Genome-Wide Analysis of DNA Methylation in Buccal Cells of Children Conceived through IVF and ICSI
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sample Preparation and DNA Methylation Extraction
2.3. Statistical Analyses
2.4. Comparison with Previous Studies
3. Results
3.1. Raw Analysis of the Buccal Cell DNA Methylation Profile of ART Children Reveals Major Variations in Cell Type Proportions
3.2. Impact of Mode of Conception, Type of Culture Medium, and Method of Fertilization at the CpG Level
3.3. Impact of the Mode of Conception, Type of Culture Medium, and Method of Fertilization at the Region Level
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adamson, G.D.; de Mouzon, J.; Chambers, G.M.; Zegers-Hochschild, F.; Mansour, R.; Ishihara, O.; Banker, M.; Dyer, S. International Committee for Monitoring Assisted Reproductive Technology: World report on assisted reproductive technology, 2011. Fertil. Steril. 2018, 110, 1067–1080. [Google Scholar] [CrossRef]
- Hansen, M.; Kurinczuk, J.J.; Bower, C.; Webb, S. Injection and in vitro Fertilization. N. Engl. J. Med. 2002, 346, 725–730. [Google Scholar] [CrossRef]
- Helmerhorst, F.M.; Perquin, D.A.M.; Donker, D.; Keirse, M.J.N.C. Perinatal outcome of singletons and twins after assisted conception: A systematic review of controlled studies. Br. Med. J. 2004, 328, 261–264. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Heilbronn, L.K. The health outcomes of human offspring conceived by assisted reproductive technologies (ART). J. Dev. Orig. Health Dis. 2017, 8, 388–402. [Google Scholar] [CrossRef]
- Guo, X.-Y.; Liu, X.M.; Jin, L.; Wang, T.T.; Ullah, K.; Sheng, J.Z.; Huang, H.F. Cardiovascular and metabolic profiles of offspring conceived by assisted reproductive technologies: A systematic review and meta-analysis. Fertil. Steril. 2017, 107, 622–631.e5. [Google Scholar] [CrossRef]
- Berntsen, S.; Söderström-Anttila, V.; Wennerholm, U.B.; Laivuori, H.; Loft, A.; Oldereid, N.B.; Romundstad, L.B.; Bergh, C.; Pinborg, A. The health of children conceived by ART: ‘The chicken or the egg?’. Hum. Reprod. Update 2019, 25, 137–158. [Google Scholar] [CrossRef] [PubMed]
- Duranthon, V.; Chavatte-Palmer, P. Long term effects of ART: What do animals tell us? Mol. Reprod. Dev. 2018, 85, 348–368. [Google Scholar] [CrossRef] [PubMed]
- Fauque, P.; De Mouzon, J.; Devaux, A.; Epelboin, S.; Gervoise-Boyer, M.J.; Levy, R.; Valentin, M.; Viot, G.; Bergère, A.; De Vienne, C.; et al. Reproductive technologies, female infertility, and the risk of imprinting-related disorders. Clin. Epigenetics 2020, 12, 191. [Google Scholar] [CrossRef]
- Argyraki, M.; Damdimopoulou, P.; Chatzimeletiou, K.; Grimbizis, G.F.; Tarlatzis, B.C.; Syrrou, M.; Lambropoulos, A. In-utero stress and mode of conception: Impact on regulation of imprinted genes, fetal development and future health. Hum. Reprod. Update 2019, 25, 777–801. [Google Scholar] [CrossRef] [PubMed]
- Bale, T.L.; Baram, T.Z.; Brown, A.S.; Goldstein, J.M.; Insel, T.R.; McCarthy, M.M.; Nemeroff, C.B.; Reyes, T.M.; Simerly, R.B.; Susser, E.S. Nestler EJ Early Life Programming and Neurodevelopmental Disorders Tracy. Biol. Psychiatry 2010, 68, 314–319. [Google Scholar] [CrossRef]
- Vaiserman, A.M. Epigenetic programming by early-life stress: Evidence from human populations. Dev. Dyn. 2015, 244, 254–265. [Google Scholar] [CrossRef]
- Kundakovic, M.; Jaric, I. The epigenetic link between prenatal adverse environments and neurodevelopmental disorders. Genes 2017, 8, 104. [Google Scholar] [CrossRef]
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef]
- Reik, W.; Romer, I.; Barton, S.C.; Surani, M.A.; Howlett, S.K.; Klose, J. Adult phenotype in the mouse can be affected by epigenetic in the early embryo. Development 1993, 119, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Murgatroyd, C.; Patchev, A.V.; Wu, Y.; Micale, V.; Bockmühl, Y.; Fischer, D.; Holsboer, F.; Wotjak, C.T.; Almeida, O.F.; Spengler, D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat. Neurosci. 2009, 12, 1559–1566. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Ibeas, P.; Heras, S.; Gómez-Redondo, I.; Planells, B.; Fernández-González, R.; Pericuesta, E.; Laguna-Barraza, R.; Pérez-Cerezales, S.; Gutiérrez-Adán, A. Embryo responses to stress induced by assisted reproductive technologies. Mol. Reprod. Dev. 2019, 86, 1292–1306. [Google Scholar] [CrossRef]
- Mantikou, E.; Youssef, M.A.; van Wely, M.; van der Veen, F.; Al-Inany, H.G.; Repping, S.; Mastenbroek, S. Embryo culture media and IVF/ICSI success rates: A systematic review. Hum. Reprod. Update 2013, 19, 210–220. [Google Scholar] [CrossRef]
- Doherty, A.S.; Mann, M.R.W.; Tremblay, K.D.; Bartolomei, M.S.; Schultz, R.M. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol. Reprod. 2000, 62, 1526–1535. [Google Scholar] [CrossRef] [PubMed]
- Khosla, S.; Dean, W.; Brown, D.; Reik, W.; Feil, R. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol. Reprod. 2001, 64, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.R.W.; Lee, S.S.; Doherty, A.S.; Verona, R.I.; Nolen, L.D.; Schultz, R.M.; Bartolomei, M.S. Selective loss of imprinting in the placenta following preimplantation development in culture. Development 2004, 131, 3727–3735. [Google Scholar] [CrossRef]
- Market-Velker, B.A.; Fernandes, A.D.; Mann, M.R.W. Side-by-side comparison of five commercial media systems in a mouse model: Suboptimal in vitro culture interferes with imprint maintenance. Biol. Reprod. 2010, 83, 938–950. [Google Scholar] [CrossRef] [PubMed]
- Carmignac, V.; Barberet, J.; Iranzo, J.; Quéré, R.; Guilleman, M.; Bourc’his, D.; Fauque, P. Effects of assisted reproductive technologies on transposon regulation in the mouse pre-implanted embryo. Hum. Reprod. 2019, 34, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Dumoulin, J.C.; Land, J.A.; Van Montfoort, A.P.; Nelissen, E.C.; Coonen, E.; Derhaag, J.G.; Schreurs, I.L.; Dunselman, G.A.; Kester, A.D.; Geraedts, J.P.; et al. Effect of in vitro culture of human embryos on birthweight of newborns. Hum. Reprod. 2010, 25, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Nelissen, E.C.; Van Montfoort, A.P.; Coonen, E.; Derhaag, J.G.; Geraedts, J.P.; Smits, L.J.; Land, J.A.; Evers, J.L.; Dumoulin, J.C. Further evidence that culture media affect perinatal outcome: Findings after transfer of fresh and cryopreserved embryos. Hum. Reprod. 2012, 27, 1966–1976. [Google Scholar] [CrossRef]
- Hassani, F.; Eftekhari-Yazdi, P.; Karimian, L.; Valojerdi, M.R.; Movaghar, B.; Fazel, M.; Fouladi, H.R.; Shabani, F.; Johansson, L. The effects of ISM1 medium on embryo quality and outcomes of IVF/ICSI cycles. Int. J. Fertil. Steril. 2013, 7, 108–115. [Google Scholar]
- Zandstra, H.; van Montfoort, A.P.A.; Dumoulin, J.C.M. Does the type of culture medium used influence birthweight of children born after IVF? Hum. Reprod. 2015, 30, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Mulder, C.L.; Wattimury, T.M.; Jongejan, A.; de Winter-Korver, C.M.; van Daalen, S.K.; Struijk, R.B.; Borgman, S.C.; Wurth, Y.; Consten, D.; van Echten-Arends, J.; et al. Comparison of DNA methylation patterns of parentally imprinted genes in placenta derived from IVF conceptions in two different culture media. Hum. Reprod. 2020, 35, 516–528. [Google Scholar] [CrossRef] [PubMed]
- Barberet, J.; Binquet, C.; Guilleman, M.; Doukani, A.; Choux, C.; Bruno, C.; Bourredjem, A.; Chapusot, C.; Bourc’his, D.; Duffourd, Y.; et al. Do assisted reproductive technologies and in vitro embryo culture influence the epigenetic control of imprinted genes and transposable elements in children? Hum. Reprod. 2021, 36, 479–492. [Google Scholar] [CrossRef]
- Mani, S.; Mainigi, M. Embryo Culture Conditions and the Epigenome. Semin. Reprod. Med. 2018, 36, 211–220. [Google Scholar] [CrossRef]
- Van Montfoort, A.P.A.; Hanssen, L.L.P.; de Sutter, P.; Viville, S.; Geraedts, J.P.M.; de Boer, P. Assisted reproduction treatment and epigenetic inheritance. Hum. Reprod. Update 2012, 18, 171–197. [Google Scholar] [CrossRef] [PubMed]
- Bruno, C.; Carmignac, V.; Netchine, I.; Choux, C.; Duffourd, Y.; Faivre, L.; Thauvin-Robinet, C.; Le Bouc, Y.; Sagot, P.; Bourc’his, D.; et al. Germline correction of an epimutation related to Silver-Russell syndrome. Hum. Mol. Genet. 2015, 24, 3314–3321. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jiang, Z.; Wang, Y.; Lin, J.; Xu, J.; Ding, G.; Huang, H. Genetic and epigenetic risks of assisted reproduction. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 44, 90–104. [Google Scholar] [CrossRef]
- Lazaraviciute, G.; Kauser, M.; Bhattacharya, S.; Haggarty, P.; Bhattacharya, S. A systematic review and meta-analysis of DNA methylation levels and imprinting disorders in children conceived by IVF/ICSI compared with children conceived spontaneously. Hum. Reprod. Update 2014, 20, 840–852. [Google Scholar] [CrossRef] [PubMed]
- Choux, C.; Binquet, C.; Carmignac, V.; Bruno, C.; Chapusot, C.; Barberet, J.; Lamotte, M.; Sagot, P.; Bourc’his, D.; Fauque, P. The epigenetic control of transposable elements and imprinted genes in newborns is affected by the mode of conception: ART versus spontaneous conception without underlying infertility. Hum. Reprod. 2018, 33, 331–340. [Google Scholar] [CrossRef]
- Chen, W.; Peng, Y.; Ma, X.; Kong, S.; Tan, S.; Wei, Y.; Zhao, Y.; Zhang, W.; Wang, Y.; Yan, L.; et al. Integrated multi-omics reveal epigenomic disturbance of assisted reproductive technologies in human offspring. EBioMedicine 2020, 61, 103076. [Google Scholar] [CrossRef] [PubMed]
- Melamed, N.; Choufani, S.; Wilkins-Haug, L.E.; Koren, G.; Weksberg, R. Comparison of genome-wide and gene-specific DNA methylation between ART and naturally conceived pregnancies. Epigenetics 2015, 10, 474–483. [Google Scholar] [CrossRef] [PubMed]
- El Hajj, N.; Haertle, L.; Dittrich, M.; Denk, S.; Lehnen, H.; Hahn, T.; Schorsch, M.; Haaf, T. DNA methylation signatures in cord blood of ICSI children. Hum. Reprod. 2017, 32, 1761–1769. [Google Scholar] [CrossRef]
- Choufani, S.; Turinsky, A.L.; Melamed, N.; Greenblatt, E.; Brudno, M.; Bérard, A.; Fraser, W.D.; Weksberg, R.; Trasler, J.; Monnier, P. Impact of assisted reproduction, infertility, sex and paternal factors on the placental DNA methylome. Hum. Mol. Genet. 2019, 28, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Gentilini, D.; Somigliana, E.; Pagliardini, L.; Rabellotti, E.; Garagnani, P.; Bernardinelli, L.; Papaleo, E.; Candiani, M.; Di Blasio, A.M.; Viganò, P. Multifactorial analysis of the stochastic epigenetic variability in cord blood confirmed an impact of common behavioral and environmental factors but not of in vitro conception. Clin. Epigenetics 2018, 10, 77. [Google Scholar] [CrossRef] [PubMed]
- Yeung, E.H.; Mendola, P.; Sundaram, R.; Zeng, X.; Guan, W.; Tsai, M.Y.; Robinson, S.L.; Stern, J.E.; Ghassabian, A.; Lawrence, D.; et al. Conception by fertility treatment and offspring deoxyribonucleic acid methylation. Fertil. Steril. 2021, 116, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Novakovic, B.; Lewis, S.; Halliday, J.; Kennedy, J.; Burgner, D.P.; Czajko, A.; Kim, B.; Sexton-Oates, A.; Juonala, M.; Hammarberg, K.; et al. Assisted reproductive technologies are associated with limited epigenetic variation at birth that largely resolves by adulthood. Nat. Commun. 2019, 10, 3922. [Google Scholar] [CrossRef] [PubMed]
- Bouillon, C.; Léandri, R.; Desch, L.; Ernst, A.; Bruno, C.; Cerf, C.; Chiron, A.; Souchay, C.; Burguet, A.; Jimenez, C.; et al. Does embryo culture medium influence the health and development of children born after in vitro fertilization? PLoS ONE 2016, 11, e0150857. [Google Scholar] [CrossRef] [PubMed]
- Phipson, B.; Maksimovic, J.; Oshlack, A. MissMethyl: An R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics 2016, 32, 286–288. [Google Scholar] [CrossRef] [PubMed]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef]
- Maksimovic, J.; Gordon, L.; Oshlack, A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012, 13, r44. [Google Scholar] [CrossRef]
- Zheng, S.C.; Webster, A.P.; Dong, D.; Feber, A.; Graham, D.G.; Sullivan, R.; Jevons, S.; Lovat, L.B.; Beck, S.; Widschwendter, M.; et al. A novel cell-type deconvolution algorithm reveals substantial contamination by immune cells in saliva, buccal and cervix. Epigenomics 2018, 10, 925–940. [Google Scholar] [CrossRef]
- Zheng, Y.; Joyce, B.T.; Liu, L.; Zhang, Z.; Kibbe, W.A.; Zhang, W.; Hou, L. Prediction of genome-wide DNA methylation in repetitive elements. Nucleic Acids Res. 2017, 45, 8697–8711. [Google Scholar] [CrossRef] [PubMed]
- Pervjakova, N.; Kasela, S.; Morris, A.P.; Kals, M.; Metspalu, A.; Lindgren, C.M.; Salumets, A.; Mägi, R. Imprinted genes and imprinting control regions show predominant intermediate methylation in adult somatic tissues. Epigenomics 2016, 8, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Zhang, X.; Huang, C.C.; Jafari, N.; Kibbe, W.A.; Hou, L.; Lin, S.M. Comparison of β-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010, 11, 587. [Google Scholar] [CrossRef]
- Peters, T.J.; Buckley, M.J.; Statham, A.L.; Pidsley, R.; Samaras, K.; Lord, R.V.; Clark, S.J.; Molloy, P.L. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 2015, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Morris, T.J.; Webster, A.P.; Yang, Z.; Beck, S.; Feber, A.; Teschendorff, A.E. ChAMP: Updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics 2017, 33, 3982–3984. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017, 45, D833–D839. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zou, D.; Li, Z.; Gao, R.; Sang, J.; Zhang, Y.; Li, R.; Xia, L.; Zhang, T.; Niu, G.; et al. EWAS Atlas: A curated knowledgebase of epigenome-wide association studies. Nucleic Acids Res. 2019, 47, D983–D988. [Google Scholar] [CrossRef] [PubMed]
- Battram, T.; Yousefi, P.; Crawford, G.; Prince, C.; Babei, M.S.; Sharp, G.; Hatcher, C.; Vega-Salas, M.J.; Khodabakhsh, S.; Whitehurst, O.; et al. The EWAS Catalog: A database of epigenome-wide association studies. OSF Prepr 2021, 2–5. [Google Scholar] [CrossRef]
- Katari, S.; Turan, N.; Bibikova, M.; Erinle, O.; Chalian, R.; Foster, M.; Gaughan, J.P.; Coutifaris, C.; Sapienza, C. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum. Mol. Genet. 2009, 18, 3769–3778. [Google Scholar] [CrossRef]
- Camprubí, C.; Iglesias-Platas, I.; Martin-Trujillo, A.; Salvador-Alarcon, C.; Rodriguez, M.A.; Barredo, D.R.; Court, F.; Monk, D. Stability of genomic imprinting and gestational-age dynamic methylation in complicated pregnancies conceived following assisted reproductive technologies. Biol. Reprod. 2013, 89, 50. [Google Scholar] [CrossRef]
- Estill, M.S.; Bolnick, J.M.; Waterland, R.A.; Bolnick, A.D.; Diamond, M.P.; Krawetz, S.A. Assisted reproductive technology alters deoxyribonucleic acid methylation profiles in bloodspots of newborn infants. Fertil. Steril. 2016, 106, 629–639.e10. [Google Scholar] [CrossRef]
- Xu, N.; Barlow, G.M.; Cui, J.; Wang, E.T.; Lee, B.; Akhlaghpour, M.; Kroener, L.; Williams, J.; Rotter, J.I.; Yii-der, I.C.; et al. Comparison of Genome-Wide and Gene-Specific DNA Methylation Profiling in First-Trimester Chorionic Villi from Pregnancies Conceived with Infertility Treatments. Reprod. Sci. 2017, 24, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Fernandez, J.E.; Loke, Y.J.; Bass-Stringer, S.; Gao, F.; Xia, Y.; Wu, H.; Lu, H.; Liu, Y.; Wang, J.; Spector, T.D.; et al. DNA methylation changes at infertility genes in newborn twins conceived by in vitro fertilisation. Genome Med. 2017, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Tobi, E.W.; Almqvist, C.; Hedman, A.; Andolf, E.; Holte, J.; Olofsson, J.I.; Wramsby, H.; Wramsby, M.; Pershagen, G.; Heijmans, B.T.; et al. DNA methylation differences at birth after conception through ART. Hum. Reprod. 2021, 36, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Penova-Veselinovic, B.; Melton, P.E.; Huang, R.C.; Yovich, J.L.; Burton, P.; Wijs, L.A.; Hart, R.J. DNA methylation patterns within whole blood of adolescents born from assisted reproductive technology are not different from adolescents born from natural conception. Hum. Reprod. 2021, 36, 2035–2049. [Google Scholar] [CrossRef]
- Caramaschi, D.; Jungius, J.; Page, C.M.; Novakovic, B.; Saffery, R.; Halliday, J.; Lewis, S.; Magnus, M.C.; London, S.J.; Håberg, S.E.; et al. Association of medically assisted reproduction with offspring cord blood DNA methylation across cohorts. Human Reprod. 2021, 36, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Van Dongen, J.; Ehli, E.A.; Jansen, R.; Van Beijsterveldt, C.E.; Willemsen, G.; Hottenga, J.J.; Kallsen, N.A.; Peyton, S.A.; Breeze, C.E.; Kluft, C.; et al. Genome-wide analysis of DNA methylation in buccal cells: A study of monozygotic twins and mQTLs. Epigenetics Chromatin 2018, 11, 54. [Google Scholar] [CrossRef]
- Guo, S.; Diep, D.; Plongthongkum, N.; Fung, H.L.; Zhang, K.; Zhang, K. Identification of methylation haplotype blocks AIDS in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat. Genet. 2017, 49, 635–642. [Google Scholar] [CrossRef]
- Niemitz, E.L.; Feinberg, A.P. Epigenetics and assisted reproductive technology: A Call for Investigation. Epigenetic Epidemiol. 2014, 74, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Breton, C.V.; Marsit, C.J.; Faustman, E.; Nadeau, K.; Goodrich, J.M.; Dolinoy, D.C.; Herbstman, J.; Holland, N.; LaSalle, J.M.; Schmidt, R.; et al. Small-magnitude effect sizes in epigenetic end points are important in children’s environmental health studies: The children’s environmental health and disease prevention research center’s epigenetics working group. Environ. Health Perspect. 2017, 125, 511–526. [Google Scholar] [CrossRef]
- Sheng, W.; Chen, L.; Wang, H.; Ma, X.; Ma, D.; Huang, G. CpG island shore methylation of ZFPM2 is identified in tetralogy of fallot samples. Pediatric Res. 2016, 80, 151–158. [Google Scholar] [CrossRef]
- Sheng, W.; Qian, Y.; Wang, H.; Ma, X.; Zhang, P.; Diao, L.; An, Q.; Chen, L.; Ma, D.; Huang, G. DNA methylation status of NKX2-5, GATA4 and HAND1 in patients with tetralogy of fallot. BMC Med. Genom. 2013, 6, 46. [Google Scholar] [CrossRef]
- Zhu, Y.; Ye, M.; Xu, H.; Gu, R.; Ma, X.; Chen, M.; Li, X.; Sheng, W.; Huang, G. Methylation status of CpG sites in the NOTCH4 promoter region regulates NOTCH4 expression in patients with tetralogy of Fallot. Mol. Med. Rep. 2020, 22, 4412–4422. [Google Scholar] [CrossRef] [PubMed]
- Tararbit, K.; Lelong, N.; Thieulin, A.C.; Houyel, L.; Bonnet, D.; Goffinet, F.; Khoshnood, B.; EPICARD Study Group. The risk for four specific congenital heart defects associated with assisted reproductive techniques: A population-based evaluation. Hum. Reprod. 2013, 28, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Giorgione, V.; Parazzini, F.; Fesslova, V.L.; Cipriani, S.O.; Candiani, M.; Inversetti, A.N.; Sigismondi, C.; Tiberio, F.; Cavoretto, P. Congenital heart defects in IVF/ICSI pregnancy: Systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 2018, 51, 33–42. [Google Scholar] [CrossRef]
- Radhakrishna, U.; Vishweswaraiah, S.; Veerappa, A.M.; Zafra, R.; Albayrak, S.; Sitharam, P.H.; Saiyed, N.M.; Mishra, N.K.; Guda, C.; Bahado-Singh, R. Newborn blood DNA epigenetic variations and signaling pathway genes associated with Tetralogy of Fallot (TOF). PLoS ONE 2018, 13, e0203893. [Google Scholar] [CrossRef]
- Halliday, J.; Lewis, S.; Kennedy, J.; Burgner, D.P.; Juonala, M.; Hammarberg, K.; Amor, D.J.; Doyle, L.W.; Saffery, R.; Ranganathan, S.; et al. Health of adults aged 22 to 35 years conceived by assisted reproductive technology. Fertil. Steril. 2019, 112, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Whitelaw, N.; Bhattacharya, S.; Hoad, G.; Horgan, G.W.; Hamilton, M.; Haggarty, P. Epigenetic status in the offspring of spontaneous and assisted conception. Hum. Reprod. 2014, 29, 1452–1458. [Google Scholar] [CrossRef]
- Santos, F.; Hyslop, L.; Stojkovic, P.; Leary, C.; Murdoch, A.; Reik, W.; Stojkovic, M.; Herbert, M.; Dean, W. Evaluation of epigenetic marks in human embryos derived from IVF and ICSI. Hum. Reprod. 2010, 25, 2387–2395. [Google Scholar] [CrossRef]
- Ghosh, J.; Coutifaris, C.; Sapienza, C.; Mainigi, M. Global DNA methylation levels are altered by modifiable clinical manipulations in assisted reproductive technologies. Clin. Epigenetics 2017, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Tierling, S.; Souren, N.Y.; Gries, J.; LoPorto, C.; Groth, M.; Lutsik, P.; Neitzel, H.; Utz-Billing, I.; Gillessen-Kaesbach, G.; Kentenich, H.; et al. Assisted reproductive technologies do not enhance the variability of DNA methylation imprints in human. J. Med. Genet. 2010, 47, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Loke, Y.J.; Galati, J.C.; Saffery, R.; Craig, J.M. Association of in vitro fertilisation (IVF) with global and IGF2/H19 methylation variation in newborn twins. J. Paediatr. Child Health 2015, 51, 21. [Google Scholar] [CrossRef]
- Vincent, R.N.; Gooding, L.D.; Louie, K.; Wong, E.C.; Ma, S. Altered DNA methylation and expression of PLAGL1 in cord blood from assisted reproductive technology pregnancies compared with natural conceptions. Fertil. Steril. 2016, 106, 739–748.e3. [Google Scholar] [CrossRef]
- Fauque, P. Ovulation induction and epigenetic anomalies. Fertil. Steril. 2013, 99, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Marshall, K.L.; Rivera, R.M. The effects of superovulation and reproductive aging on the epigenome of the oocyte and embryo. Mol. Reprod. Dev. 2018, 85, 90–105. [Google Scholar] [CrossRef]
- Lowe, R.; Gemma, C.; Beyan, H.; Hawa, M.I.; Bazeos, A.; Leslie, R.D.; Montpetit, A.; Rakyan, V.K.; Ramagopalan, S.V. Buccals are likely to be a more informative surrogate tissue than blood for epigenome-wide association studies. Epigenetics 2013, 8, 445–454. [Google Scholar] [CrossRef]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, 3156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cui, Y.; Zhou, Z.; Sha, J.; Li, Y.; Liu, J. Altered global gene expressions of human placentae subjected to assisted reproductive technology treatments. Placenta 2010, 31, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Fauque, P.; Ripoche, M.A.; Tost, J.; Journot, L.; Gabory, A.; Busato, F.; Le Digarcher, A.; Mondon, F.; Gut, I.; Jouannet, P.; et al. Modulation of imprinted gene network in placenta results in normal development of in vitro manipulated mouse embryos. Hum. Mol. Genet. 2010, 19, 1779–1790. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ahmadi, H.; Fathi, F.; Karimi, H.; Amidi, F.; Mehdinejadiani, S.; Moeini, A.; Hoseini, S.; Sobhani, A. Altered TH1, TH2, TH17 balance in assisted reproductive technology conceived mice. J. Reprod. Immunol. 2020, 139, 103117. [Google Scholar] [CrossRef] [PubMed]
- Karimi, H.; Mahdavi, P.; Fakhari, S.; Faryabi, M.R.; Esmaeili, P.; Banafshi, O.; Mohammadi, E.; Fathi, F.; Mokarizadeh, A. Altered helper T cell-mediated immune responses in male mice conceived through in vitro fertilization. Reprod. Toxicol. 2017, 69, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Nelissen, E.C.M.; van Montfoort, A.P.A.; Dumoulin, J.C.M.; Evers, J.L.H. Epigenetics and the placenta. Hum. Reprod. Update 2011, 17, 397–417. [Google Scholar] [CrossRef] [PubMed]
- Choux, C.; Carmignac, V.; Bruno, C.; Sagot, P.; Vaiman, D.; Fauque, P. The placenta: Phenotypic and epigenetic modifications induced by Assisted Reproductive Technologies throughout pregnancy. Clin. Epigenetics 2015, 7, 87. [Google Scholar] [CrossRef]
- Mansell, G.; Gorrie-Stone, T.J.; Bao, Y.; Kumari, M.; Schalkwyk, L.S.; Mill, J.; Hannon, E. Guidance for DNA methylation studies: Statistical insights from the Illumina EPIC array. BMC Genom. 2019, 20, 366. [Google Scholar] [CrossRef] [PubMed]
- Bush, N.R.; Edgar, R.D.; Park, M.; MacIsaac, J.L.; McEwen, L.M.; Adler, N.E.; Essex, M.J.; Kobor, M.S.; Boyce, W.T. The biological embedding of early-life socioeconomic status and family adversity in children’s genome-wide DNA methylation. Epigenomics 2018, 10, 1445–1461. [Google Scholar] [CrossRef] [PubMed]

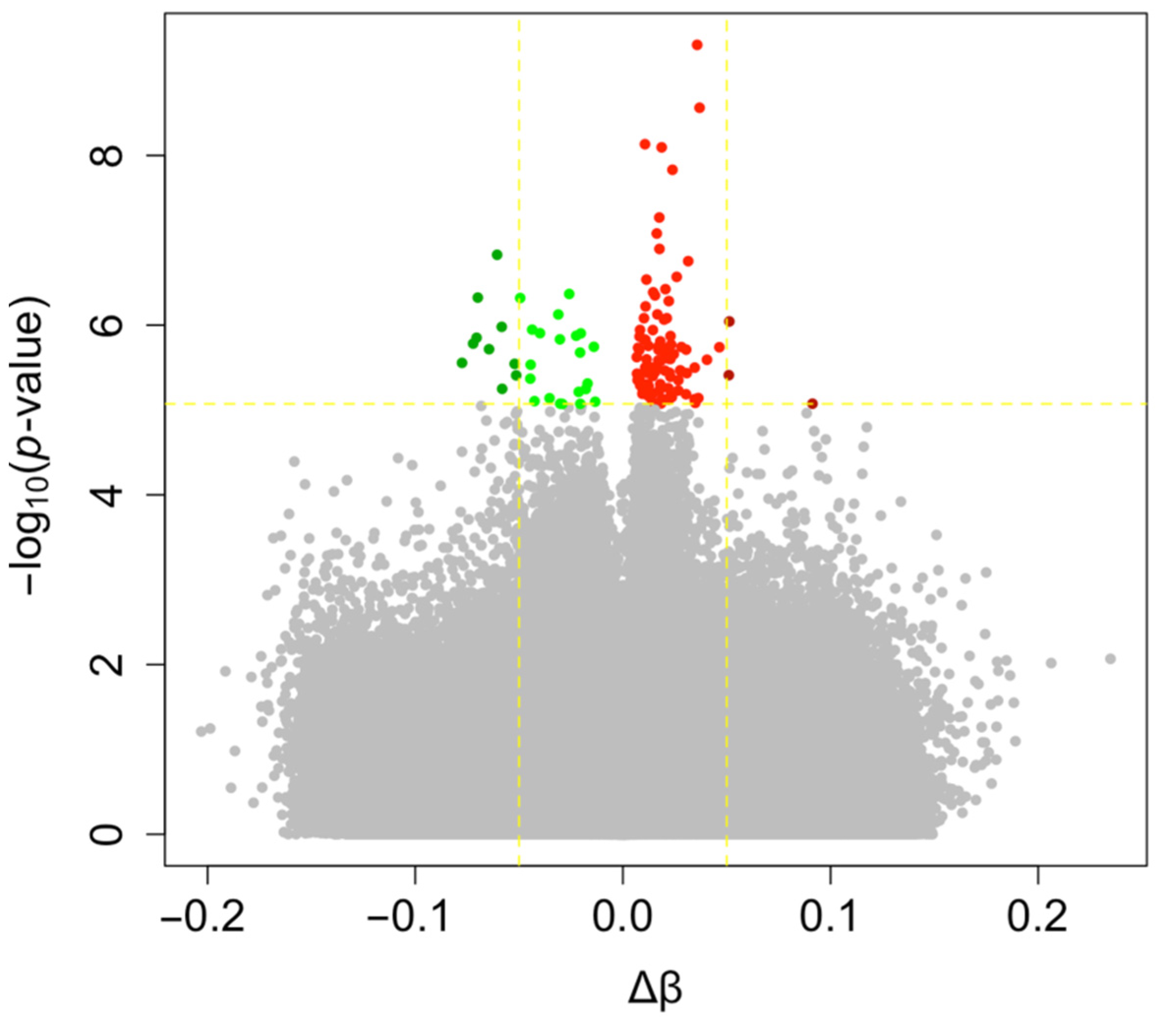
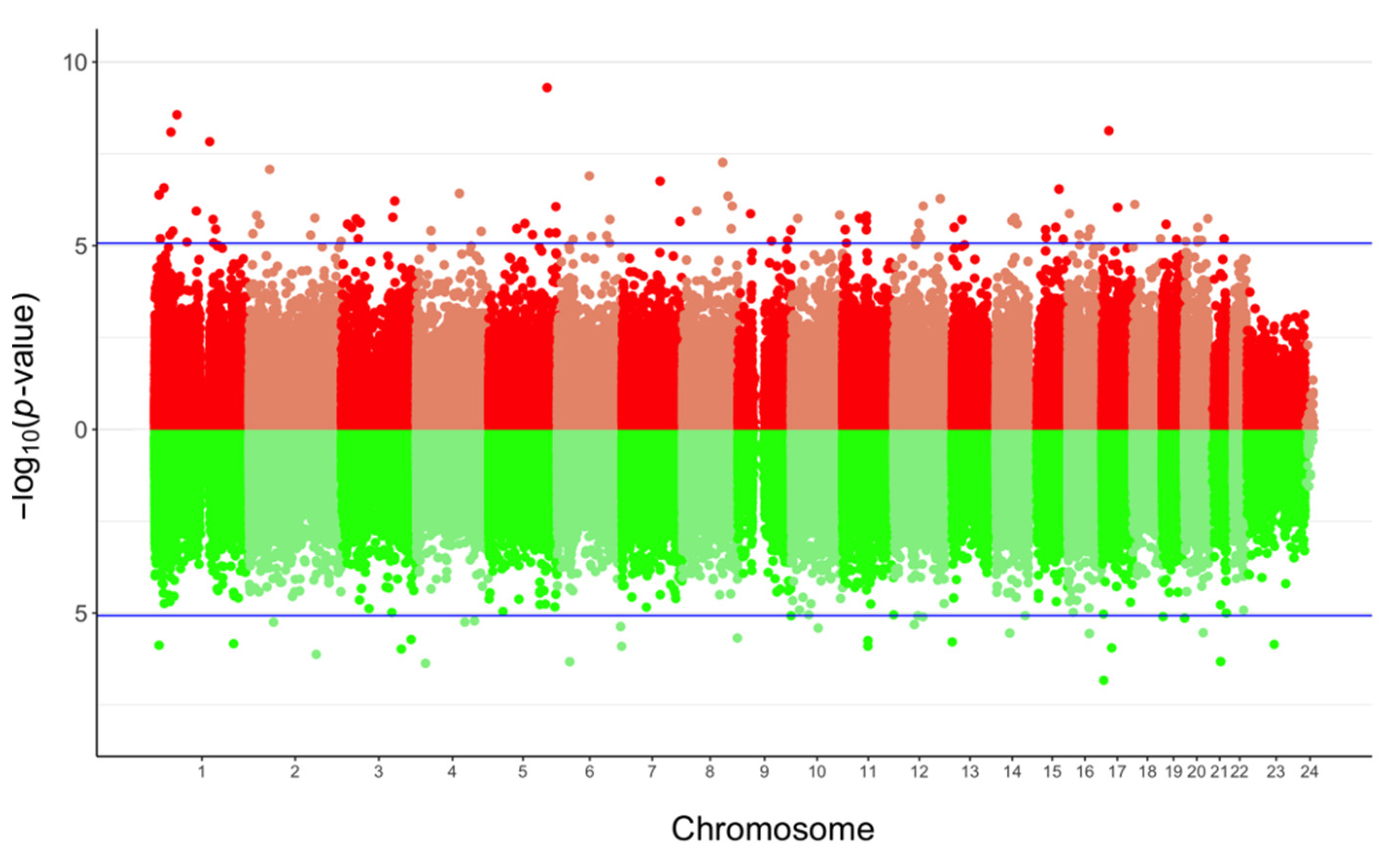
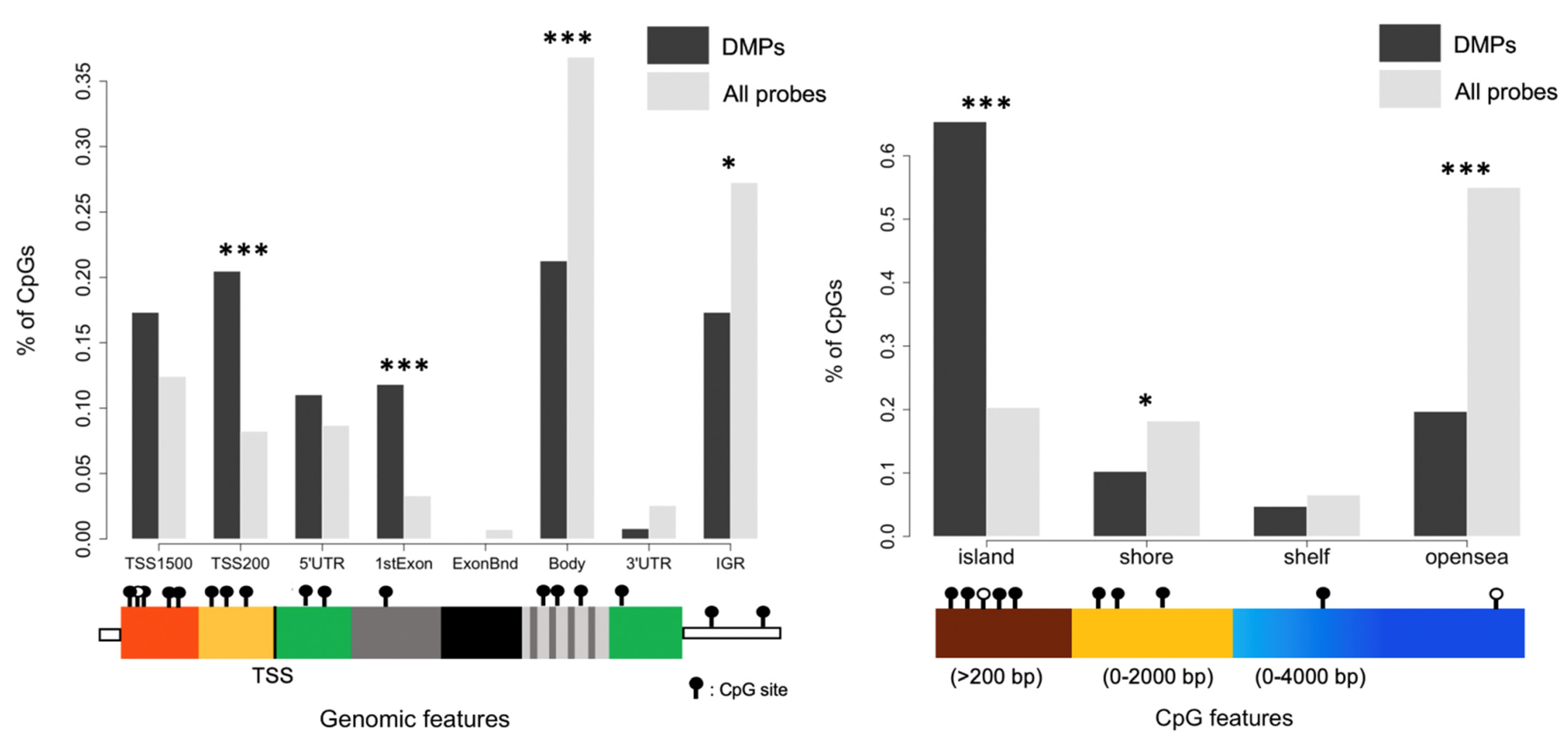
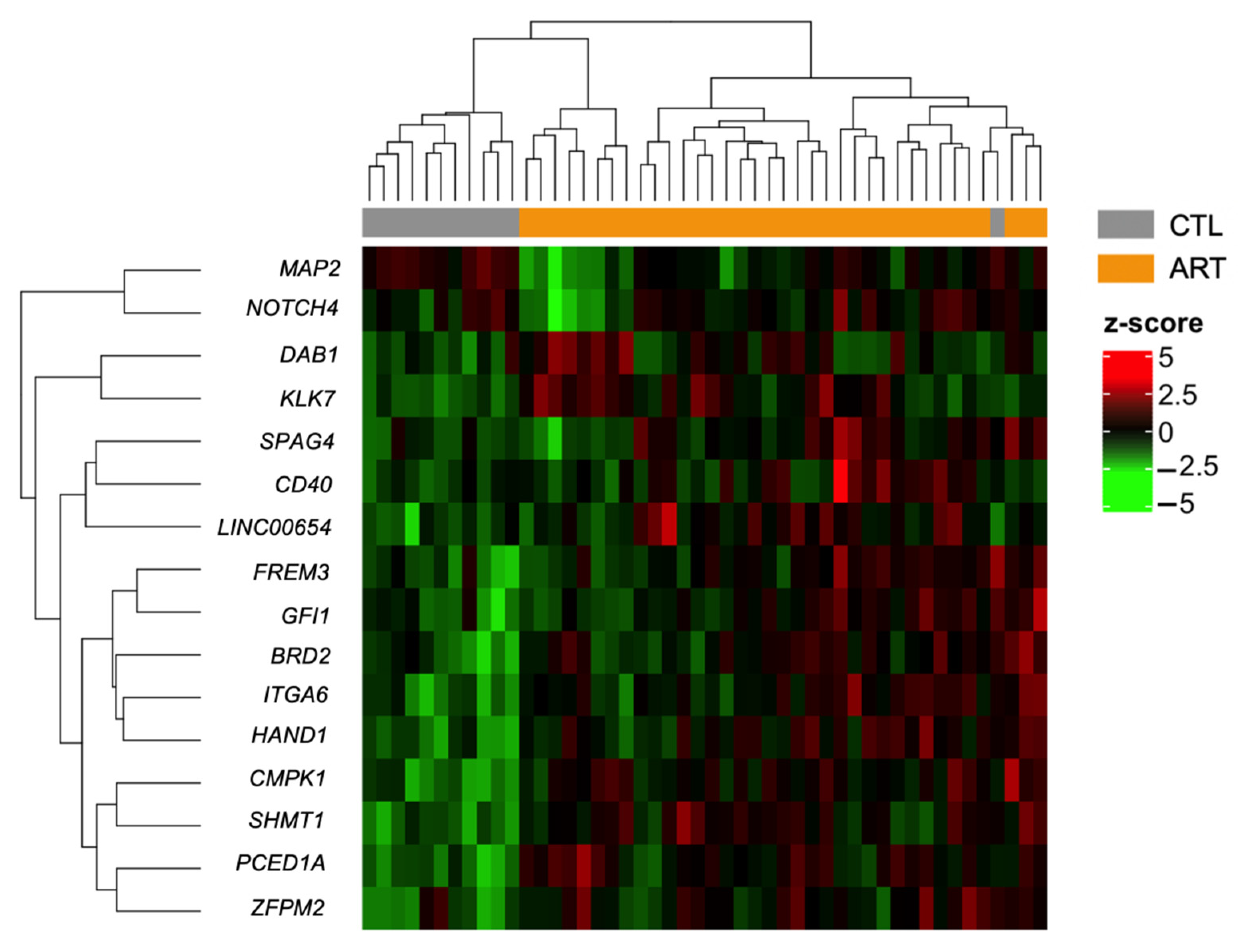
| Probe ID | p-Value | Δβ | Genomic Feature | CpG Feature | Gene |
|---|---|---|---|---|---|
| cg25587535 | 0.0004 | 0.036 | TSS200 | island | HAND1 |
| cg26311208 | 0.0010 | 0.037 | TSS200 | island | DAB1 |
| cg18958584 | 0.0015 | 0.011 | 1stExon | island | SHMT1 |
| cg24877558 | 0.0015 | 0.019 | TSS1500 | island | FOXJ3 |
| cg18788524 | 0.0022 | 0.024 | 1stExon | island | SEC22B |
| cg17154315 | 0.0067 | 0.018 | 5′UTR | island | ZFPM2 |
| cg02079951 | 0.0088 | 0.016 | Body | island | ASB3 |
| cg00270497 | 0.0117 | 0.018 | TSS200 | island | RIPPLY2 |
| cg13593809 | 0.0122 | −0.061 | Body | shore | LOC101559451 |
| cg19767562 | 0.0130 | 0.031 | Body | island | TFR2 |
| cg04856657 | 0.0179 | 0.026 | TSS200 | island | PNRC2 |
| cg23727043 | 0.0179 | 0.011 | TSS1500 | island | ADAMTS7 |
| cg05700616 | 0.0197 | −0.026 | IGR | opensea | PPARGC1A |
| cg11857246 | 0.0197 | 0.015 | 5′UTR | island | MAD2L2 |
| cg14427382 | 0.0197 | −0.070 | Body | shore | LOC100294145 |
| cg16866373 | 0.0197 | 0.015 | 5′UTR | island | CCN3 |
| cg19306866 | 0.0197 | −0.049 | IGR | opensea | KRTAP6-2 |
| ch.4.113910337F | 0.0197 | 0.020 | IGR | opensea | ANK2 |
| cg00243897 | 0.0203 | 0.022 | TSS200 | island | HPD |
| cg12110529 | 0.0223 | 0.011 | IGR | island | ZBTB38 |
| Location (hg19) | Number of Probes | FDR | Maximum Difference | Mean Difference | Gene | Genomic Feature |
|---|---|---|---|---|---|---|
| chr19:51486901-51487968 | 14 | 4.49 × 10−22 | 0.084 | 0.038 | KLK7 | covers exons |
| chr20:34204902-34205488 | 7 | 2.81 × 10−20 | 0.062 | 0.030 | SPAG4 | covers exons |
| chr20:5485144-5486007 | 7 | 2.86 × 10−20 | 0.127 | 0.094 | LINC00654 | overlaps exon upstream |
| chr5:153857468-153858102 | 7 | 1.10 × 10−16 | 0.038 | 0.011 | HAND1 | overlaps exon upstream |
| chr20:44746392-44747351 | 9 | 2.19 × 10−12 | 0.103 | 0.064 | CD40 | covers exons |
| chr1:92949813-92950575 | 20 | 1.34 × 10−11 | 0.028 | 0.007 | GFI1 | inside intron |
| chr2:173292579-173292636 | 2 | 8.09 × 10−11 | 0.017 | 0.013 | ITGA6 | inside exon |
| chr4:144621270-144621385 | 3 | 1.34 × 10−10 | 0.017 | 0.013 | FREM3 | inside exon |
| chr1:59012392-59012820 | 11 | 1.84 × 10−10 | 0.035 | 0.001 | DAB1 | overlaps exon upstream |
| chr1:47799827-47800167 | 3 | 3.11 × 10−10 | 0.015 | 0.011 | CMPK1 | inside intron |
| chr20:2820742-2821472 | 14 | 8.23 × 10−10 | 0.019 | 0.008 | PCED1A | covers exons |
| chr8:106331160-106331166 | 2 | 9.47 × 10−10 | 0.018 | 0.008 | ZFPM2 | inside exon |
| chr2:210444075-210444270 | 6 | 1.17 × 10−09 | −0.067 | −0.043 | MAP2 | inside intron |
| chr17:18266764-18266775 | 2 | 1.51 × 10−09 | 0.011 | 0.007 | SHMT1 | inside exon |
| chr6:32164503-32164801 | 7 | 1.88 × 10−09 | 0.053 | 0.034 | NOTCH4 | overlaps exon downstream |
| chr6:32939777-32940054 | 10 | 1.88 × 10−09 | 0.020 | 0.004 | BRD2 | inside exon |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ducreux, B.; Frappier, J.; Bruno, C.; Doukani, A.; Guilleman, M.; Simon, E.; Martinaud, A.; Bourc’his, D.; Barberet, J.; Fauque, P. Genome-Wide Analysis of DNA Methylation in Buccal Cells of Children Conceived through IVF and ICSI. Genes 2021, 12, 1912. https://doi.org/10.3390/genes12121912
Ducreux B, Frappier J, Bruno C, Doukani A, Guilleman M, Simon E, Martinaud A, Bourc’his D, Barberet J, Fauque P. Genome-Wide Analysis of DNA Methylation in Buccal Cells of Children Conceived through IVF and ICSI. Genes. 2021; 12(12):1912. https://doi.org/10.3390/genes12121912
Chicago/Turabian StyleDucreux, Bastien, Jean Frappier, Céline Bruno, Abiba Doukani, Magali Guilleman, Emmanuel Simon, Aurélie Martinaud, Déborah Bourc’his, Julie Barberet, and Patricia Fauque. 2021. "Genome-Wide Analysis of DNA Methylation in Buccal Cells of Children Conceived through IVF and ICSI" Genes 12, no. 12: 1912. https://doi.org/10.3390/genes12121912
APA StyleDucreux, B., Frappier, J., Bruno, C., Doukani, A., Guilleman, M., Simon, E., Martinaud, A., Bourc’his, D., Barberet, J., & Fauque, P. (2021). Genome-Wide Analysis of DNA Methylation in Buccal Cells of Children Conceived through IVF and ICSI. Genes, 12(12), 1912. https://doi.org/10.3390/genes12121912






