Clinical Spectrum and Genetic Diagnosis of 54 Consecutive Patients Aged 0–25 with Bilateral Cataracts
Abstract
:1. Introduction
2. Materials and Methods
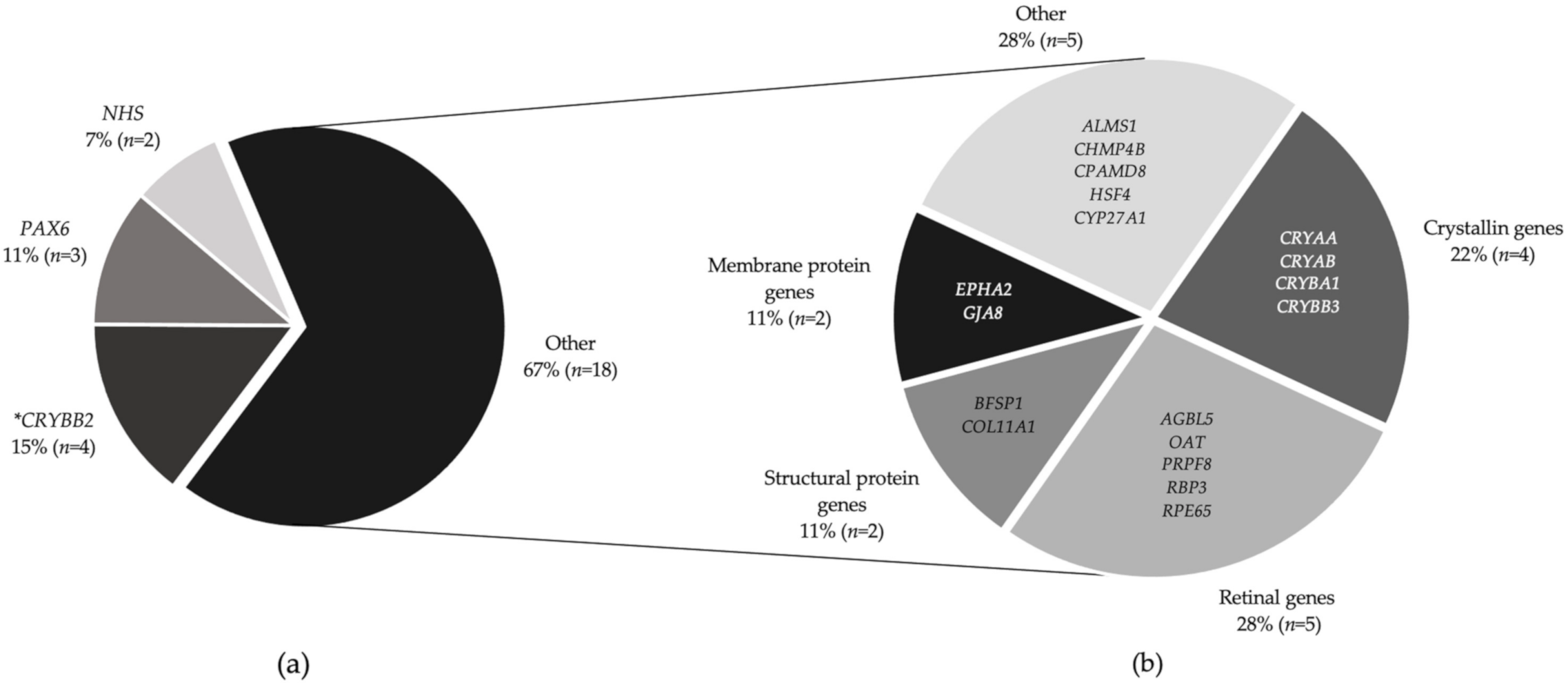
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Report on Vision. Available online: https://www.who.int/publications/i/item/world-report-on-vision (accessed on 17 November 2020).
- Sheeladevi, S.; Lawrenson, J.G.; Fielder, A.R.; Suttle, C.M. Global prevalence of childhood cataract: A systematic review. Eye 2016, 30, 1160–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahi, J.S.; Dezateux, C. Congenital and infantile cataract in the United Kingdom: Underlying or associated factors. British Congenital Cataract Interest Group. Investig. Ophthalmol. Vis. Sci. 2000, 41, 2108–2114. [Google Scholar]
- Patel, A.; Hayward, J.D.; Tailor, V.; Nyanhete, R.; Ahlfors, H.; Gabriel, C.; Jannini, T.B.; Abbou-Rayyah, Y.; Henderson, R.; Nischal, K.K.; et al. The oculome panel test: Next-generation sequencing to diagnose a diverse range of genetic developmental eye disorders. Ophthalmology 2019, 126, 888–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichi, F.; Lembo, A.; Serafino, M.; Nucci, P. Genetics of congenital cataract. Dev. Ophthalmol. 2016, 57, 1–14. [Google Scholar] [CrossRef]
- Shiels, A.; Bennett, T.M.; Hejtmancik, J.F. Cat-Map: Putting cataract on the map. Mol. Vis. 2010, 16, 2007–2015. [Google Scholar]
- Bell, S.J.; Oluonye, N.; Harding, P.; Moosajee, M. Congenital cataract: A guide to genetic and clinical management. Ther. Adv. Rare Dis. 2020, 1. [Google Scholar] [CrossRef]
- Hejtmancik, J.F. Congenital cataracts and their molecular genetics. Semin. Cell Dev. Biol. 2008, 19, 134–149. [Google Scholar] [CrossRef] [Green Version]
- Shiels, A.; Hejtmancik, J.F. Molecular genetics of cataract. Prog. Mol. Biol. Transl. Sci. 2015, 134, 203–218. [Google Scholar]
- Bremond-Gignac, D. Congenital aniridia in children. Rev. Prat. 2019, 69, 67–70. [Google Scholar]
- Lee, H.; Khan, R.; O’Keefe, M. Aniridia: Current pathology and management. Acta Ophthalmol. 2008, 86, 708–715. [Google Scholar] [CrossRef]
- Nelson, L.B.; Spaeth, G.L.; Nowinski, T.S.; Margo, C.E.; Jackson, L. Aniridia. A review. Surv. Ophthalmol. 1984, 28, 621–642. [Google Scholar] [CrossRef]
- Fishman, G.A.; Anderson, R.J.; Lourenco, P. Prevalence of posterior subcapsular lens opacities in patients with retinitis pigmentosa. Br. J. Ophthalmol. 1985, 69, 263–266. [Google Scholar] [CrossRef]
- Murakami, Y.; Yoshida, N.; Ikeda, Y.; Nakatake, S.; Fujiwara, K.; Notomi, S.; Nabeshima, T.; Nakao, S.; Hisatomi, T.; Enaida, H.; et al. Relationship between aqueous flare and visual function in retinitis pigmentosa. Am. J. Ophthalmol. 2015, 159, 958–963.e951. [Google Scholar] [CrossRef]
- Fujiwara, K.; Ikeda, Y.; Murakami, Y.; Funatsu, J.; Nakatake, S.; Tachibana, T.; Yoshida, N.; Nakao, S.; Hisatomi, T.; Yoshida, S.; et al. Risk factors for posterior subcapsular cataract in retinitis pigmentosa. Investig. Opthalmol. Vis. Sci. 2017, 58, 2534–2537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, R.L.; O’Sullivan, J.; Ashworth, J.; Bhaskar, S.; Williams, S.; Biswas, S.; Kehdi, E.; Ramsden, S.C.; Clayton-Smith, J.; Black, G.C.; et al. Personalized diagnosis and management of congenital cataract by next-generation sequencing. Ophthalmology 2014, 121, 2124–2137.e2. [Google Scholar] [CrossRef]
- Lenassi, E.; Clayton-Smith, J.; Douzgou, S.; Ramsden, S.C.; Ingram, S.; Hall, G.; Hardcastle, C.L.; Fletcher, T.A.; Taylor, R.L.; Ellingford, J.M.; et al. Clinical utility of genetic testing in 201 preschool children with inherited eye disorders. Genet. Med. 2019, 22, 745–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Black, G.C.; MacEwen, C.; Lotery, A. The integration of genomics into clinical ophthalmic services in the UK. Eye 2019, 34, 993–996. [Google Scholar] [CrossRef]
- European Society of Human. Provision of genetic services in Europe: Current practices and issues. Eur. J. Hum. Genet. 2003, 11 (Suppl. 2), S2–S4. [Google Scholar] [CrossRef] [Green Version]
- Turnbull, C.; Scott, R.H.; Thomas, E.; Jones, L.; Murugaesu, N.; Pretty, F.B.; Halai, D.; Baple, E.; Craig, C.; Hamblin, A.; et al. The 100 000 Genomes Project: Bringing whole genome sequencing to the NHS. BMJ 2018, 361, k1687. [Google Scholar] [CrossRef] [Green Version]
- Genomics Services. Available online: https://www.rcophth.ac.uk/wp-content/uploads/2020/03/Genomics-Services-Guidance-2020.pdf (accessed on 17 November 2020).
- NHS Long Term Plan. Available online: https://www.longtermplan.nhs.uk/wp-content/uploads/2019/08/nhs-long-term-plan-version-1.2.pdf (accessed on 17 November 2020).
- Musleh, M.; Hall, G.; Lloyd, I.C.; Gillespie, R.L.; Waller, S.; Douzgou, S.; Clayton-Smith, J.; Kehdi, E.; Black, G.C.; Ashworth, J. Diagnosing the cause of bilateral paediatric cataracts: Comparison of standard testing with a next-generation sequencing approach. Eye 2016, 30, 1175–1181. [Google Scholar] [CrossRef] [Green Version]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef] [PubMed]
- Jaganathan, K.; Panagiotopoulou, S.K.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting splicing from primary sequence with deep learning. Cell 2019, 176, 535–548.e24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bendl, J.; Musil, M.; Štourač, J.; Zendulka, J.; Desprès, P.; Brezovsky, J. PredictSNP2: A unified platform for accurately evaluating SNP effects by exploiting the different characteristics of variants in distinct genomic regions. PLoS Comput. Biol. 2016, 12, e1004962. [Google Scholar] [CrossRef] [PubMed]
- Sim, N.-L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yates, A.; Achuthan, P.; Akanni, W.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef]
- Astuti, G.D.N.; Arno, G.; Hull, S.; Pierrache, L.; Venselaar, H.; Carss, K.; Raymond, F.L.; Collin, R.W.J.; Faradz, S.M.H.; Born, L.I.V.D.; et al. Mutations in AGBL5, encoding α-tubulin deglutamylase, are associated with autosomal recessive retinitis pigmentosa. Investig. Opthalmol. Vis. Sci. 2016, 57, 6180–6187. [Google Scholar] [CrossRef] [Green Version]
- Jackson, D.; Malka, S.; Harding, P.; Palma, J.; Dunbar, H.; Moosajee, M. Molecular diagnostic challenges for non-retinal developmental eye disorders in the United Kingdom. Am. J. Med. Genet. Part. C Semin. Med. Genet. 2020, 184, 578–589. [Google Scholar] [CrossRef]
- Méjécase, C.; Malka, S.; Guan, Z.; Slater, A.; Arno, G.; Moosajee, M. Practical guide to genetic screening for inherited eye diseases. Ther. Adv. Ophthalmol. 2020, 12. [Google Scholar] [CrossRef]
- Shiels, A.; Hejtmancik, J.F. Mutations and mechanisms in congenital and age-related cataracts. Exp. Eye Res. 2017, 156, 95–102. [Google Scholar] [CrossRef] [Green Version]
- Shiels, A.; Hejtmancik, J.F. Genetics of human cataract. Clin. Genet. 2013, 84, 120–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souzeau, E.; Rudkin, A.K.; Dubowsky, A.; Casson, R.J.; Muecke, J.S.; Mancel, E.; Whiting, M.; Mills, R.A.; Burdon, K.P.; Craig, J.E. PAX6 molecular analysis and genotype–phenotype correlations in families with aniridia from Australasia and Southeast Asia. Mol. Vis. 2018, 24, 261–273. [Google Scholar] [PubMed]
- Zhai, Y.; Li, J.; Yu, W.; Zhu, S.; Yu, Y.; Wu, M.; Sun, G.; Gong, X.; Yao, K. Targeted exome sequencing of congenital cataracts related genes: Broadening the mutation spectrum and genotype-phenotype correlations in 27 Chinese Han families. Sci. Rep. 2017, 7, 1219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, F.; Luo, Y.; Wu, J.; Gao, C.; Liu, X.; Mei, H.; Zhou, X. The mutation spectrum in familial versus sporadic congenital cataract based on next-generation sequencing. BMC Ophthalmol. 2020, 20, 361. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Chen, G.; Cao, X.; Zhang, Y. Cerebrotendinous xanthomatosis: A comprehensive review of pathogenesis, clinical manifestations, diagnosis, and management. Orphanet J. Rare Dis. 2014, 9, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amador, M.D.M.; Masingue, M.; Debs, R.; Lamari, F.; Perlbarg, V.; Roze, E.; Degos, B.; Mochel, F. Treatment with chenodeoxycholic acid in cerebrotendinous xanthomatosis: Clinical, neurophysiological, and quantitative brain structural outcomes. J. Inherit. Metab. Dis. 2018, 41, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Foster, A.; Gilbert, C.; Rahi, J.S. Epidemiology of cataract in childhood: A global perspective. J. Cataract. Refract. Surg. 1997, 23 (Suppl. 1), 601–604. [Google Scholar] [CrossRef]
- De Jong, E.P.; Vossen, A.C.; Walther, F.J.; Lopriore, E. How to use...neonatal TORCH testing. Arch. Dis. Child. Educ. Pract. Ed. 2013, 98, 93–98. [Google Scholar] [CrossRef]
- Stone, E.M. Genetic testing for inherited eye disease. Arch. Ophthalmol. 2007, 125, 205–212. [Google Scholar] [CrossRef] [Green Version]

| Family ID | Gene | Disease Name | Zygosity | Transcript | Base Change | Amino Acid Change | Variant Type | Inh. | ACGS Variant Classification | In Silico Prediction Scores | PhastCons Conservation Score | GnomAD | Co-Segregation | Cons. | FH | Family/Patient Reported Before | Variant Prior Reported Phenotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PAX6 | Aniridia | Het | NM_000280.4 | c.2T > G | p. ? | Start codon lost | AD | Pathogenic | CADD 27.7; PredictSNP Neutral (63%) | 1 | * | N | N | Y | - | Aniridia |
| 2 | COL11A1 | Stickler syndrome, type II | Het | NM_001854.3 | c.2755-2A > G▲ | - | Non-coding (splice) | AD | Likely Pathogenic | CADD 35; SpliceAI 1.00 (acceptor loss); PredictSNP Deleterious (68%) | 1 | * | De novo ◊ | N | N | - | - |
| 3 | CPAMD8 | Anterior segment dysgenesis 8 | Hom | NM_015692.3 | c.4351T > C▲ | p. (Ser1451Pro) | Missense | AR | VUS | CADD 24.9; PredictSNP Deleterious (87%); PolyPhen-2 Probably damaging (1.000) | 1 | * | Y ▪ | Y | N | - | - |
| 4 | OAT | Gyrate atrophy of choroid and retina | Compound het | NM_000274.4 | c.596C > A; c.1250C > T | p. Pro199Gln; p. Pro417Leu | Missense | AR | Pathogenic; Pathogenic | CADD 27.1 / 29.0; PredictSNP Deleterious (82% / 87%); PolyPhen-2 Probably damaging (0.992 / 1.000) | 1; 1 | 1.414 × 10−5; 2.833 × 10−5 | N | N | N | - | Gyrate Atrophy |
| 5 | EPHA2◆ | Cataract 6, multiple types | Het | NM_004431.3 | c.1751C > T | p. Pro584Leu | Missense | AD | Likely Pathogenic | CADD 25.3; PredictSNP Deleterious (87%); PolyPhen-2 Probably damaging (0.997) | 1 | * | Y—variant in two affected relatives | N | Y | [28] | Congenital cataract |
| 7 | AGBL5 | Retinitis pigmentosa 75 | Hom | NM_021831.5 | c.323C > G▲ | p. Pro108Arg | Missense | AR | Likely Pathogenic | CADD 25.7; PredictSNP Deleterious (87%); PolyPhen-2 Probably damaging (1.000) | 1 | * | Y ▪ | Y | Sibling only | [30] | - |
| 8 | RPE65 | Leber congenital amaurosis 2 | Hom | NM_000329.3 | c.1067dupA | p. Asn356LysfsTer9 | Frameshift (duplication) | AR | Pathogenic | PredictSNP; SIFT Damaging (0.858) | 1 | * | Y ▪ | Y | N | - | Leber congenital amaurosis 2 |
| 9 | CRYBB2 | Cataract 3, multiple types | Het | NM_000496.2 | c.230G > A▲ | p. (Gly77Asp) | Missense | AD | VUS | CADD 27.3; PredictSNP Deleterious (87%); PolyPhen-2 Probably damaging (1.000) | 1 | * | Y—variant in affected mother | N | Y | - | - |
| 10 | HSF4 | Cataract 5, multiple types | Het | NM_001040667.2 | c.360+1G > A▲ | - | Missense | AD | Likely Pathogenic | CADD 35; SpliceAI 0.98 (donor loss); PredictSNP Deleterious (89%) | 1 | * | Y—variant in affected father | N | Y | [31] | - |
| 12 | RBP3 | Retinitis pigmentosa 66 | Hom | NM_002900.2 | c.832_834delTTC | p. (Phe278del) | In-frame deletion | AR | VUS | PredictSNP; SIFT Damaging (0.894) | 1 | 2.173 × 10−5 | N | N | N | - | - |
| 13 | PAX6 | Aniridia | Het | NM_000280.4 | c.718C > T | p. Arg240Ter | Nonsense | AD | Likely Pathogenic | CADD 37; PredictSNP Deleterious (79%) | 1 | * | N | N | N | - | Aniridia |
| 16 | CRYAA | Cataract 9, multiple types | Het | NM_000394.2 | c.346C > G▲ | p. (Arg116Gly) | Missense | AD | Likely Pathogenic | CADD 28.4; PredictSNP Deleterious (87%); PolyPhen-2 Probably damaging (1.000) | 1 | * | Y—variant in 3+ affected family members | N | Y | - | - |
| 19 | CRYBB3 | Cataract 22 | Het | NM_004076.4 | c.466G > A | p. (Gly156Arg) | Missense | AD | Likely Pathogenic | CADD 27.3; PredictSNP Deleterious (87%); PolyPhen-2 Probably damaging (1.000) | 1 | 3.19 × 10−5 | N | N | N | [31] | Congenital cataract |
| 22 | CRYBB2 | Cataract 3, multiple types | Het | NM_000496.2 | c.355G > A | p. (Gly119Arg) | Missense | AD | Likely Pathogenic | CADD 29.6; PredictSNP Deleterious (81%); PolyPhen-2 Probably damaging (1.000) | 1 | * | De novo ◊ | N | N | [31] | Cataract 3 |
| 23 | BFSP1 | Cataract 33, multiple types | Het | NM_001195.4 | c.957-3C > G▲ | - | Non-coding (splice) | AD | VUS | CADD 24.6; SpliceAI 0.76 (acceptor loss); PredictSNP Deleterious (97%) | 0.992 | * | Y—variant in affected father | N | Y | - | - |
| 24 | PRPF8 | Retinitis pigmentosa 13 | Het | NM_006445.3 | c.5804G > A | p. (Arg1935His) | Missense | AD | Likely Pathogenic | CADD 28.3; PredictSNP Deleterious (87%); PolyPhen-2 Probably damaging (1.000) | 1 | * | De novo ◊ | N | N | - | Retinitis Pigmentosa |
| 25 | PAX6 | Aniridia | Het | NM_000280.4 | c.1184-1G > C | - | Non-coding (splice) | AD | Pathogenic | CADD 34; SpliceAI 0.97 (acceptor loss); PredictSNP Deleterious (89%) | 1 | * | N | N | Y | - | - |
| 26 | ALMS1 | Alstrom syndrome | Compound het | NM_015120.4 | c.4569dup; c.10975C > T | p. (Tyr1524LeufsTer5); p. (Arg3659Ter) | Nonsense; Frameshift (duplication) | AR | Pathogenic; Pathogenic | Frameshift—SIFT Damaging (0.858); Nonsense—PredictSNP2 Neutral (60%) | 0; 0 | *; 4.02 × 10−6 | Y—variants in trans | N | N | - | Alstrom syndrome |
| 27 | NHS | Cataract 40, X-linked | Hemi | NM_198270.3 | c.245dup | p. (Pro83AlafsTer100) | Nonsense | XR | Pathogenic | PredictSNP ; SIFT Damaging (0.579) | 0.945 | * | N | N | Y | [31] | - |
| 28 | CRYBB2 | Cataract 3, multiple types | Het | NM_000496.2 | c.463C > T | p. (Gln155Ter) | Nonsense | AD | Pathogenic | CADD 45; PredictSNP N/A (low confidence) | 1 | * | Y—variant in 4 affected relatives | N | Y | [31] | Congenital cataract |
| 34 | GJA8 | Cataract 1, multiple types | Het | NM_005267.4 | c.134G > T | p. (Trp45Leu) | Missense | AD | Likely Pathogenic | CADD 28.2; PredictSNP Deleterious (82%); PolyPhen-2 Probably damaging (1.000) | 1 | * | De novo ◊ | N | N | [31] | Cataract |
| 37 | NHS | Cataract 40, X-linked | Hemi | NM_001291867.2 | c.1625del | p. Pro542LeufsTer35 | Nonsense | XR | Likely Pathogenic | PredictSNP; SIFT Damaging (0.858) | 0.979 | * | Y—unaffected mother is carrier | N | N | - | - |
| 40 | CRYBB2 | Cataract 3, multiple types | Het | NM_000496.2 | c.355G > A | p. (Gly119Arg) | Missense | AD | Likely Pathogenic | CADD 29.6; PredictSNP Deleterious (81%); PolyPhen-2 Probably damaging (1.000) | 1 | * | Y—variant absent in unaffected mother, present in affected sister | N | Y | - | Cataract 3 |
| 41 | CHMP4B | Cataract 31, multiple types | Het | NM_176812.4 | c.481G > C | p. (Glu161Gln) | Frameshift deletion | AD | Likely Pathogenic | CADD 33; PredictSNP Deleterious (87%); PolyPhen-2 Probably damaging (1.000) | 1 | * | Y—variant in affected father | N | Y | [31,32] | Cataract 31 |
| 42 | CRYBA1 | Cataract 10, multiple types | Het | NM_005208.4 | c.215+5G > C▲ | - | Non-coding (splice) | AD | VUS | CADD 27.9; SpliceAI 0.98 (donor loss); PredictSNP Deleterious (97%) | 1 | * | Y—variant in unaffected mother (non-penetrance—maternal relatives affected) | N | Y | - | - |
| 43 | CYP27A1 | Cerebrotendinous xanthomatosis | Hom | NM_000784.3 | c.1420C > T▲ | p. (Arg474Trp) | Missense | AR | Likely Pathogenic | CADD 29.4; PredictSNP Deleterious (87%); PolyPhen-2 Probably damaging (1.000) | 0.984 | 2.396 × 10−5 | N | Y | N | - | Cerebrotendinous xanthomatosis |
| 44 | CRYAB | Cataract 16, multiple type | Het | NM_001885.2 | c.358A > G | p. (Arg120Gly) | Missense | AD | Pathogenic | CADD 26.7; PredictSNP Deleterious (87%); PolyPhen-2 Probably damaging (1.000) | 1 | * | Y—variant in 2 affected relatives | N | Y | - | Myofibrillar Myopathy |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bell, S.; Malka, S.; Lloyd, I.C.; Moosajee, M. Clinical Spectrum and Genetic Diagnosis of 54 Consecutive Patients Aged 0–25 with Bilateral Cataracts. Genes 2021, 12, 131. https://doi.org/10.3390/genes12020131
Bell S, Malka S, Lloyd IC, Moosajee M. Clinical Spectrum and Genetic Diagnosis of 54 Consecutive Patients Aged 0–25 with Bilateral Cataracts. Genes. 2021; 12(2):131. https://doi.org/10.3390/genes12020131
Chicago/Turabian StyleBell, Suzannah, Samantha Malka, Ian Christopher Lloyd, and Mariya Moosajee. 2021. "Clinical Spectrum and Genetic Diagnosis of 54 Consecutive Patients Aged 0–25 with Bilateral Cataracts" Genes 12, no. 2: 131. https://doi.org/10.3390/genes12020131
APA StyleBell, S., Malka, S., Lloyd, I. C., & Moosajee, M. (2021). Clinical Spectrum and Genetic Diagnosis of 54 Consecutive Patients Aged 0–25 with Bilateral Cataracts. Genes, 12(2), 131. https://doi.org/10.3390/genes12020131








