Developmental Effects of (Pre-)Gestational Diabetes on Offspring: Systematic Screening Using Omics Approaches
Abstract
:1. (Pre-)Gestational Diabetes Mellitus and Omics: A Brief Introduction
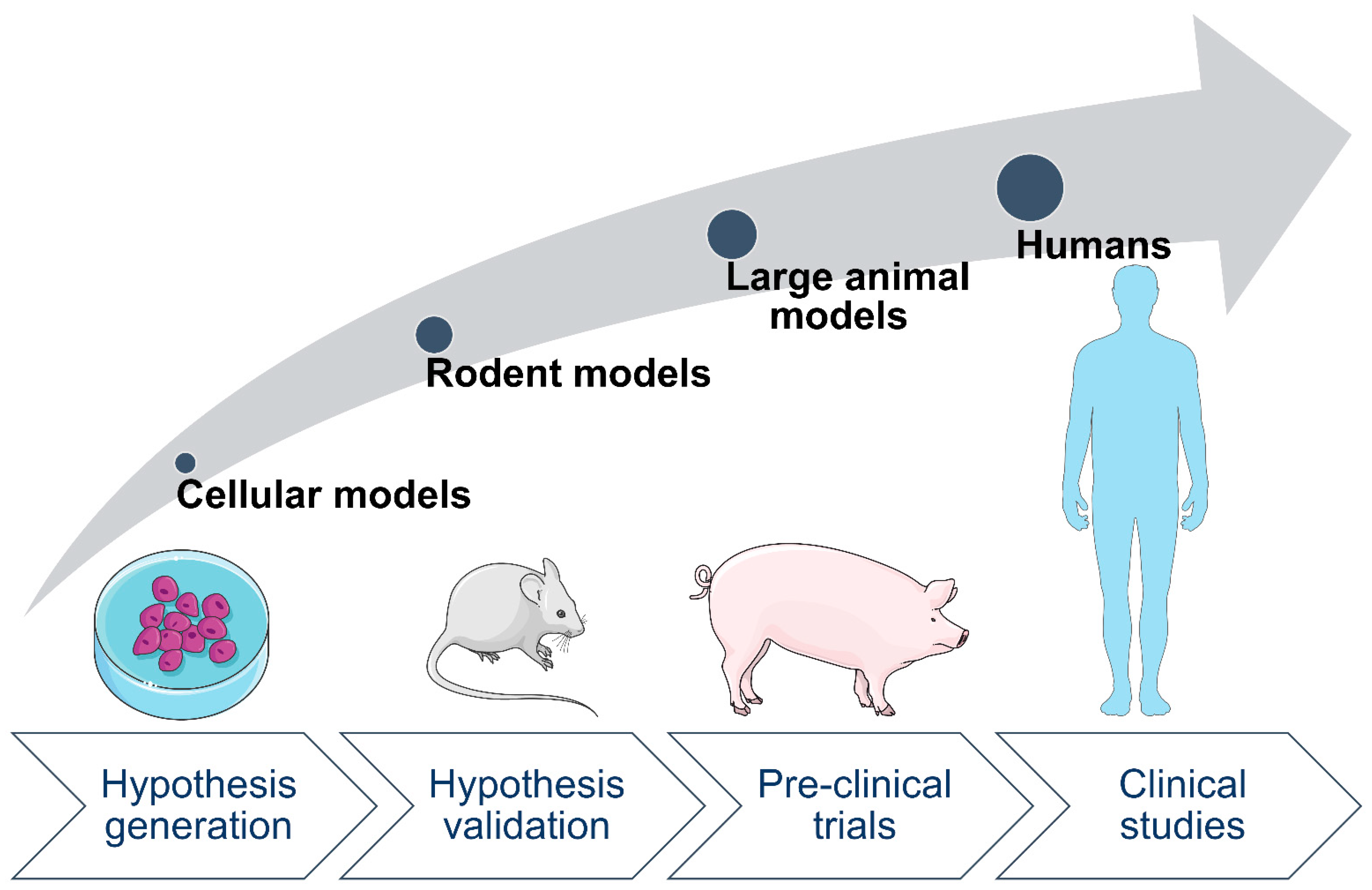
2. Animal Models Are Valuable for Studying Effects of Maternal Diabetes on Offspring
3. Common Tissues and Biofluids for Studying Effects of Maternal Diabetes on Offspring
4. (Epi)Genetic Factors Affecting Offspring Outcomes after Exposure to Maternal Diabetes
| Maternal Characteristics | Bio-Specimen | Major Findings in Offspring | Reference |
|---|---|---|---|
| GDM | Fetal-side placenta | Reduced methylation level of LEP, contributing to cord blood leptin level regulation | [61] |
| GDM | Fetal-side placenta | Increased LEP methylation | [60] |
| GDM | Fetal-side placenta | Altered methylation of PPARGC1A mediating the association between maternal hyperglycemia and cord blood leptin levels | [63] |
| GDM | Fetal-side placenta | DNA methylation profile of ADIPOQ was associated with maternal glucose status | [67] |
| GDM | Fetal-side placenta | Reduced LPL methylation | [69] |
| GDM | Fetal-side placenta | Epivariation near the LPL locus correlated with anthropometric parameters (birth weight, mid-childhood weight, fat mass) of children at age 5 years | [70] |
| GDM | Fetal-side placenta | Reduced SLC6A4 DNA methylation | [71] |
| GDM | Cord blood and chorionic villi | Decreased MEST methylation | [77] |
| GDM | Cord blood | Altered methylation of the OR2L13 promoter (a gene associated with autism spectrum disorder) and of the gene body of CYP2E1 (which is upregulated in type 1 and type 2 diabetes) | [13] |
| GDM | Cord blood | Differentially methylated genes associated with type 1 diabetes mellitus, immune MHC, and neuron development | [76] |
| GDM | Cord blood | Decreased LEP methylation; association with increased cord blood leptin levels | [62] |
| GDM | Peripheral blood | Differentially methylated genes associated with type 2 diabetes, obesity, diabetic nephropathy or coronary heart disease | [82] |
| GDM | Peripheral blood mononuclear cells | Differential methylation of several genes known to be associated with cardiometabolic traits; | [79] |
| GDM | Peripheral blood | Accelerated epigenetic aging associated with cardiometabolic risk factors | [78] |
| GDM | Peripheral blood | Methylation of SH3PXD2A was associated with multiple adiposity-related outcomes, including BMI, waist circumference, and circulating leptin levels | [80] |
5. Transcriptomic Changes in Offspring after Exposure to Maternal Diabetes
| Species | Maternal Characteristics | Bio-Specimen | Major Findings in Offspring | Reference |
|---|---|---|---|---|
| Human | GDM | HUVEC | Increased mRNA levels of genes coding for growth factors linked to insulin sensing and to the extracellular matrix | [87] |
| Human | Type 1 diabetes | Umbilical cord | Altered expression of genes involved in vascular development, vessel wall integrity, and vascular function | [88] |
| Rat | STZ-induced diabetes | Heart | Altered expression of Ahsp and Kel; possible relation to polycythemia | [99] |
| Mouse | STZ-induced diabetes | Heart | Altered expression of Cd36 and Ldha induced by maternal diabetes plus haploinsufficiency of Hif1a | [97] |
| Mouse | STZ-induced diabetes | Brain | Dysregulation of genes in frontal cortex related to forebrain development; dysregulation of neurodevelopment and immune-related genes in the striatum | [94] |
| Mouse | Diet-induced diabetes | Brain | Altered expression of genes related to inflammatory and neurodevelopmental processes | [95] |
| Rat | Infusion model of localized hyperglycemia | Islets | Dysregulation of genes associated to diabetes mellitus, inflammation and cell-death pathways | [89] |
| Mouse | STZ-induced diabetes | Liver | Differential expression of genes related to “FOXO signaling pathway” and “PPAR signaling pathway” in male offspring, and of genes related to “AMPK signaling pathway”, “fatty acid metabolism pathway”, and “PPAR signaling pathway” in female offspring | [90] |
6. Proteomic Changes in Offspring after Exposure to Maternal Diabetes
7. Metabolomic Changes in Offspring after Exposure to Maternal Diabetes
| Species | Maternal Characteristics | Bio-Specimen | Major Findings in Offspring | Reference |
|---|---|---|---|---|
| Human | GDM | Blood | Concentrations of lysine, putrescine, guanidinoacetate, and hexadecanedioate were negatively correlated with maternal hyperglycemia | [127] |
| Human | GDM | Cord Blood | Phosphatidylcholine acyl-alkyl C 32:1 and proline levels were associated with maternal GDM | [117] |
| Human | GDM | Blood | Association of the phospholipid metabolic pattern with higher adiposity, impaired insulin sensitivity and altered adipocytokines across the adolescent transition, among girls exposed to in utero GDM | [114] |
| Human | GDM | Blood | Intergenerational correlation of meta-bolites (carnitine, PC ae C34:3, taurine, creatinine, proline, SM-(OH) C14:1) between women with GDM and offspring 8 years after birth | [115] |
| Human | GDM | Cord blood | Elevated concentrations of Pro, Met, Ile, Leu, Ala and Phe; potentially, increased Gln-to-Glu conversion | [118] |
| Human | GDM and PGDM | Cord blood | Altered concentrations of metabolites of carbohydrate and choline metabolism | [123] |
| Human | GDM and overweight/obesity | Cord blood | Alteration of metabolites associated with anthropometric changes in newborn children, which were not detected longitudinally | [116] |
| Human | GDM and overweight/obesity | Cord blood | Elevated total hexoses; decreased levels of free carnitine, acyl carnitines, long-chain non-esterified fatty acids, phospholipids, specific Krebs cycle metabolites, and β-oxidation markers in cord blood but not in maternal blood | [112] |
| Human | Hyperglycemia | Cord blood | Concentrations of 3-hydroxybutyrate and its carnitine ester, glycerol and medium chain carnitine esters correlated with maternal 1h glucose levels | [122] |
| Human | GDM | Urine and meconium | No difference in urine; evidence for disrupted metabolic pathways, including lipid, amino acid, and purine metabolism from meconium analysis; argininosuccinic acid, methyladenosine, methylguanosine, aurodeoxycholic acid, glycocholic acid, hydroxyindoleacetylglycine, oxotrihydroxyleukotriene B4, tetrahydrodipicolinate, and DHAP (8:0) suggested as markers for GDM-induced disorders | [124] |
| Human | Type 1 diabetes | Serum | No significant associations between maternal type 1 diabetes and metabolite concentrations in offspring | [121] |
| Pig | Mutant insulin C93S causing hyperglycemia | Plasma | Increased concentrations of lysine, α-aminoadipic acid and phospholipids; biochemical evidence for an increased mitochondrial import of fatty acids for β-oxidation | [130] |
| Rat | GDM | Liver | Increased levels of diacylglycerol and reduced levels of phosphatidylethanolamine | [129] |
| Mouse | IR | Liver | Altered concentrations of the 16:1n7 lipid family; at 6 months of age a trend towards increased triglyceride species, while phospholipids were significantly reduced | [128] |
8. Microbiomics and Nutriomics Studies Addressing GDM Effects on Offspring
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 16S rRNA | 16S ribosomal RNA |
| 2 hPG | 2-h plasma glucose |
| Ala | Alanine |
| ABCA1 | ATP-binding cassette transporter A1 |
| AC | Acyl-carnitine |
| ADIPOQ | Adiponectin |
| AF | Amniotic fluid |
| AGT | Angiotensinogen |
| Ahsp | α haemoglobin stabilizing protein |
| AMPK | AMP-activated protein kinase |
| APOA1 | Apolipoprotein A-I |
| APOD | Apolipoprotein D |
| APOM | Apolipoprotein M |
| ARHGEF11 | Rho guanine nucleotide exchange factor 11 |
| BMI | Body mass index |
| CEPT | Cholesteryl ester transfer protein |
| ChiP | Chromatin immunoprecipitation assay |
| CNYP2 | Canopy FGF signaling regulator 2 |
| CP | Ceruloplasmin |
| CpG | Cytosine-guanine dinucleotide |
| CVD | Cardiovascular disorder |
| CYP2E1 | Cytochrome P450 Family 2 Subfamily E Member 1 |
| DAZAP1 | Azoospermia Associated Protein 1 |
| DEG | Differentially expressed genes |
| DNA | Deoxyribonucleic acid |
| EFSOCH | The Exeter Family Study of Childhood Health |
| EGR1 | Early growth response 1 |
| ELISA | Enzyme-linked immunosorbent assay |
| EPOCH | Exploring Perinatal Outcomes among Children |
| FIA-ESI-MS/MS | Flow injection analysis-electrospray ionization-tandem mass spectrometry |
| FOXO1 | Forkhead box protein O1 |
| FPG | Fasting plasma glucose |
| FSH | Follicle-stimulating hormone |
| GDM | Gestational diabetes mellitus |
| Gln | Glutamine |
| Glu | Glutamate |
| GWAS | Genome-wide association study |
| HAPO | The Hyperglycemia and Adverse Pregnancy Outcome |
| HFHS | High fat, high sucrose diet |
| HFS | High fat diet and sucrose diet |
| HIF1 | Hypoxia-inducible factor 1 |
| HRG | Histidine-rich glycoprotein |
| HUVEC | Human umbilical vein endothelial cells |
| Ile | Isoleucine |
| IPA | Ingenuity Pathway Analysis |
| IR | Insulin resistance |
| IRS1-het | Haploinsufficient for insulin receptor substrate-1 |
| Kel | Kell metallo-endopeptidase |
| KNG1 | Kinogen-1 |
| LCAT | Lecithin–cholesterol acyltransferase |
| LC-MS/MS | Liquid Chromatography with tandem Mass Spectrometry |
| LDL | Low-density lipoprotein |
| Leu | Leucine |
| LF | Low fat diet |
| LGA | Large for gestational age |
| LPL | Lipoprotein lipase |
| LUM | Lumican |
| LV | Left ventricle |
| MEST | Mesoderm Specific Transcript |
| Met | Methionine |
| MHC | Major Histocompatibility Complex |
| MIA | Maternal immune activation |
| mRNA | Messenger RNA |
| NEFA | Non-esterified fatty acid |
| NGS | Next generation sequencing |
| NMR | Nuclear magnetic resonance |
| OGTT | Oral glucose tolerance test |
| OR2L13 | Olfactory Receptor Family 2 Subfamily L Member 13 |
| ORM2 | α-1-acid glycoprotein 2 |
| PACE | Pregnancy and Childhood epigenetics consortium |
| PCK1 | Phosphoenolpyruvate Carboxykinase 1 |
| PGDM | Pregestational diabetes mellitus |
| Phe | Phenylalanine |
| PL | Phospholipid |
| PLG | Plasminogen |
| PLTP | Phospholipid transfer protein |
| PPAR | Peroxisome proliferator-activated receptor |
| PPARGC1α | Peroxisome proliferator-activated receptor-γ, co-activator 1, α |
| Pro | Proline |
| RDH16 | Retinol Dehydrogenase 16 |
| RNA | Ribonucleic acid |
| SETP7 | Septin 7 |
| SH3PXD2A | SH3 And PX Domains 2A |
| SLC6A4 | Serotonin transporter gene |
| SNP | Single nucleotide polymorphism |
| SOD2 | Superoxide dismutase 2 |
| SRSF2 | Serine and Arginine Rich Splicing Factor 2 |
| STZ | Streptozotocin |
| TF | Serotransferrin |
| UHPLC-MS | Ultra-high-performance liquid chromatography Mass spectrometry |
| Vegfa | Vascular endothelial growth factor A |
| WB | Western blot |
| WGCNA | Weighted Correlation Network Analysis |
References
- Hadden, D.R.; McLaughlin, C. Normal and abnormal maternal metabolism during pregnancy. Semin. Fetal Neonatal Med. 2009, 14, 66–71. [Google Scholar] [CrossRef]
- Herrera, E. Metabolic adaptations in pregnancy and their implications for the availability of substrates to the fetus. Eur. J. Clin. Nutr. 2000, 54, S47–S51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, Z.; Liu, F.; Li, S. Metabolic Adaptations in Pregnancy: A Review. Ann. Nutr. Metab. 2017, 70, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Parrettini, S.; Caroli, A.; Torlone, E. Nutrition and Metabolic Adaptations in Physiological and Complicated Pregnancy: Focus on Obesity and Gestational Diabetes. Front. Endocrinol. 2020, 11, 611929. [Google Scholar] [CrossRef] [PubMed]
- Holme, A.M.; Roland, M.C.; Lorentzen, B.; Michelsen, T.M.; Henriksen, T. Placental glucose transfer: A human in vivo study. PLoS ONE 2015, 10, e0117084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butte, N.F. Carbohydrate and lipid metabolism in pregnancy: Normal compared with gestational diabetes mellitus. Am. J. Clin. Nutr. 2000, 71, S1256–S1261. [Google Scholar] [CrossRef]
- Di Cianni, G.; Miccoli, R.; Volpe, L.; Lencioni, C.; Del Prato, S. Intermediate metabolism in normal pregnancy and in gestational diabetes. Diabetes Metab. Res. Rev. 2003, 19, 259–270. [Google Scholar] [CrossRef]
- Kampmann, U.; Knorr, S.; Fuglsang, J.; Ovesen, P. Determinants of Maternal Insulin Resistance during Pregnancy: An Updated Overview. J. Diabetes Res. 2019, 2019, 5320156. [Google Scholar] [CrossRef] [Green Version]
- Ernst, S.; Demirci, C.; Valle, S.; Velazquez-Garcia, S.; Garcia-Ocana, A. Mechanisms in the adaptation of maternal beta-cells during pregnancy. Diabetes Manag. 2011, 1, 239–248. [Google Scholar] [CrossRef] [Green Version]
- Napso, T.; Yong, H.E.J.; Lopez-Tello, J.; Sferruzzi-Perri, A.N. The Role of Placental Hormones in Mediating Maternal Adaptations to Support Pregnancy and Lactation. Front. Physiol. 2018, 9, 1091. [Google Scholar] [CrossRef]
- Herrera, E.; Desoye, G. Maternal and fetal lipid metabolism under normal and gestational diabetic conditions. Horm. Mol. Biol. Clin. Investig. 2016, 26, 109–127. [Google Scholar] [CrossRef]
- Buchanan, T.A.; Xiang, A.; Kjos, S.L.; Watanabe, R. What is gestational diabetes? Diabetes Care 2007, 30 (Suppl. 2), S105–S111. [Google Scholar] [CrossRef] [Green Version]
- Howe, C.G.; Cox, B.; Fore, R.; Jungius, J.; Kvist, T.; Lent, S.; Miles, H.E.; Salas, L.A.; Rifas-Shiman, S.; Starling, A.P.; et al. Maternal Gestational Diabetes Mellitus and Newborn DNA Methylation: Findings From the Pregnancy and Childhood Epigenetics Consortium. Diabetes Care 2020, 43, 98–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Mayo, R.M.; Chatry, A.; Hu, G. Gestational Diabetes Mellitus: Its Epidemiology and Implication beyond Pregnancy. Curr. Epidemiol. Rep. 2016, 3, 1–11. [Google Scholar] [CrossRef]
- DeSisto, C.L.; Kim, S.Y.; Sharma, A.J. Prevalence estimates of gestational diabetes mellitus in the United States, Pregnancy Risk Assessment Monitoring System (PRAMS), 2007–2010. Prev. Chronic Dis. 2014, 11, E104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacks, D.A.; Hadden, D.R.; Maresh, M.; Deerochanawong, C.; Dyer, A.R.; Metzger, B.E.; Lowe, L.P.; Coustan, D.R.; Hod, M.; Oats, J.J.; et al. Frequency of gestational diabetes mellitus at collaborating centers based on IADPSG consensus panel-recommended criteria: The Hyperglycemia and Adverse Pregnancy Outcome (HAPO) Study. Diabetes Care 2012, 35, 526–528. [Google Scholar] [CrossRef] [Green Version]
- American Diabetes Association. Diagnosis and Classification of Diabetes Mellitus. Diabetes Care 2014, 37, S81–S90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravnsborg, T.; Svaneklink, S.; Andersen, L.L.T.; Larsen, M.R.; Jensen, D.M.; Overgaard, M. First-trimester proteomic profiling identifies novel predictors of gestational diabetes mellitus. PLoS ONE 2019, 14, e0214457. [Google Scholar] [CrossRef]
- Damm, P.; Houshmand-Oeregaard, A.; Kelstrup, L.; Lauenborg, J.; Mathiesen, E.R.; Clausen, T.D. Gestational diabetes mellitus and long-term consequences for mother and offspring: A view from Denmark. Diabetologia 2016, 59, 1396–1399. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Palacios, M.; Ruiz-Alcaraz, A.J.; Sanchez-Campillo, M.; Larque, E. Role of Insulin in Placental Transport of Nutrients in Gestational Diabetes Mellitus. Ann. Nutr. Metab. 2017, 70, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, J. Diabetes and pregnancy; blood sugar of newborn infants during fasting and glucose administration. Ugeskr Laeger 1952, 114, 685. [Google Scholar]
- Ornoy, A.; Reece, E.A.; Pavlinkova, G.; Kappen, C.; Miller, R.K. Effect of maternal diabetes on the embryo, fetus, and children: Congenital anomalies, genetic and epigenetic changes and developmental outcomes. Birth Defects Res. C Embryo Today 2015, 105, 53–72. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.F.; Kuo, C.F.; Chiou, M.J.; Chang, S.H. Maternal and fetal outcomes of pregnant women with type 1 diabetes, a national population study. Oncotarget 2017, 8, 80679–80687. [Google Scholar] [CrossRef] [Green Version]
- Broughton, C.; Douek, I. An overview of the management of diabetes from pre-conception, during pregnancy and in the postnatal period. Clin. Med. 2019, 19, 399–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitanchez, D.; Yzydorczyk, C.; Simeoni, U. What neonatal complications should the pediatrician be aware of in case of maternal gestational diabetes? World J. Diabetes 2015, 6, 734–743. [Google Scholar] [CrossRef]
- Kawakita, T.; Bowers, K.; Hazrati, S.; Zhang, C.; Grewal, J.; Chen, Z.; Sun, L.; Grantz, K.L. Increased Neonatal Respiratory Morbidity Associated with Gestational and Pregestational Diabetes: A Retrospective Study. Am. J. Perinatol. 2017, 34, 1160–1168. [Google Scholar] [CrossRef]
- Boghossian, N.S.; Yeung, E.; Albert, P.S.; Mendola, P.; Laughon, S.K.; Hinkle, S.N.; Zhang, C. Changes in diabetes status between pregnancies and impact on subsequent newborn outcomes. Am. J. Obstet. Gynecol. 2014, 210, 431.e1-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forsbach-Sanchez, G.; Vasquez-Lara, J.; Hernandez-Herrera, R.; Tamez-Perez, H.E. Neonatal morbidity associated to gestational diabetes. A descriptive study on 74 patients. Rev. Med. Inst. Mex. Seguro Soc. 2008, 46, 141–144. [Google Scholar]
- Karczewski, K.J.; Snyder, M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Huhn, E.A.; Rossi, S.W.; Hoesli, I.; Gobl, C.S. Controversies in Screening and Diagnostic Criteria for Gestational Diabetes in Early and Late Pregnancy. Front. Endocrinol. 2018, 9, 696. [Google Scholar] [CrossRef] [Green Version]
- Albl, B.; Haesner, S.; Braun-Reichhart, C.; Streckel, E.; Renner, S.; Seeliger, F.; Wolf, E.; Wanke, R.; Blutke, A. Tissue Sampling Guides for Porcine Biomedical Models. Toxicol. Pathol. 2016, 44, 414–420. [Google Scholar] [CrossRef]
- Pasek, R.C.; Gannon, M. Advancements and challenges in generating accurate animal models of gestational diabetes mellitus. Am. J. Physiol. Endocrinol. Metab. 2013, 305, E1327–E1338. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J.E. Developmental Programming of Obesity and Diabetes in Mouse, Monkey, and Man in 2018: Where are We Headed? Diabetes 2018, 67, 2137–2151. [Google Scholar] [CrossRef] [Green Version]
- Zettler, S.; Renner, S.; Kemter, E.; Hinrichs, A.; Klymiuk, N.; Backman, M.; Riedel, E.O.; Mueller, C.; Streckel, E.; Braun-Reichhart, C.; et al. A decade of experience with genetically tailored pig models for diabetes and metabolic research. Anim. Reprod. 2020, 17, e20200064. [Google Scholar] [CrossRef]
- Renner, S.; Blutke, A.; Clauss, S.; Deeg, C.A.; Kemter, E.; Merkus, D.; Wanke, R.; Wolf, E. Porcine models for studying complications and organ crosstalk in diabetes mellitus. Cell Tissue Res. 2020, 380, 341–378. [Google Scholar] [CrossRef] [PubMed]
- Renner, S.; Dobenecker, B.; Blutke, A.; Zöls, S.; Wanke, R.; Ritzmann, M.; Wolf, E. Comparative aspects of rodent and nonrodent animal models for mechanistic and translational diabetes research. Theriogenology 2016, 86, 406–421. [Google Scholar] [CrossRef]
- Litten-Brown, J.C.; Corson, A.M.; Clarke, L. Porcine models for the metabolic syndrome, digestive and bone disorders: A general overview. Animal 2010, 4, 899–920. [Google Scholar] [CrossRef] [Green Version]
- Souza, R.T.; Mayrink, J.; Leite, D.F.; Costa, M.L.; Calderon, I.M.; Rocha Filho, E.A.; Vettorazzi, J.; Feitosa, F.E.; Cecatti, J.G.; Preterm, S.S.G. Metabolomics applied to maternal and perinatal health: A review of new frontiers with a translation potential. Clinics 2019, 74, e894. [Google Scholar] [CrossRef]
- Chu, A.H.Y.; Tint, M.T.; Chang, H.F.; Wong, G.; Yuan, W.L.; Tull, D.; Nijagal, B.; Narayana, V.K.; Meikle, P.J.; Chang, K.T.E.; et al. High placental inositol content associated with suppressed pro-adipogenic effects of maternal glycaemia in offspring: The GUSTO cohort. Int. J. Obes. 2021, 45, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Lizárraga, D.; García-Gasca, A. The Placenta as a Target of Epigenetic Alterations in Women with Gestational Diabetes Mellitus and Potential Implications for the Offspring. Epigenomes 2021, 5, 13. [Google Scholar] [CrossRef]
- Valencia-Ortega, J.; Saucedo, R.; Sánchez-Rodríguez, M.A.; Cruz-Durán, J.G.; Martínez, E.G.R. Epigenetic Alterations Related to Gestational Diabetes Mellitus. Int. J. Mol. Sci. 2021, 22, 9462. [Google Scholar] [CrossRef]
- Collins, F.S.; Green, E.D.; Guttmacher, A.E.; Guyer, M.S. A vision for the future of genomics research. Nature 2003, 422, 835–847. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Derraik, J.G.B.; Maessen, S.E.; Gibbins, J.D.; Cutfield, W.S.; Lundgren, M.; Ahlsson, F. Large-for-gestational-age phenotypes and obesity risk in adulthood: A study of 195,936 women. Sci. Rep. 2020, 10, 2157. [Google Scholar] [CrossRef] [Green Version]
- Boney, C.M.; Verma, A.; Tucker, R.; Vohr, B.R. Metabolic syndrome in childhood: Association with birth weight, maternal obesity, and gestational diabetes mellitus. Pediatrics 2005, 115, e290–e296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, L.; Nilsson, I.A.K.; Gissler, M.; Lavebratt, C. Associations of Maternal Diabetes and Body Mass Index With Offspring Birth Weight and Prematurity. JAMA Pediatr. 2019, 173, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Knight, B.; Shields, B.M.; Hattersley, A.T. The Exeter Family Study of Childhood Health (EFSOCH): Study protocol and methodology. Paediatr. Perinat. Epidemiol. 2006, 20, 172–179. [Google Scholar] [CrossRef]
- Coustan, D.R.; Lowe, L.P.; Metzger, B.E. The hyperglycemia and adverse pregnancy outcome (HAPO) study: Can we use the results as a basis for change? J. Matern. Fetal Neonatal Med. 2010, 23, 204–209. [Google Scholar] [CrossRef]
- Hughes, A.E.; Nodzenski, M.; Beaumont, R.N.; Talbot, O.; Shields, B.M.; Scholtens, D.M.; Knight, B.A.; Lowe, W.L., Jr.; Hattersley, A.T.; Freathy, R.M. Fetal Genotype and Maternal Glucose Have Independent and Additive Effects on Birth Weight. Diabetes 2018, 67, 1024–1029. [Google Scholar] [CrossRef] [Green Version]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef]
- Feil, R.; Fraga, M.F. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef] [Green Version]
- Tirado-Magallanes, R.; Rebbani, K.; Lim, R.; Pradhan, S.; Benoukraf, T. Whole genome DNA methylation: Beyond genes silencing. Oncotarget 2017, 8, 5629–5637. [Google Scholar] [CrossRef] [Green Version]
- Finer, S.; Mathews, C.; Lowe, R.; Smart, M.; Hillman, S.; Foo, L.; Sinha, A.; Williams, D.; Rakyan, V.K.; Hitman, G.A. Maternal gestational diabetes is associated with genome-wide DNA methylation variation in placenta and cord blood of exposed offspring. Hum. Mol. Genet. 2015, 24, 3021–3029. [Google Scholar] [CrossRef] [PubMed]
- Moen, G.H.; Sommer, C.; Prasad, R.B.; Sletner, L.; Groop, L.; Qvigstad, E.; Birkeland, K.I. MECHANISMS IN ENDOCRINOLOGY: Epigenetic modifications and gestational diabetes: A systematic review of published literature. Eur. J. Endocrinol. 2017, 176, R247–R267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franzago, M.; Fraticelli, F.; Stuppia, L.; Vitacolonna, E. Nutrigenetics, epigenetics and gestational diabetes: Consequences in mother and child. Epigenetics 2019, 14, 215–235. [Google Scholar] [CrossRef] [Green Version]
- Słupecka-Ziemilska, M.; Wychowański, P.; Puzianowska-Kuznicka, M. Gestational Diabetes Mellitus Affects Offspring's Epigenome. Is There a Way to Reduce the Negative Consequences? Nutrients 2020, 12, 2792. [Google Scholar] [CrossRef] [PubMed]
- Chu, A.H.Y.; Godfrey, K.M. Gestational Diabetes Mellitus and Developmental Programming. Ann. Nutr. Metab. 2020, 76 (Suppl. 3), 4–15. [Google Scholar] [CrossRef]
- Dluski, D.F.; Wolinska, E.; Skrzypczak, M. Epigenetic Changes in Gestational Diabetes Mellitus. Int. J. Mol. Sci. 2021, 22, 7649. [Google Scholar] [CrossRef] [PubMed]
- Lesseur, C.; Armstrong, D.A.; Paquette, A.G.; Li, Z.; Padbury, J.F.; Marsit, C.J. Maternal obesity and gestational diabetes are associated with placental leptin DNA methylation. Am. J. Obstet. Gynecol. 2014, 211, 654.e1–654.e9. [Google Scholar] [CrossRef] [Green Version]
- Gagné-Ouellet, V.; Breton, E.; Thibeault, K.; Fortin, C.-A.; Cardenas, A.; Guérin, R.; Perron, P.; Hivert, M.-F.; Bouchard, L. Mediation Analysis Supports a Causal Relationship between Maternal Hyperglycemia and Placental DNA Methylation Variations at the Leptin Gene Locus and Cord Blood Leptin Levels. Int. J. Mol. Sci. 2020, 21, 329. [Google Scholar] [CrossRef] [Green Version]
- Allard, C.; Desgagné, V.; Patenaude, J.; Lacroix, M.; Guillemette, L.; Battista, M.C.; Doyon, M.; Ménard, J.; Ardilouze, J.L.; Perron, P.; et al. Mendelian randomization supports causality between maternal hyperglycemia and epigenetic regulation of leptin gene in newborns. Epigenetics 2015, 10, 342–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Côté, S.; Gagné-Ouellet, V.; Guay, S.-P.; Allard, C.; Houde, A.-A.; Perron, P.; Baillargeon, J.-P.; Gaudet, D.; Guérin, R.; Brisson, D.; et al. PPARGC1α gene DNA methylation variations in human placenta mediate the link between maternal hyperglycemia and leptin levels in newborns. Clin. Epigenetics 2016, 8, 72. [Google Scholar] [CrossRef] [Green Version]
- Friedman, J. Leptin at 20: An overview. J. Endocrinol. 2014, 223. [Google Scholar] [CrossRef] [Green Version]
- Flenkenthaler, F.; Ländström, E.; Shashikadze, B.; Backman, M.; Blutke, A.; Philippou-Massier, J.; Renner, S.; Hrabe de Angelis, M.; Wanke, R.; Blum, H.; et al. Differential Effects of Insulin-Deficient Diabetes Mellitus on Visceral vs. Subcutaneous Adipose Tissue—Multi-omics Insights From the Munich MIDY Pig Model. Front. Med. 2021, 8. [Google Scholar] [CrossRef]
- Mantzoros, C.S.; Rifas-Shiman, S.L.; Williams, C.J.; Fargnoli, J.L.; Kelesidis, T.; Gillman, M.W. Cord blood leptin and adiponectin as predictors of adiposity in children at 3 years of age: A prospective cohort study. Pediatrics 2009, 123, 682–689. [Google Scholar] [CrossRef] [Green Version]
- Bouchard, L.; Hivert, M.-F.; Guay, S.-P.; St-Pierre, J.; Perron, P.; Brisson, D. Placental adiponectin gene DNA methylation levels are associated with mothers' blood glucose concentration. Diabetes 2012, 61, 1272–1280. [Google Scholar] [CrossRef] [Green Version]
- Houde, A.-A.; Hivert, M.-F.; Bouchard, L. Fetal epigenetic programming of adipokines. Adipocyte 2013, 2, 41–46. [Google Scholar] [CrossRef] [Green Version]
- Houde, A.A.; St-Pierre, J.; Hivert, M.F.; Baillargeon, J.P.; Perron, P.; Gaudet, D.; Brisson, D.; Bouchard, L. Placental lipoprotein lipase DNA methylation levels are associated with gestational diabetes mellitus and maternal and cord blood lipid profiles. J. Dev. Orig. Health Dis. 2014, 5, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Gagné-Ouellet, V.; Houde, A.-A.; Guay, S.-P.; Perron, P.; Gaudet, D.; Guérin, R.; Jean-Patrice, B.; Hivert, M.-F.; Brisson, D.; Bouchard, L. Placental lipoprotein lipase DNA methylation alterations are associated with gestational diabetes and body composition at 5 years of age. Epigenetics 2017, 12, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Blazevic, S.; Horvaticek, M.; Kesic, M.; Zill, P.; Hranilovic, D.; Ivanisevic, M.; Desoye, G.; Stefulj, J. Epigenetic adaptation of the placental serotonin transporter gene (SLC6A4) to gestational diabetes mellitus. PLoS ONE 2017, 12, e0179934. [Google Scholar] [CrossRef] [Green Version]
- Muller, C.L.; Anacker, A.M.J.; Veenstra-VanderWeele, J. The serotonin system in autism spectrum disorder: From biomarker to animal models. Neuroscience 2016, 321, 24–41. [Google Scholar] [CrossRef] [Green Version]
- Rowland, J.; Wilson, C.A. The association between gestational diabetes and ASD and ADHD: A systematic review and meta-analysis. Sci. Rep. 2021, 11, 5136. [Google Scholar] [CrossRef]
- Felix, J.F.; Joubert, B.R.; Baccarelli, A.A.; Sharp, G.C.; Almqvist, C.; Annesi-Maesano, I.; Arshad, H.; Baïz, N.; Bakermans-Kranenburg, M.J.; Bakulski, K.M.; et al. Cohort Profile: Pregnancy And Childhood Epigenetics (PACE) Consortium. Int. J. Epidemiol. 2018, 47, 22–23. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Lu, J.; Xie, W.; Lu, X.; Liang, Y.; Li, M.; Wang, Z.; Huang, X.; Tang, M.; Pfaff, D.W.; et al. Maternal diabetes induces autism-like behavior by hyperglycemia-mediated persistent oxidative stress and suppression of superoxide dismutase 2. Proc. Natl. Acad. Sci. USA 2019, 116, 23743–23752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, X.; Liu, F.; Zhang, H.; Kan, M.; Wang, T.; Dong, M.; Liu, Y. Genome-wide DNA methylation profiling in infants born to gestational diabetes mellitus. Diabetes Res. Clin. Pract. 2018, 142, 10–18. [Google Scholar] [CrossRef] [PubMed]
- El Hajj, N.; Pliushch, G.; Schneider, E.; Dittrich, M.; Muller, T.; Korenkov, M.; Aretz, M.; Zechner, U.; Lehnen, H.; Haaf, T. Metabolic programming of MEST DNA methylation by intrauterine exposure to gestational diabetes mellitus. Diabetes 2013, 62, 1320–1328. [Google Scholar] [CrossRef] [Green Version]
- Shiau, S.; Wang, L.; Liu, H.; Zheng, Y.; Drong, A.; Joyce, B.T.; Wang, J.; Li, W.; Leng, J.; Shen, Y.; et al. Prenatal gestational diabetes mellitus exposure and accelerated offspring DNA methylation age in early childhood. Epigenetics 2021, 16, 186–195. [Google Scholar] [CrossRef] [PubMed]
- West, N.A.; Kechris, K.; Dabelea, D. Exposure to Maternal Diabetes in Utero and DNA Methylation Patterns in the Offspring. Immunometabolism 2013, 1, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Yang, I.V.; Zhang, W.; Davidson, E.J.; Fingerlin, T.E.; Kechris, K.; Dabelea, D. Epigenetic marks of in utero exposure to gestational diabetes and childhood adiposity outcomes: The EPOCH study. Diabet. Med. 2018, 35, 612–620. [Google Scholar] [CrossRef]
- Crume, T.L.; Ogden, L.; West, N.A.; Vehik, K.S.; Scherzinger, A.; Daniels, S.; McDuffie, R.; Bischoff, K.; Hamman, R.F.; Norris, J.M.; et al. Association of exposure to diabetes in utero with adiposity and fat distribution in a multiethnic population of youth: The Exploring Perinatal Outcomes among Children (EPOCH) Study. Diabetologia 2011, 54, 87–92. [Google Scholar] [CrossRef] [Green Version]
- Hjort, L.; Martino, D.; Grunnet, L.G.; Naeem, H.; Maksimovic, J.; Olsson, A.H.; Zhang, C.; Ling, C.; Olsen, S.F.; Saffery, R.; et al. Gestational diabetes and maternal obesity are associated with epigenome-wide methylation changes in children. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Goldman, D. Role of RNA modifications in brain and behavior. Genes Brain Behav. 2018, 17, e12444. [Google Scholar] [CrossRef] [Green Version]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marguerat, S.; Bahler, J. RNA-seq: From technology to biology. Cell Mol. Life Sci. 2010, 67, 569–579. [Google Scholar] [CrossRef] [Green Version]
- Sozoniuk, M.; Chrobak, L.; Kowalczyk, K.; Kankofer, M. Is it useful to use several “omics” for obtaining valuable results? Mol. Biol. Rep. 2019, 46. [Google Scholar] [CrossRef] [Green Version]
- Ambra, R.; Manca, S.; Palumbo, M.C.; Leoni, G.; Natarelli, L.; De Marco, A.; Consoli, A.; Pandolfi, A.; Virgili, F. Transcriptome analysis of human primary endothelial cells (HUVEC) from umbilical cords of gestational diabetic mothers reveals candidate sites for an epigenetic modulation of specific gene expression. Genomics 2014, 103, 337–348. [Google Scholar] [CrossRef] [Green Version]
- Koskinen, A.; Lehtoranta, L.; Laiho, A.; Laine, J.; Kaapa, P.; Soukka, H. Maternal diabetes induces changes in the umbilical cord gene expression. Placenta 2015, 36, 767–774. [Google Scholar] [CrossRef]
- Casasnovas, J.; Jo, Y.; Rao, X.; Xuei, X.; Brown, M.E.; Kua, K.L. High glucose alters fetal rat islet transcriptome and induces progeny islet dysfunction. J. Endocrinol. 2019, 240, 309–323. [Google Scholar] [CrossRef] [Green Version]
- Inoguchi, Y.; Ichiyanagi, K.; Ohishi, H.; Maeda, Y.; Sonoda, N.; Ogawa, Y.; Inoguchi, T.; Sasaki, H. Poorly controlled diabetes during pregnancy and lactation activates the Foxo1 pathway and causes glucose intolerance in adult offspring. Sci. Rep. 2019, 9, 10181. [Google Scholar] [CrossRef] [Green Version]
- Backman, M.; Flenkenthaler, F.; Blutke, A.; Dahlhoff, M.; Landstrom, E.; Renner, S.; Philippou-Massier, J.; Krebs, S.; Rathkolb, B.; Prehn, C.; et al. Multi-omics insights into functional alterations of the liver in insulin-deficient diabetes mellitus. Mol. Metab. 2019, 26, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Page, K.A.; Luo, S.; Wang, X.; Chow, T.; Alves, J.; Buchanan, T.A.; Xiang, A.H. Children Exposed to Maternal Obesity or Gestational Diabetes Mellitus During Early Fetal Development Have Hypothalamic Alterations that Predict Future Weight Gain. Diabetes Care 2019, 42, 1473–1480. [Google Scholar] [CrossRef] [PubMed]
- Camprubi Robles, M.; Campoy, C.; Garcia Fernandez, L.; Lopez-Pedrosa, J.M.; Rueda, R.; Martin, M.J. Maternal Diabetes and Cognitive Performance in the Offspring: A Systematic Review and Meta-Analysis. PLoS ONE 2015, 10, e0142583. [Google Scholar] [CrossRef]
- Aviel-Shekler, K.; Hamshawi, Y.; Sirhan, W.; Getselter, D.; Srikanth, K.D.; Malka, A.; Piran, R.; Elliott, E. Gestational diabetes induces behavioral and brain gene transcription dysregulation in adult offspring. Transl. Psychiatry 2020, 10, 412. [Google Scholar] [CrossRef] [PubMed]
- Money, K.M.; Barke, T.L.; Serezani, A.; Gannon, M.; Garbett, K.A.; Aronoff, D.M.; Mirnics, K. Gestational diabetes exacerbates maternal immune activation effects in the developing brain. Mol. Psychiatry 2018, 23, 1920–1928. [Google Scholar] [CrossRef]
- Ke, Q.; Costa, M. Hypoxia-inducible factor-1 (HIF-1). Mol. Pharmacol. 2006, 70, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Cerychova, R.; Bohuslavova, R.; Papousek, F.; Sedmera, D.; Abaffy, P.; Benes, V.; Kolar, F.; Pavlinkova, G. Adverse effects of Hif1a mutation and maternal diabetes on the offspring heart. Cardiovasc. Diabetol. 2018, 17, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Arah, O.A.; Liew, Z.; Cnattingius, S.; Olsen, J.; Sorensen, H.T.; Qin, G.; Li, J. Maternal diabetes during pregnancy and early onset of cardiovascular disease in offspring: Population based cohort study with 40 years of follow-up. BMJ 2019, 367, l6398. [Google Scholar] [CrossRef] [Green Version]
- Preston, C.C.; Larsen, T.D.; Eclov, J.A.; Louwagie, E.J.; Gandy, T.C.T.; Faustino, R.S.; Baack, M.L. Maternal High Fat Diet and Diabetes Disrupts Transcriptomic Pathways That Regulate Cardiac Metabolism and Cell Fate in Newborn Rat Hearts. Front. Endocrinol. 2020, 11, 570846. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopylov, A.; Papysheva, O.; Gribova, I.; Kotaysch, G.; Kharitonova, L.; Mayatskaya, T.; Sokerina, E.; Kaysheva, A.; Morozov, S. Molecular pathophysiology of diabetes mellitus during pregnancy with antenatal complications. Sci. Rep. 2020, 10, 19641. [Google Scholar] [CrossRef] [PubMed]
- Miao, Z.; Wang, J.; Wang, F.; Liu, L.; Ding, H.; Shi, Z. Comparative proteomics of umbilical vein blood plasma from normal and gestational diabetes mellitus patients reveals differentially expressed proteins associated with childhood obesity. Proteom. Clin. Appl. 2016, 10, 1122–1131. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Xu, G.F.; Jiang, Y.; Zhu, H.; Sun, L.J.; Peng, R.; Luo, Q. Comparative proteomic analysis of maternal peripheral plasma and umbilical venous plasma from normal and gestational diabetes mellitus pregnancies. Medicine 2018, 97, e12232. [Google Scholar] [CrossRef]
- Kumar, T.R.; Wang, Y.; Lu, N.; Matzuk, M.M. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nat. Genet. 1997, 15, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.L.; Talton, O.O.; Ganesan, S.; Schulz, L.C.; Keating, A.F. Developmental origins of ovarian disorder: Impact of maternal lean gestational diabetes on the offspring ovarian proteome in micedagger. Biol. Reprod. 2019, 101, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Bouatra, S.; Aziat, F.; Mandal, R.; Guo, A.C.; Wilson, M.R.; Knox, C.; Bjorndahl, T.C.; Krishnamurthy, R.; Saleem, F.; Liu, P.; et al. The human urine metabolome. PLoS ONE 2013, 8, e73076. [Google Scholar] [CrossRef] [Green Version]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted metabolomics. Curr. Protoc. Mol. Biol. 2012, 98, 30–32. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Francis, E.; Hu, G.; Chen, L. Metabolomic profiling of women with gestational diabetes mellitus and their offspring: Review of metabolomics studies. J. Diabetes Complicat. 2018, 32, 512–523. [Google Scholar] [CrossRef] [Green Version]
- Berglund, S.K.; Garcia-Valdes, L.; Torres-Espinola, F.J.; Segura, M.T.; Martinez-Zaldivar, C.; Aguilar, M.J.; Agil, A.; Lorente, J.A.; Florido, J.; Padilla, C.; et al. Maternal, fetal and perinatal alterations associated with obesity, overweight and gestational diabetes: An observational cohort study (PREOBE). BMC Public Health 2016, 16, 207. [Google Scholar] [CrossRef] [Green Version]
- Shokry, E.; Marchioro, L.; Uhl, O.; Bermudez, M.G.; Garcia-Santos, J.A.; Segura, M.T.; Campoy, C.; Koletzko, B. Impact of maternal BMI and gestational diabetes mellitus on maternal and cord blood metabolome: Results from the PREOBE cohort study. Acta Diabetol. 2019, 56, 421–430. [Google Scholar] [CrossRef]
- Dube, M.C.; Morisset, A.S.; Tchernof, A.; Weisnagel, S.J. Cord blood C-peptide levels relate to the metabolic profile of women with and without gestational diabetes. Acta Obstet. Gynecol. Scand. 2012, 91, 1469–1473. [Google Scholar] [CrossRef]
- Perng, W.; Ringham, B.M.; Smith, H.A.; Michelotti, G.; Kechris, K.M.; Dabelea, D. A prospective study of associations between in utero exposure to gestational diabetes mellitus and metabolomic profiles during late childhood and adolescence. Diabetologia 2020, 63, 296–312. [Google Scholar] [CrossRef]
- Ott, R.; Pawlow, X.; Weiss, A.; Hofelich, A.; Herbst, M.; Hummel, N.; Prehn, C.; Adamski, J.; Romisch-Margl, W.; Kastenmuller, G.; et al. Intergenerational Metabolomic Analysis of Mothers with a History of Gestational Diabetes Mellitus and Their Offspring. Int. J. Mol. Sci. 2020, 21, 9647. [Google Scholar] [CrossRef]
- Shokry, E.; Marchioro, L.; Uhl, O.; Bermudez, M.G.; Garcia-Santos, J.A.; Segura, M.T.; Campoy, C.; Koletzko, B. Transgenerational cycle of obesity and diabetes: Investigating possible metabolic precursors in cord blood from the PREOBE study. Acta Diabetol. 2019, 56, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.P.; Reichetzeder, C.; Prehn, C.; von Websky, K.; Slowinski, T.; Chen, Y.P.; Yin, L.H.; Kleuser, B.; Yang, X.S.; Adamski, J.; et al. Fetal Serum Metabolites Are Independently Associated with Gestational Diabetes Mellitus. Cell Physiol. Biochem. 2018, 45, 625–638. [Google Scholar] [CrossRef] [PubMed]
- Cetin, I.; de Santis, M.S.N.; Taricco, E.; Radaelli, T.; Teng, C.; Ronzoni, S.; Spada, E.; Milani, S.; Pardi, G. Maternal and fetal amino acid concentrations in normal pregnancies and in pregnancies with gestational diabetes mellitus. Am. J. Obstet. Gynecol. 2005, 192, 610–617. [Google Scholar] [CrossRef]
- Ziegler, A.G.; Meier-Stiegen, F.; Winkler, C.; Bonifacio, E. Prospective evaluation of risk factors for the development of islet autoimmunity and type 1 diabetes during puberty—TEENDIAB: Study design. Pediatr. Diabetes 2012, 13, 419–424. [Google Scholar] [CrossRef]
- Ziegler, A.G.; Hummel, M.; Schenker, M.; Bonifacio, E. Autoantibody appearance and risk for development of childhood diabetes in offspring of parents with type 1 diabetes: The 2-year analysis of the German BABYDIAB Study. Diabetes 1999, 48, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Pitchika, A.; Jolink, M.; Winkler, C.; Hummel, S.; Hummel, N.; Krumsiek, J.; Kastenmuller, G.; Raab, J.; Kordonouri, O.; Ziegler, A.G.; et al. Associations of maternal type 1 diabetes with childhood adiposity and metabolic health in the offspring: A prospective cohort study. Diabetologia 2018, 61, 2319–2332. [Google Scholar] [CrossRef] [Green Version]
- Lowe, W.L., Jr.; Bain, J.R.; Nodzenski, M.; Reisetter, A.C.; Muehlbauer, M.J.; Stevens, R.D.; Ilkayeva, O.R.; Lowe, L.P.; Metzger, B.E.; Newgard, C.B.; et al. Maternal BMI and Glycemia Impact the Fetal Metabolome. Diabetes Care 2017, 40, 902–910. [Google Scholar] [CrossRef] [Green Version]
- Walejko, J.M.; Chelliah, A.; Keller-Wood, M.; Wasserfall, C.; Atkinson, M.; Gregg, A.; Edison, A.S. Diabetes Leads to Alterations in Normal Metabolic Transitions of Pregnancy as Revealed by Time-Course Metabolomics. Metabolites 2020, 10, 350. [Google Scholar] [CrossRef]
- Peng, S.; Zhang, J.; Liu, L.; Zhang, X.; Huang, Q.; Alamdar, A.; Tian, M.; Shen, H. Newborn meconium and urinary metabolome response to maternal gestational diabetes mellitus: A preliminary case-control study. J. Proteome Res. 2015, 14, 1799–1809. [Google Scholar] [CrossRef]
- Graca, G.; Goodfellow, B.J.; Barros, A.S.; Diaz, S.; Duarte, I.F.; Spagou, K.; Veselkov, K.; Want, E.J.; Lindon, J.C.; Carreira, I.M.; et al. UPLC-MS metabolic profiling of second trimester amniotic fluid and maternal urine and comparison with NMR spectral profiling for the identification of pregnancy disorder biomarkers. Mol. Biosyst. 2012, 8, 1243–1254. [Google Scholar] [CrossRef]
- Underwood, M.A.; Gilbert, W.M.; Sherman, M.P. Amniotic Fluid: Not Just Fetal Urine Anymore. J. Perinatol. 2005, 25, 341–348. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Ge, J.; Li, X.; Jiao, R.; Li, Y.; Quan, H.; Li, J.; Guo, Q.; Wang, W. Integrated metabolome analysis reveals novel connections between maternal fecal metabolome and the neonatal blood metabolome in women with gestational diabetes mellitus. Sci. Rep. 2020, 10, 3660. [Google Scholar] [CrossRef] [PubMed]
- Isganaitis, E.; Woo, M.; Ma, H.; Chen, M.; Kong, W.; Lytras, A.; Sales, V.; Decoste-Lopez, J.; Lee, K.J.; Leatherwood, C.; et al. Developmental programming by maternal insulin resistance: Hyperinsulinemia, glucose intolerance, and dysregulated lipid metabolism in male offspring of insulin-resistant mice. Diabetes 2014, 63, 688–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, T.J.; Fonseca, M.A.; Campbell, K.E.; Moyce, B.L.; Cole, L.K.; Hatch, G.M.; Doucette, C.A.; Klein, J.; Aliani, M.; Dolinsky, V.W. Maternal obesity characterized by gestational diabetes increases the susceptibility of rat offspring to hepatic steatosis via a disrupted liver metabolome. J. Physiol. 2015, 593, 3181–3197. [Google Scholar] [CrossRef] [Green Version]
- Renner, S.; Martins, A.S.; Streckel, E.; Braun-Reichhart, C.; Backman, M.; Prehn, C.; Klymiuk, N.; Bahr, A.; Blutke, A.; Landbrecht-Schessl, C.; et al. Mild maternal hyperglycemia in INS (C93S) transgenic pigs causes impaired glucose tolerance and metabolic alterations in neonatal offspring. Dis. Model. Mech. 2019, 12. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.S. Microbiomics: Were we all wrong before? Periodontology 2000 2021, 85, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Walker, R.W.; Clemente, J.C.; Peter, I.; Loos, R.J.F. The prenatal gut microbiome: Are we colonized with bacteria in utero? Pediatr. Obes. 2017, 12 (Suppl. 1), 3–17. [Google Scholar] [CrossRef] [Green Version]
- Han, S.; Ellberg, C.C.; Olomu, I.N.; Vyas, A.K. Gestational microbiome: Metabolic perturbations and developmental programming. Reproduction 2021, 162, R85–R98. [Google Scholar] [CrossRef]
- Ponzo, V.; Ferrocino, I.; Zarovska, A.; Amenta, M.B.; Leone, F.; Monzeglio, C.; Rosato, R.; Pellegrini, M.; Gambino, R.; Cassader, M.; et al. The microbiota composition of the offspring of patients with gestational diabetes mellitus (GDM). PLoS ONE 2019, 14, e0226545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soderborg, T.K.; Carpenter, C.M.; Janssen, R.C.; Weir, T.L.; Robertson, C.E.; Ir, D.; Young, B.E.; Krebs, N.F.; Hernandez, T.L.; Barbour, L.A.; et al. Gestational Diabetes Is Uniquely Associated With Altered Early Seeding of the Infant Gut Microbiota. Front. Endocrinol. 2020, 11. [Google Scholar] [CrossRef]
- Hu, J.; Nomura, Y.; Bashir, A.; Fernandez-Hernandez, H.; Itzkowitz, S.; Pei, Z.; Stone, J.; Loudon, H.; Peter, I. Diversified microbiota of meconium is affected by maternal diabetes status. PLoS ONE 2013, 8, e78257. [Google Scholar] [CrossRef] [Green Version]
- Su, M.; Nie, Y.; Shao, R.; Duan, S.; Jiang, Y.; Wang, M.; Xing, Z.; Sun, Q.; Liu, X.; Xu, W. Diversified gut microbiota in newborns of mothers with gestational diabetes mellitus. PLoS ONE 2018, 13, e0205695. [Google Scholar] [CrossRef]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Crusell, M.K.W.; Hansen, T.H.; Nielsen, T.; Allin, K.H.; Rühlemann, M.C.; Damm, P.; Vestergaard, H.; Rørbye, C.; Jørgensen, N.R.; Christiansen, O.B.; et al. Comparative Studies of the Gut Microbiota in the Offspring of Mothers With and Without Gestational Diabetes. Front. Cell. Infect. Microbiol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Hasan, S.; Aho, V.; Pereira, P.; Paulin, L.; Koivusalo, S.B.; Auvinen, P.; Eriksson, J.G. Gut microbiome in gestational diabetes: A cross-sectional study of mothers and offspring 5 years postpartum. Acta Obstet. Gynecol. Scand. 2018, 97, 38–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crusell, M.K.W.; Hansen, T.H.; Nielsen, T.; Allin, K.H.; Rühlemann, M.C.; Damm, P.; Vestergaard, H.; Rørbye, C.; Jørgensen, N.R.; Christiansen, O.B.; et al. Gestational diabetes is associated with change in the gut microbiota composition in third trimester of pregnancy and postpartum. Microbiome 2018, 6, 89. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Takahashi, S.; Saito, K. Omics and integrated omics for the promotion of food and nutrition science. J. Tradit. Complement. Med. 2011, 1, 25–30. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.; Li, X. Advances in Research on Diabetes by Human Nutriomics. Int. J. Mol. Sci. 2019, 20, 5375. [Google Scholar] [CrossRef] [Green Version]
- Soh, S.E.; Tint, M.T.; Gluckman, P.D.; Godfrey, K.M.; Rifkin-Graboi, A.; Chan, Y.H.; Stünkel, W.; Holbrook, J.D.; Kwek, K.; Chong, Y.S.; et al. Cohort profile: Growing Up in Singapore Towards healthy Outcomes (GUSTO) birth cohort study. Int. J. Epidemiol. 2014, 43, 1401–1409. [Google Scholar] [CrossRef]

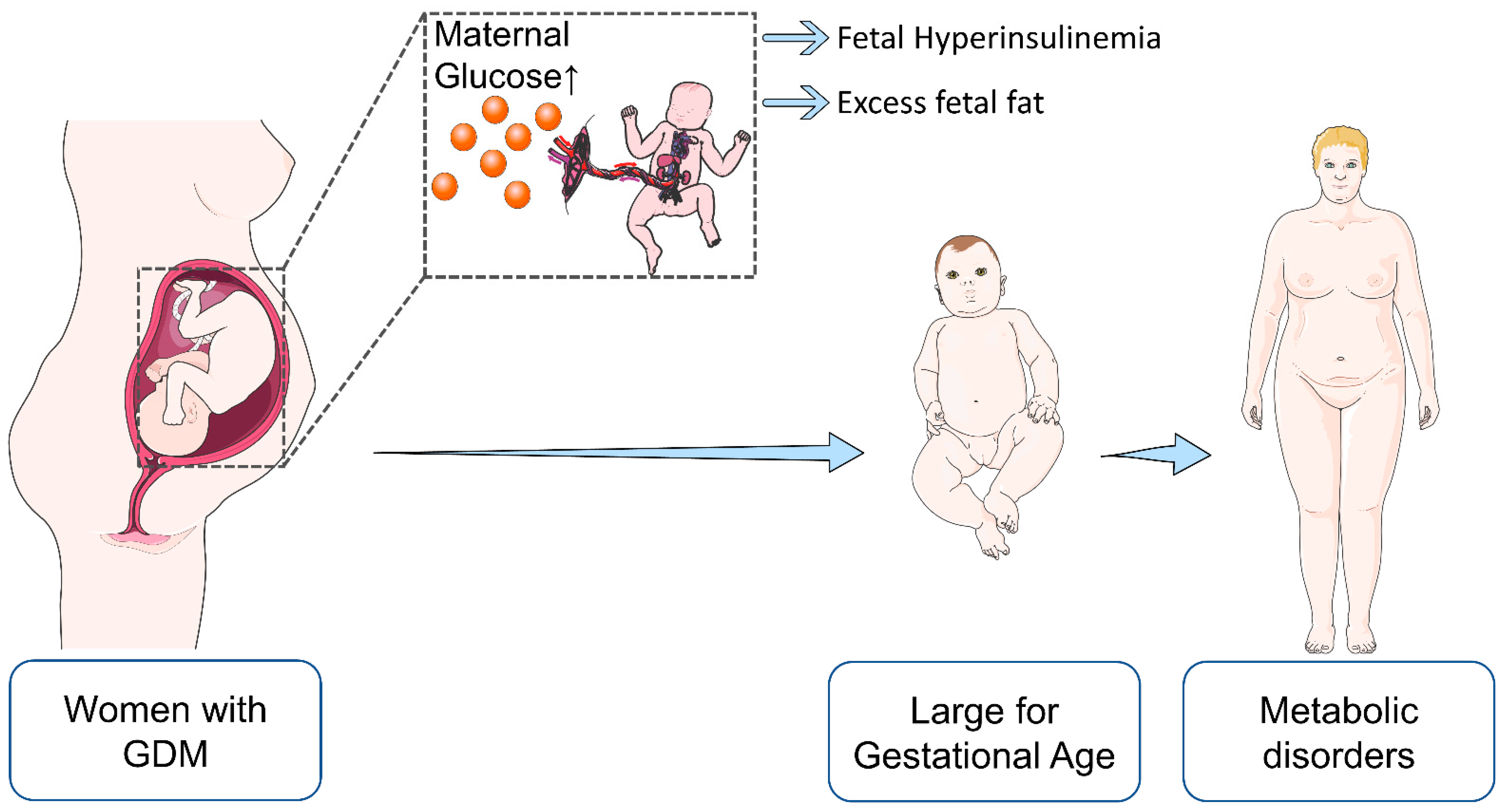
| Maternal | Fetal/Offspring |
|---|---|
| Pre-eclampsia | Intrauterine death |
| Cesarean section | Congenital malformations |
| Labor complications | Macrosomia |
| Pre-term delivery | Polycythemia and hyperbilirubinemia |
| Postpartum hemorrhage | Respiratory distress syndrome |
| Recurrent GDM | Insulin resistance Metabolic syndrome |
| Type 2 diabetes | Type 2 diabetes |
| Complications of type 2 diabetes (cardiovascular disease, nephorpathy, neuropathy, retinopathy) | Complications of type 2 diabetes (cardiovascular disease, nephropathy, neuropathy, retinopathy) |
| weight gain/obesity | Weight gain/obesity |
| Species | Maternal Characteristics | Bio-Specimen | Major Findings in Offspring | Reference |
|---|---|---|---|---|
| Human | GDM, type 1 diabetes, type 2 diabetes | Cord blood | Altered abundance of APOM, CP, PLG, AGT, KNG1, APOA1, ORM2, TF, HRG, APOD, LUM; processes such as inflammation, extracellular matrix remodeling, lipid metabolism, etc. mainly affected | [102] |
| Human | GDM | Umbilical venous plasma | Altered abundance of CEPT and APOM; FSH as upstream regulator of the differentially abundant proteins | [104] |
| Human | GDM | Umbilical venous plasma | Altered abundance of PLTP and LCAT (related to abnormal glucose and lipid metabolism) and ARHGEF11 (known to influence embryo development) | [103] |
| Mouse | Diet-induced diabetes | Ovaries | Altered abundance of CNPY2, DAZAP1, SEPT7, and SRSF2; potential impact on fertility and oocyte quality of offspring in later life | [106] |
| Maternal Characteristics | Bio-Specimen | Major Findings in Offspring | Reference |
|---|---|---|---|
| GDM | Feces | GDM alone or together with maternal overweight/obesity influences infant microbiota in a way that set the stage for future risks of inflammatory and metabolic disease | [136] |
| GDM | Feces | Glycemic regulation in late pregnancy is linked with relatively modest variation in the gut microbiota composition of the offspring at age 1 week and 9 months; lower richness of the gut microbiota in GDM neonates compared with neonates born to mothers without GDM | [140] |
| GDM | Feces | Increased relative abundance of pro-inflammatory taxa, in particular Escherichia and Parabacteroides | [135] |
| GDM | Feces | Increased abundance of Anaerotruncus genus | [141] |
| Type 2 PGDM GDM | Meconium | Enrichment of the meconium microbiome for the same bacterial taxa as reported in the fecal microbiome of adult diabetic patients | [137] |
| Species | Maternal Characteristics | Major Findings in Offspring | Reference |
|---|---|---|---|
| Human | Hyperglycemia | Positive correlation of maternal glycemia and fetal birth weight/abdominal adiposity in the case of low, but not high, placental inositol content | [39] |
| Rat | GDM | Metabolic disturbances in liver of offspring from gestational diabetic dams worsened upon a high-fat diet; no protective effect of a low-fat diet against metabolic changes (obesity, hepatic steatosis, insulin resistance) | [129] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shashikadze, B.; Flenkenthaler, F.; Stöckl, J.B.; Valla, L.; Renner, S.; Kemter, E.; Wolf, E.; Fröhlich, T. Developmental Effects of (Pre-)Gestational Diabetes on Offspring: Systematic Screening Using Omics Approaches. Genes 2021, 12, 1991. https://doi.org/10.3390/genes12121991
Shashikadze B, Flenkenthaler F, Stöckl JB, Valla L, Renner S, Kemter E, Wolf E, Fröhlich T. Developmental Effects of (Pre-)Gestational Diabetes on Offspring: Systematic Screening Using Omics Approaches. Genes. 2021; 12(12):1991. https://doi.org/10.3390/genes12121991
Chicago/Turabian StyleShashikadze, Bachuki, Florian Flenkenthaler, Jan B. Stöckl, Libera Valla, Simone Renner, Elisabeth Kemter, Eckhard Wolf, and Thomas Fröhlich. 2021. "Developmental Effects of (Pre-)Gestational Diabetes on Offspring: Systematic Screening Using Omics Approaches" Genes 12, no. 12: 1991. https://doi.org/10.3390/genes12121991
APA StyleShashikadze, B., Flenkenthaler, F., Stöckl, J. B., Valla, L., Renner, S., Kemter, E., Wolf, E., & Fröhlich, T. (2021). Developmental Effects of (Pre-)Gestational Diabetes on Offspring: Systematic Screening Using Omics Approaches. Genes, 12(12), 1991. https://doi.org/10.3390/genes12121991






