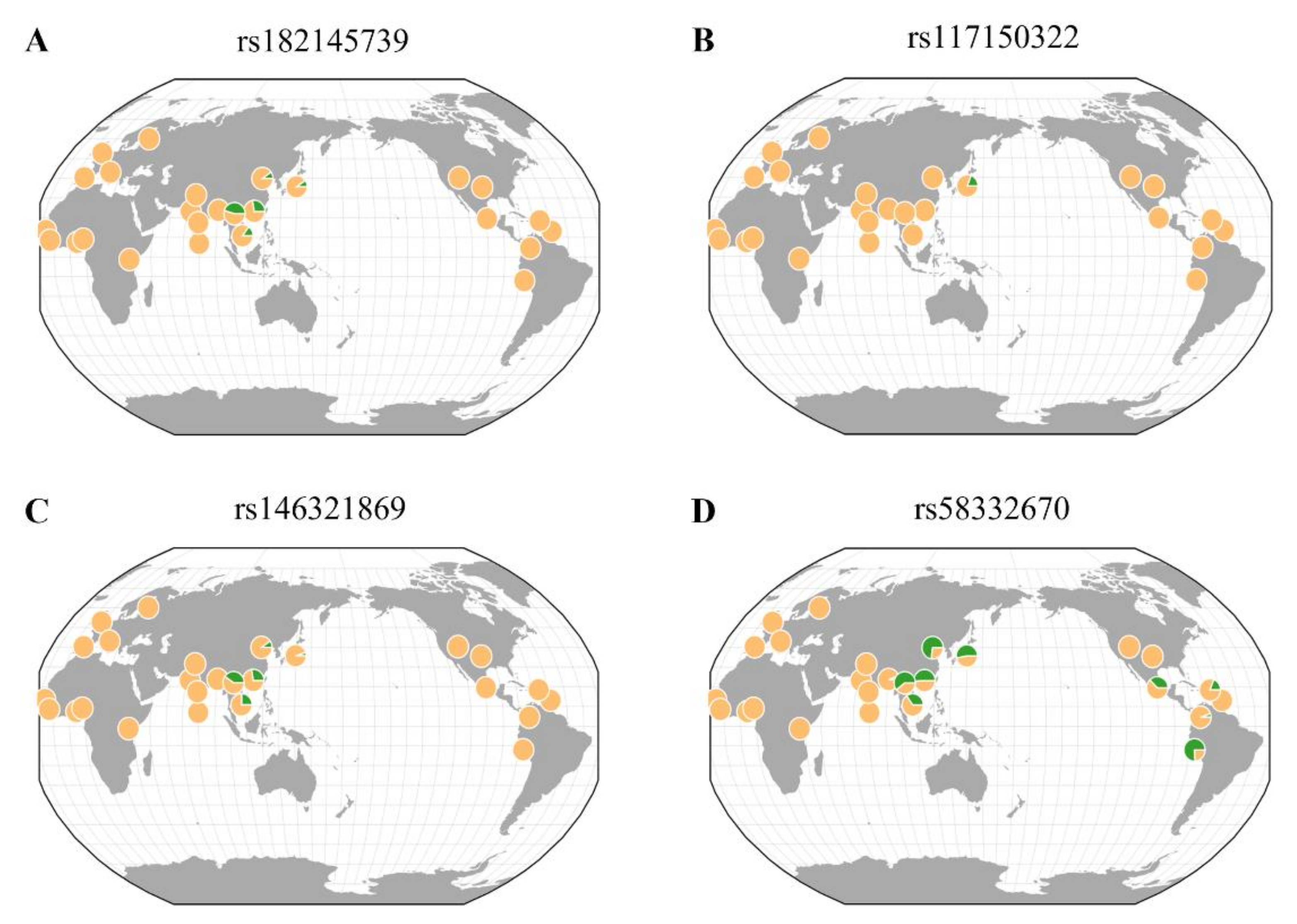
3.4. Geographical Distribution of Rare Variants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parmar, M.S. Chronic renal disease. BMJ 2002, 325, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Levey, A.; Atkins, R.; Coresh, J.; Cohen, E.; Collins, A.; Eckardt, K.-U.; Nahas, M.; Jaber, B.; Jadoul, M.; Levin, A. Chronic kidney disease as a global public health problem: Approaches and initiatives–a position statement from Kidney Disease Improving Global Outcomes. Kidney Int. 2007, 72, 247–259. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.M. Inflammatory mediators and renal fibrosis. Adv. Exp. Med. Biol. 2019, 1165, 381–406. [Google Scholar] [PubMed]
- Center for Disease Control and Prevention (CDC). Chronic Kidney Disease in the United States. 2021. Available online: https://www.cdc.gov/kidneydisease/publications-resources/CKD-national-facts.html (accessed on 20 September 2021).
- Wouters, O.J.; O’donoghue, D.J.; Ritchie, J.; Kanavos, P.G.; Narva, A.S. Early chronic kidney disease: Diagnosis, management and models of care. Nat. Rev. Nephrol. 2015, 11, 491–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krol, E.; Rutkowski, B.; Czarniak, P.; Kraszewska, E.; Lizakowski, S.; Szubert, R.; Czekalski, S.; Sulowicz, W.; Wiecek, A. Early detection of chronic kidney disease: Results of the PolNef study. Am. J. Nephrol. 2009, 29, 264–273. [Google Scholar] [PubMed] [Green Version]
- Köttgen, A.; Raffler, J.; Sekula, P.; Kastenmüller, G. Genome-wide association studies of metabolite concentrations (mGWAS): Relevance for nephrology. Semin. Nephrol. 2018, 38, 151–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Metabolomics for biomarker discovery: Moving to the clinic. Biomed. Res. Int. 2015, 2015, 354671. [Google Scholar] [CrossRef]
- Graham, S.F.; Chevallier, O.P.; Roberts, D.; Holscher, C.; Elliott, C.T.; Green, B.D. Investigation of the human brain metabolome to identify potential markers for early diagnosis and therapeutic targets of Alzheimer’s disease. Anal. Chem. 2013, 85, 1803–1811. [Google Scholar] [CrossRef]
- Zhao, Y.Y. Metabolomics in chronic kidney disease. Clin. Chim. Acta. 2013, 422, 59–69. [Google Scholar] [CrossRef]
- Levey, A.S.; Becker, C.; Inker, L.A. Glomerular filtration rate and albuminuria for detection and staging of acute and chronic kidney disease in adults: A systematic review. JAMA 2015, 313, 837–846. [Google Scholar] [CrossRef] [Green Version]
- Toffaletti, J.G. Clarifying the Confusion of GFRs, Creatinine, and Cystatin C. 2018. Available online: https://acutecaretesting.org/ (accessed on 9 September 2021).
- Perrone, R.D.; Madias, N.E.; Levey, A.S. Serum creatinine as an index of renal function: New insights into old concepts. Clin. Chem. 1992, 38, 1933–1953. [Google Scholar] [CrossRef]
- Chen, Y.; Guillemin, G.J. Kynurenine pathway metabolites in humans: Disease and healthy States. Int. J. Tryptophan Res. 2009, 2, IJTR.S2097. [Google Scholar] [CrossRef] [Green Version]
- Schefold, J.C.; Zeden, J.P.; Fotopoulou, C.; von Haehling, S.; Pschowski, R.; Hasper, D.; Volk, H.D.; Schuett, C.; Reinke, P. Increased indoleamine 2,3-dioxygenase (IDO) activity and elevated serum levels of tryptophan catabolites in patients with chronic kidney disease: A possible link between chronic inflammation and uraemic symptoms. Nephrol. Dial. Transplant. 2009, 24, 1901–1908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mbongue, J.C.; Nicholas, D.A.; Torrez, T.W.; Kim, N.S.; Firek, A.F.; Langridge, W.H. The role of indoleamine 2, 3-dioxygenase in immune suppression and autoimmunity. Vaccines 2015, 3, 703–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohib, K.; Guan, Q.; Diao, H.; Du, C.; Jevnikar, A.M. Proapoptotic activity of indoleamine 2,3-dioxygenase expressed in renal tubular epithelial cells. Am. J. Physiol.-Renal Physiol. 2007, 293, F801–F812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, K.; Fujigaki, S.; Heyes, M.P.; Shibata, K.; Takemura, M.; Fujii, H.; Wada, H.; Noma, A.; Seishima, M. Mechanism of increases in L-kynurenine and quinolinic acid in renal insufficiency. Am. J. Physiol. Renal. Physiol. 2000, 279, F565–F572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawlak, D.; Tankiewicz, A.; Mysliwiec, P.; Buczko, W. Tryptophan metabolism via the kynurenine pathway in experimental chronic renal failure. Nephron 2002, 90, 328–335. [Google Scholar] [CrossRef]
- Aregger, F.; Uehlinger, D.E.; Fusch, G.; Bahonjic, A.; Pschowski, R.; Walter, M.; Schefold, J.C. Increased urinary excretion of kynurenic acid is associated with non-recovery from acute kidney injury in critically ill patients. BMC Nephrol. 2018, 19, 44. [Google Scholar] [CrossRef]
- Lee, H.; Jang, H.B.; Yoo, M.G.; Park, S.I.; Lee, H.J. Amino acid metabolites associated with chronic kidney disease: An Eight-Year Follow-Up Korean Epidemiology Study. Biomedicines 2020, 8, 222. [Google Scholar] [CrossRef]
- Canadas-Garre, M.; Anderson, K.; Cappa, R.; Skelly, R.; Smyth, L.J.; McKnight, A.J.; Maxwell, A.P. Genetic Susceptibility to Chronic Kidney Disease—Some More Pieces for the Heritability Puzzle. Front. Genet. 2019, 10, 453. [Google Scholar] [CrossRef] [PubMed]
- MacCluer, J.W.; Scavini, M.; Shah, V.O.; Cole, S.A.; Laston, S.L.; Voruganti, V.S.; Paine, S.S.; Eaton, A.J.; Comuzzie, A.G.; Tentori, F.; et al. Heritability of measures of kidney disease among Zuni Indians: The Zuni Kidney Project. Am. J. Kidney Dis. 2010, 56, 289–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tin, A.; Kottgen, A. Genome-Wide Association Studies of CKD and Related Traits. Clin. J. Am. Soc. Nephrol. 2020, 15, 1643–1656. [Google Scholar] [CrossRef] [PubMed]
- Boger, C.A.; Heid, I.M. Chronic kidney disease: Novel insights from genome-wide association studies. Kidney Blood Press. Res. 2011, 34, 225–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Brief. Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef]
- Liu, L.; Kiryluk, K. Insights into CKD from Metabolite GWAS. J. Am. Soc. Nephrol. 2018, 29, 1349–1351. [Google Scholar] [CrossRef]
- Kim, Y.; Han, B.G.; KoGES Group. Cohort Profile: The Korean Genome and Epidemiology Study (KoGES) Consortium. Int. J. Epidemiol. 2017, 46, 1350. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-S.; Xu, T.; Lee, Y.; Kim, N.-H.; Kim, Y.-J.; Kim, J.-M.; Cho, S.Y.; Kim, K.-Y.; Nam, M.; Adamski, J. Identification of putative biomarkers for type 2 diabetes using metabolomics in the Korea Association REsource (KARE) cohort. Metabolomics 2016, 12, 178. [Google Scholar] [CrossRef]
- Cho, Y.S.; Go, M.J.; Kim, Y.J.; Heo, J.Y.; Oh, J.H.; Ban, H.-J.; Yoon, D.; Lee, M.H.; Kim, D.-J.; Park, M. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet. 2009, 41, 527–534. [Google Scholar] [CrossRef]
- Howie, B.N.; Donnelly, P.; Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009, 5, e1000529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Mor, A.; Kalaska, B.; Pawlak, D. Kynurenine pathway in chronic kidney disease: What’s old, what’s new, and what’s next? Int. J. Tryptophan Res. 2020, 13, 1178646920954882. [Google Scholar] [CrossRef]
- Pawlak, K.; Kowalewska, A.; Pawlak, D.; Mysliwiec, M. Kynurenine and its metabolites—Kynurenic acid and anthranilic acid are associated with soluble endothelial adhesion molecules and oxidative status in patients with chronic kidney disease. Am. J. Med. Sci. 2009, 338, 293–300. [Google Scholar] [CrossRef]
- Rhee, E.P.; Clish, C.B.; Ghorbani, A.; Larson, M.G.; Elmariah, S.; McCabe, E.; Yang, Q.; Cheng, S.; Pierce, K.; Deik, A.; et al. A combined epidemiologic and metabolomic approach improves CKD prediction. J. Am. Soc. Nephrol. 2013, 24, 1330–1338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goek, O.N.; Prehn, C.; Sekula, P.; Romisch-Margl, W.; Doring, A.; Gieger, C.; Heier, M.; Koenig, W.; Wang-Sattler, R.; Illig, T.; et al. Metabolites associate with kidney function decline and incident chronic kidney disease in the general population. Nephrol. Dial. Transplant. 2013, 28, 2131–2138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Liu, J.; Tu, Y.; Zhao, Z.; Qu, J.; Chen, K.; Chen, Y.; Sun, Y.; Zhao, H.; Deng, Y.; et al. RSU-1 interaction with prohibitin-2 links cell-extracellular matrix detachment to downregulation of ERK signaling. J. Biol. Chem. 2021, 296, 100109. [Google Scholar] [CrossRef]
- Najy, A.J.; Won, J.J.; Movilla, L.S.; Kim, H.-R.C. Differential tumorigenic potential and matriptase activation between PDGF B versus PDGF D in prostate cancer. Mol. Cancer Res. 2012, 10, 1087–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eby, M.T.; Jasmin, A.; Kumar, A.; Sharma, K.; Chaudhary, P.M. TAJ, a novel member of the tumor necrosis factor receptor family, activates the c-Jun N-terminal kinase pathway and mediates caspase-independent cell death. J. Biol. Chem. 2000, 275, 15336–15342. [Google Scholar] [CrossRef] [Green Version]
- Kurtzeborn, K.; Kwon, H.N.; Kuure, S. MAPK/ERK signaling in regulation of renal differentiation. Int. J. Mol. Sci. 2019, 20, 1779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hida, M.; Omori, S.; Awazu, M. ERK and p38 MAP kinase are required for rat renal development. Kidney Int. 2002, 61, 1252–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ihermann-Hella, A.; Lume, M.; Miinalainen, I.J.; Pirttiniemi, A.; Gui, Y.; Peranen, J.; Charron, J.; Saarma, M.; Costantini, F.; Kuure, S. Mitogen-activated protein kinase (MAPK) pathway regulates branching by remodeling epithelial cell adhesion. PLoS Genet. 2014, 10, e1004193. [Google Scholar] [CrossRef] [Green Version]
- Fujigaki, H.; Saito, K.; Fujigaki, S.; Takemura, M.; Sudo, K.; Ishiguro, H.; Seishima, M. The signal transducer and activator of transcription 1alpha and interferon regulatory factor 1 are not essential for the induction of indoleamine 2,3-dioxygenase by lipopolysaccharide: Involvement of p38 mitogen-activated protein kinase and nuclear factor-kappaB pathways, and synergistic effect of several proinflammatory cytokines. J. Biochem. 2006, 139, 655–662. [Google Scholar] [PubMed]
- Beaulieu, J.M.; Espinoza, S.; Gainetdinov, R.R. Dopamine receptors–IUPHAR Review 13. Br. J. Pharmacol. 2015, 172, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Armando, I.; Villar, V.A.; Jose, P.A. Dopamine and renal function and blood pressure regulation. Compr. Physiol. 2011, 1, 1075–1117. [Google Scholar] [PubMed]
- Harris, R.C.; Zhang, M.Z. Dopamine, the kidney, and hypertension. Curr. Hypertens. Rep. 2012, 14, 138–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pat, B.; Yang, T.; Kong, C.; Watters, D.; Johnson, D.W.; Gobe, G. Activation of ERK in renal fibrosis after unilateral ureteral obstruction: Modulation by antioxidants. Kidney Int. 2005, 67, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Reznichenko, A.; Snieder, H.; van den Born, J.; de Borst, M.H.; Damman, J.; van Dijk, M.C.; van Goor, H.; Hepkema, B.G.; Hillebrands, J.-L.; Leuvenink, H.G. CUBN as a novel locus for end-stage renal disease: Insights from renal transplantation. PLoS ONE 2012, 7, e36512. [Google Scholar] [CrossRef] [Green Version]
- Qian, H.; Kowalski, M.H.; Kramer, H.J.; Tao, R.; Lash, J.P.; Stilp, A.M.; Cai, J.; Li, Y.; Franceschini, N. Genome-wide association of kidney traits in hispanics/latinos using dense imputed whole-genome sequencing data: The hispanic community health study/study of latinos. Circ. Genom. Precis. Med. 2020, 13, e002891. [Google Scholar] [CrossRef] [PubMed]
- Ostendorf, T.; van Roeyen, C.R.; Peterson, J.D.; Kunter, U.; Eitner, F.; Hamad, A.J.; Chan, G.; Jia, X.C.; Macaluso, J.; Gazit-Bornstein, G.; et al. A fully human monoclonal antibody (CR002) identifies PDGF-D as a novel mediator of mesangioproliferative glomerulonephritis. J. Am. Soc. Nephrol. 2003, 14, 2237–2247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muhanna, N.; Doron, S.; Wald, O.; Horani, A.; Eid, A.; Pappo, O.; Friedman, S.L.; Safadi, R. Activation of hepatic stellate cells after phagocytosis of lymphocytes: A novel pathway of fibrogenesis. Hepatology 2008, 48, 963–977. [Google Scholar] [CrossRef] [Green Version]
- Charni Chaabane, S.; Coomans de Brachene, A.; Essaghir, A.; Velghe, A.; Lo Re, S.; Stockis, J.; Lucas, S.; Khachigian, L.M.; Huaux, F.; Demoulin, J.B. PDGF-D expression is down-regulated by TGFbeta in fibroblasts. PLoS ONE 2014, 9, e108656. [Google Scholar] [CrossRef] [Green Version]
- Free, R.B.; Hazelwood, L.A.; Spalding, H.N.; Cabrera, D.M.; Sibley, D.R. Sorting nexin-25, a novel member of the dopamine receptor signalplex, up-regulates D1 and D2 dopamine receptor expression in HEK293 cells. FASEB J. 2007, 21, A423. [Google Scholar] [CrossRef]
- Yang, J.; Armando, I.; Jones, J.; Zeng, C.; Jose, P.; Villar, V. Sorting nexins: New determinants for the development of hypertension. Ann. Clin. Exp. Hypertens. 2014, 2, 1008. [Google Scholar]
- Huang, J.; Kong, Y.; Xie, C.; Zhou, L. Stem/progenitor cell in kidney: Characteristics, homing, coordination, and maintenance. Stem. Cell Res. Ther. 2021, 12, 197. [Google Scholar] [CrossRef] [PubMed]
- Chou, Y.H.; Pan, S.Y.; Yang, C.H.; Lin, S.L. Stem cells and kidney regeneration. J. Formos. Med. Assoc. 2014, 113, 201–209. [Google Scholar] [CrossRef] [Green Version]
- Schutgens, F.; Rookmaaker, M.B.; Blokzijl, F.; van Boxtel, R.; Vries, R.; Cuppen, E.; Verhaar, M.C.; Clevers, H. Troy/TNFRSF19 marks epithelial progenitor cells during mouse kidney development that continue to contribute to turnover in adult kidney. Proc. Natl. Acad. Sci. USA 2017, 114, E11190–E11198. [Google Scholar] [CrossRef] [Green Version]
- Grynberg, K.; Ma, F.Y.; Nikolic-Paterson, D.J. The JNK signaling pathway in renal fibrosis. Front. Physiol. 2017, 8, 829. [Google Scholar] [CrossRef]
- Mei, J.; Li, M.Q.; Ding, D.; Li, D.J.; Jin, L.P.; Hu, W.G.; Zhu, X.Y. Indoleamine 2,3-dioxygenase-1 (IDO1) enhances survival and invasiveness of endometrial stromal cells via the activation of JNK signaling pathway. Int. J. Clin. Exp. Pathol. 2013, 6, 431–444. [Google Scholar]
- Opitz, C.A.; Litzenburger, U.M.; Opitz, U.; Sahm, F.; Ochs, K.; Lutz, C.; Wick, W.; Platten, M. The indoleamine-2,3-dioxygenase (IDO) inhibitor 1-methyl-D-tryptophan upregulates IDO1 in human cancer cells. PLoS ONE 2011, 6, e19823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Characteristics | Quantitative Trait Analysis | Case–Control Analysis for CKD | ||
|---|---|---|---|---|
| Controls | Cases | p-Value * | ||
| Number of participants | 2579 | 1550 | 264 | |
| Gender [men (%)] | 1218 (47.23) | 789 (50.90) | 81 (30.68) | <0.001 |
| Age (M years ± SD) | 57.10 ± 9.05 | 54.98 ± 8.64 | 65.72 ± 6.53 | <0.001 |
| Height (M cm ± SD) | 159.55 ± 9.16 | 160.55 ± 8.98 | 155.42 ± 8.28 | <0.001 |
| Weight (M kg ± SD) | 62.63 ± 10.36 | 62.30 ± 10.41 | 60.88 ± 9.53 | 0.042 |
| BMI (M kg/m2 ± SD) | 24.56 ± 3.23 | 24.11 ± 3.09 | 25.20 ± 3.47 | <0.001 |
| eGFR (mL/min/1.73 m2) | 75.58 ± 11.92 | 78.68 ± 9.69 | 55.24 ± 9.21 | <0.001 |
| Creatinine (mg/dL) | 0.98 ± 0.20 | 0.96 ± 0.14 | 1.18 ± 0.42 | <0.001 |
| BUN (mg/dL) | 15.69 ± 4.26 | 15.33 ± 3.92 | 17.91 ± 5.45 | <0.001 |
| No. | SNP | Nearest Gene | Chromosome Position | Minor Allele | MAF | Function | IDO Activity | eGFR | CKD | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β ± S.E. | p-Value | β ± S.E | p-Value | OR (95% CI) | p-Value | |||||||
| 1 | rs59178336 | RSU1 | 10:16822091 | C | 0.095 | Intron | 0.26 ± 0.049 | 9.41 × 10−8 | −0.59 ± 0.50 | 0.235 | 1.47 (1.02–2.14) | 0.041 |
| 2 | rs10469937 | CCDC85A | 2:56629317 | C | 0.435 | - | −0.14 ± 0.028 | 1.03 × 10−6 | 0.35 ± 0.29 | 0.228 | 0.91 (0.73–1.14) | 0.407 |
| 3 | rs7588698 | HDAC4 | 2:240041896 | A | 0.055 | Intron | 0.30 ± 0.062 | 1.42 × 10−6 | −0.78 ± 0.64 | 0.225 | 1.43 (0.88–2.32) | 0.145 |
| 4 | rs2513735 | PDGFD | 11:104081184 | T | 0.065 | - | 0.27 ± 0.057 | 2.39 × 10−6 | −1.45 ± 0.58 | 0.012 | 1.08 (0.70–1.67) | 0.730 |
| 5 | rs6730950 | RTN4 | 2:55386276 | C | 0.149 | - | 0.19 ± 0.040 | 2.90 × 10−6 | −0.75 ± 0.41 | 0.065 | 1.06 (0.78–1.43) | 0.721 |
| 6 | rs1094818 | WARS2 | 1:119238523 | G | 0.078 | - | 0.25 ± 0.053 | 3.49 × 10−6 | −0.95 ± 0.54 | 0.080 | 1.34 (0.91–1.96) | 0.140 |
| 7 | rs78549225 | RBFOX1 | 16:6969406 | G | 0.119 | Intron | 0.21 ± 0.044 | 3.50 × 10−6 | −0.66 ± 0.46 | 0.148 | 1.24 (0.88–1.74) | 0.222 |
| 8 | rs78259836 | SNX25 | 4:186261649 | A | 0.105 | Intron | 0.21 ± 0.046 | 3.54 × 10−6 | −1.07 ± 0.47 | 0.024 | 1.12 (0.79–1.57) | 0.522 |
| 9 | rs7237751 | LOC107984031 | 18:47255785 | G | 0.139 | Upstream | 0.19 ± 0.041 | 4.42 × 10−6 | −0.86 ± 0.42 | 0.043 | 1.27 (0.93–1.74) | 0.137 |
| 10 | rs12226572 | UBASH3B | 11:122648650 | A | 0.054 | Intron | 0.29 ± 0.062 | 4.54 × 10−6 | −1.05 ± 0.64 | 0.100 | 1.64 (1.07–2.51) | 0.025 |
| 11 | rs143090547 | PLPP1 | 5:54753484 | T | 0.056 | Intron | 0.28 ± 0.062 | 5.00 × 10−6 | −1.23 ± 0.63 | 0.051 | 1.37 (0.86–2.19) | 0.190 |
| 12 | rs17608925 | ORMDL3 | 17:38082831 | C | 0.066 | Intron | 0.26 ± 0.056 | 5.84 × 10−6 | −0.01 ± 0.58 | 0.989 | 1.36 (0.90–2.05) | 0.147 |
| 13 | rs73192989 | RBM19 | 12:114580187 | T | 0.080 | - | 0.23 ± 0.051 | 6.77 × 10−6 | −0.75 ± 0.53 | 0.155 | 1.00 (0.67–1.50) | 0.989 |
| 14 | rs199564331 | BRINP3 | 1:190127911 | D | 0.113 | Intron | 0.20 ± 0.045 | 9.02 × 10−6 | −0.56 ± 0.47 | 0.226 | 1.27 (0.90–1.80) | 0.174 |
| 15 | rs3773884 | MME | 3:154859650 | G | 0.052 | Intron | 0.28 ± 0.063 | 9.33 × 10−6 | −0.52 ± 0.64 | 0.418 | 1.28 (0.81–2.01) | 0.291 |
| No. | SNP | Nearest Gene | Chromosome Position | Minor Allele | MAF | HWE p-Value | Function | IDO Activity | eGFR | CKD | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β ± S.E | p-Value | β ± S.E | p-Value | OR (95% CI) | p-Value | ||||||||
| 1 | rs182145739 | LOC105377444 | 4:138651320 | T | 0.011 | 1 | Intron | 0.82 ± 0.13 | 8.46 × 10−10 | −1.08 ± 1.36 | 0.427 | 2.63 (1.15–6.03) | 0.022 |
| 2 | rs149763281 | SLC24A2 | 9:20104936 | C | 0.011 | 0.277 | Intron | 0.78 ± 0.13 | 3.88 × 10−9 | −1.68 ± 1.36 | 0.218 | 0.95 (0.34–2.72) | 0.936 |
| 3 | rs117150322 | TNFRSF19 | 13:24120841 | A | 0.009 | 0.191 | - | 0.83 ± 0.15 | 9.89 × 10−9 | −3.22 ± 1.49 | 0.031 | 1.01 (0.30–3.38) | 0.994 |
| 4 | rs188289326 | CACNA2D3 | 3:54867936 | A | 0.010 | 1 | Intron | 0.81 ± 0.14 | 1.02 × 10−8 | −2.34 ± 1.45 | 0.107 | 1.20 (0.49–2.96) | 0.690 |
| 5 | rs146321869 | LOC101928535 | 11:106108552 | G | 0.009 | 0.183 | - | 0.83 ± 0.15 | 1.16 × 10−8 | −3.34 ± 1.51 | 0.026 | 1.88 (0.67–5.26) | 0.232 |
| 6 | rs337828 | ARSB | 5:78196735 | G | 0.009 | 1 | Intron | 0.81 ± 0.15 | 3.55 × 10−8 | −1.53 ± 1.51 | 0.310 | 1.50 (0.55–4.14) | 0.431 |
| 7 | rs58332670 | FSTL5 | 4:163207867 | C | 0.035 | 0.549 | - | 0.42 ± 0.08 | 3.58 × 10−8 | −2.24 ± 0.79 | 4.32 × 10−3 | 1.43 (0.80–2.57) | 0.231 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-R.; Jin, H.-S.; Eom, Y.-B. Metabolite Genome-Wide Association Study for Indoleamine 2,3-Dioxygenase Activity Associated with Chronic Kidney Disease. Genes 2021, 12, 1905. https://doi.org/10.3390/genes12121905
Kim H-R, Jin H-S, Eom Y-B. Metabolite Genome-Wide Association Study for Indoleamine 2,3-Dioxygenase Activity Associated with Chronic Kidney Disease. Genes. 2021; 12(12):1905. https://doi.org/10.3390/genes12121905
Chicago/Turabian StyleKim, Hye-Rim, Hyun-Seok Jin, and Yong-Bin Eom. 2021. "Metabolite Genome-Wide Association Study for Indoleamine 2,3-Dioxygenase Activity Associated with Chronic Kidney Disease" Genes 12, no. 12: 1905. https://doi.org/10.3390/genes12121905
APA StyleKim, H.-R., Jin, H.-S., & Eom, Y.-B. (2021). Metabolite Genome-Wide Association Study for Indoleamine 2,3-Dioxygenase Activity Associated with Chronic Kidney Disease. Genes, 12(12), 1905. https://doi.org/10.3390/genes12121905







