Butanol Tolerance of Lactiplantibacillus plantarum: A Transcriptome Study
Abstract
:1. Introduction
2. Materials and Methods
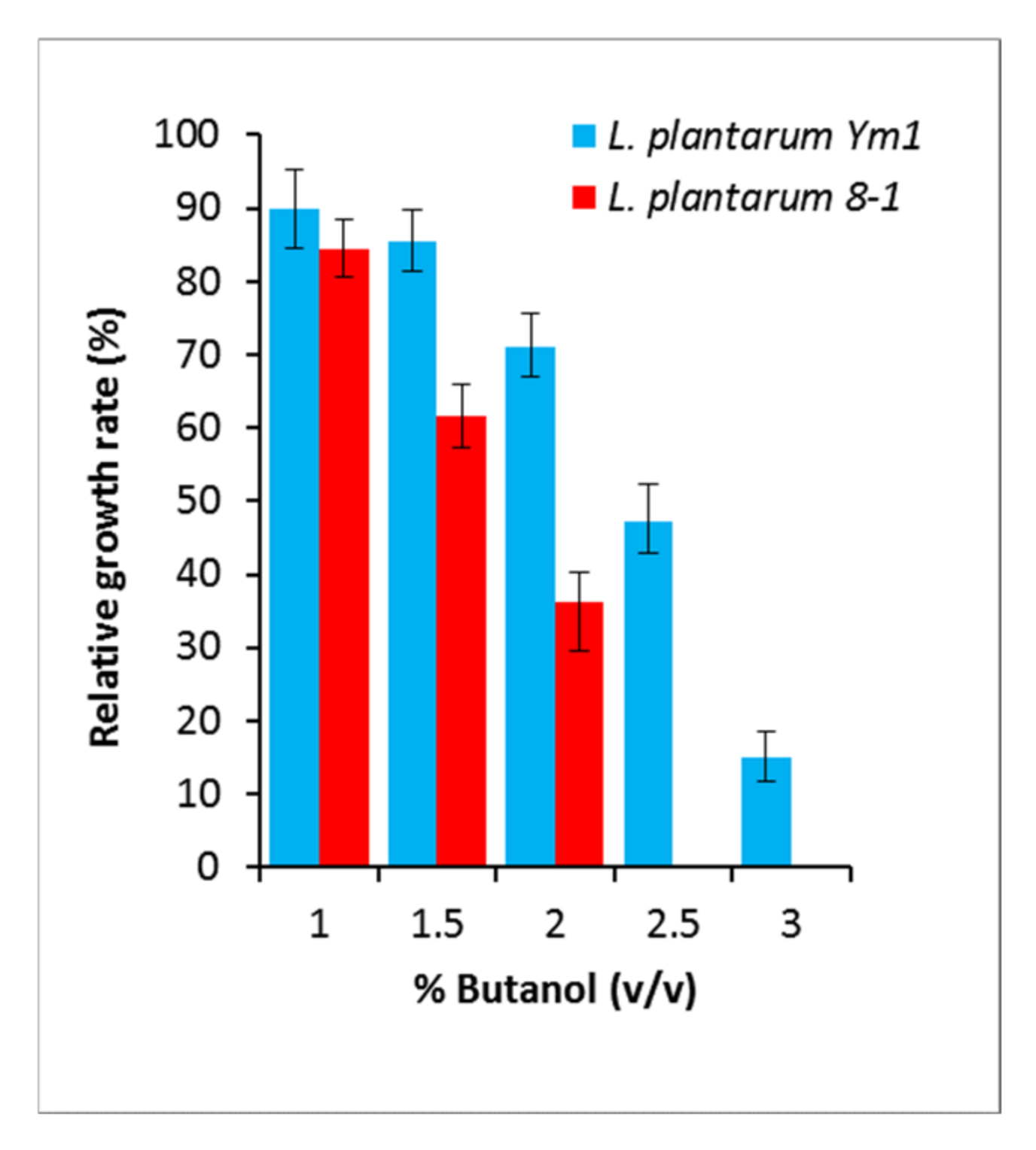
2.1. Bacterial Strains, Media, and Butanol Tolerance Assay
2.2. Analytical Methods
2.3. RNA Isolation, Libraries Construction and Sequencing
2.4. Bioinformatics Analysis
3. Results and Discussion
3.1. Selection of Butanol-Tolerant and Butanol-Sensitive Strains
3.2. Overview of Differentially Expressed Genes (DEGs)
3.3. Cell Envelope and Cell Membrane Alterations
3.4. Membrane Transport
3.5. Chaperones and Chaperonins Expression
3.6. Pyruvate Metabolism
3.7. Nucleotide Metabolism
3.8. Glycerol Metabolism
3.9. Amino Acid Metabolism
3.10. Translation
3.11. Transcription Factors and Regulators
3.12. Electron Transport
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pugazhendhi, A.; Mathimani, T.; Varjani, S.; Rene, E.R.; Kumare, G.; Kim, S.-H.; Ponnusamyg, V.K.; Yooni, J.-J. Biobutanol as a promising liquid fuel for the future—Recent updates and perspectives. Fuel 2019, 253, 637–646. [Google Scholar] [CrossRef]
- Vees, C.A.; Neuendorf, C.S.; Pflügl, S. Towards continuous industrial bioprocessing with solventogenic and acetogenic clostridia: Challenges, progress and perspectives. J. Ind. Microbiol. Biotechnol. 2020, 47, 753–787. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Lu, C.; Chen, C.; Yang, S. Biological production of butanol and higher alcohols. In Bioprocessing Technologies in Biorefinery for Sustainable Production of Fuels, Chemicals, and Polymers; Yang, S.-T., El-Enshasy, H.A., Thongchul, N., Eds.; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2013; pp. 235–261. [Google Scholar]
- Li, S.; Huang, L.; Ke, C.; Pang, Z.; Liu, L. Pathway dissection, regulation, engineering and application: Lessons learned from biobutanol production by solventogenic clostridia. Biotechnol. Biofuels 2020, 13, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peabody, G.L.; Kao, K.C. Recent progress in biobutanol tolerance in microbial systems with an emphasis on Clostridium. FEMS Microbiol. Lett. 2016, 363, fnw017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panahi, H.K.S.; Dehhaghi, M.; Kinder, J.; Ezeji, T. A review on green liquid fuels for the transportation sector: A prospect of microbial solutions to climate change. Biofuel Res. J. 2019, 6, 995–1024. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, E.; Pilizota, T.; Davies, P.R.; McVey, A.; French, C.E. Characterization of the effects of n-butanol on the cell envelope of E. coli. Appl. Microbiol. Biotechnol. 2016, 100, 9653–9659. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Zhang, M.; Guo, X.; Li, W.; Lu, D. The biological mechanisms of butanol tolerance and the application of solvent-tolerant bacteria for environmental protection. J. Chem. Technol. Biotechnol. 2020, 95, 1290–1297. [Google Scholar] [CrossRef]
- Formanek, J.; Mackie, R.; Blaschek, H.P. Enhanced butanol production by Clostridium beijerinckii BA101 grown in semidefined P2 medium containing 6 percent maltodextrin or glucose. Appl. Environ. Microbiol. 1997, 63, 2306–2310. [Google Scholar] [CrossRef] [Green Version]
- Li, S.-B.; Qian, Y.; Liang, Z.-W.; Guo, Y.; Zhao, M.-M.; Pang, Z.-W. Enhanced butanol production from cassava with Clostridium acetobutylicum by genome shuffling. World J. Microbiol. Biotechnol. 2016, 32, 53. [Google Scholar] [CrossRef]
- Jang, Y.S.; Malaviya, A.; Lee, S.Y. Acetone–butanol–ethanol production with high productivity using Clostridium acetobutylicum BKM19. Biotechnol. Bioeng. 2016, 110, 1646–1653. [Google Scholar] [CrossRef]
- Borden, J.R.; Papoutsakis, E.T. Dynamics of genomic-library enrichment and identification of solvent tolerance genes for Clostridium acetobutylicum. Appl. Environ. Microbiol. 2007, 73, 3061–3068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atsumi, S.; Wu, T.Y.; Machado, I.M.; Huang, W.C.; Chen, P.Y.; Pellegrini, M.; Liao, J.C. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol. Syst. Biol. 2010, 6, 449. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhao, J.; Yu, L.; Tang, I.-C.; Xue, C.; Yang, S.T. Engineering Clostridium acetobutylicum with a histidine kinase knockout for enhanced n-butanol tolerance and production. Appl. Microbiol. Biotechnol. 2015, 99, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, T.; Maeda, T.; Furusawa, C. Understanding and engineering alcohol-tolerant bacteria using OMICS technology. World J. Microbiol. Biotechnol. 2018, 34, 157. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Xue, T.; Ma, Y.; Zhang, J.; Wang, Z.; Hong, J.; Hui, L.; Qiao, J.; Song, H.; Zhang, M. Identification of functional butanol-tolerant genes from Escherichia coli mutants derived from error-prone PCR-based whole-genome shuffling. Biotechnol. Biofuels 2019, 12, 73. [Google Scholar] [CrossRef]
- Sedlar, K.; Kolek, J.; Gruber, M.; Jureckova, K.; Branska, B.; Csaba, G.; Vasylkivska, M.; Zimmer, R.; Patakova, P.; Provaznik, I. A transcriptional response of Clostridium beijerinckii NRRL B-598 to a butanol shock. Biotechnol. Biofuels 2019, 12, 243. [Google Scholar] [CrossRef] [Green Version]
- Tomas, C.A.; Welker, N.E.; Papoutsakis, E.T. Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell’s transcriptional program. Appl. Environ. Microbiol. 2003, 69, 4951–4965. [Google Scholar] [CrossRef] [Green Version]
- Tomas, C.A.; Beamish, J.; Papoutsakis, E.T. Transcriptional analysis of butanol stress and tolerance in Clostridium acetobutylicum. J. Bacteriol. 2004, 186, 2006–2018. [Google Scholar] [CrossRef] [Green Version]
- Mogk, A.; Homuth, G.; Scholz, C.; Kim, L.; Schmid, F.X.; Schumann, W. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 1997, 16, 4579–4590. [Google Scholar] [CrossRef] [Green Version]
- Desmond, C.; Fitzgerald, G.F.; Stanton, C.; Ross, R.P. Improved stress tolerance of GroESL-overproducing Lactococcus lactis and probiotic Lactobacillus paracasei NFBC 338. Appl. Environ. Microbiol. 2004, 70, 5929–5936. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Bonilla, P.; Zhang, J.; Wang, Y.; Blersch, D.; de-Bashan, L.-E.; Guo, L.; Wang, Y. Enhancing the tolerance of Clostridium saccharoperbutylacetonicum to lignocellulosic-biomass-derived inhibitors for efficient biobutanol production by overexpressing efflux pumps genes from Pseudomonas putida. Bioresour. Technol. 2020, 312, 123532. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Lang, N.; Zhang, L.; Wu, H.; Jiang, W.; Gu, Y. A novel regulatory pathway consisting of a two-component system and an ABC-type transporter contributes to butanol tolerance in Clostridium acetobutylicum. Appl. Microbiol. Biotechnol. 2020, 104, 5011–5023. [Google Scholar] [CrossRef] [PubMed]
- Li, J.S.; Barber, C.C.; Herman, N.A.; Cai, W.; Zafrir, E.; Du, Y.; Zhu, X.; Skyrud, W.; Zhang, W. Investigation of secondary metabolism in the industrial butanol hyper-producer Clostridium saccharoperbutylacetonicum N1-4. J. Ind. Microbiol. Biotechnol. 2020, 47, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, D.R.; Leonard, E.; Yoon, S.H.; Tseng, H.C.; Yuan, C.; Prather, K.L. Engineering alternative butanol production platforms in heterologous bacteria. Metab. Eng. 2009, 11, 262–273. [Google Scholar] [CrossRef] [Green Version]
- Berezina, O.V.; Zakharova, N.V.; Brandt, A.; Yarotsky, S.V.; Schwarz, W.H.; Zverlov, V.V. Reconstructing the clostridial n-butanol metabolic pathway in Lactobacillus brevis. Appl. Microbiol. Biotechnol. 2010, 87, 635–646. [Google Scholar] [CrossRef]
- Liu, S.; Bischoff, K.M.; Qureshi, N.; Hughes, S.R.; Rich, J.O. Functional expression of the thiolase gene thl from Clostridium beijerinckii P260 in Lactococcus lactis and Lactobacillus buchneri. N. Biotechnol. 2010, 27, 283–288. [Google Scholar] [CrossRef]
- Si, T.; Luo, Y.; Xiao, H.; Zhao, H. Utilizing an endogenous pathway for 1-butanol production in Saccharomyces cerevisiae. Metab. Eng. 2014, 22, 60–68. [Google Scholar] [CrossRef]
- Shi, S.; Si, T.; Liu, Z.; Zhang, H.; Ang, E.L.; Zhao, H. Metabolic engineering of a synergistic pathway for n-butanol production in Saccharomyces cerevisiae. Sci. Rep. 2016, 6, 25675. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Yang, Y.-M.; Beck, D.A.C.; Wang, Q.-W.; Chen, W.-J.; Yang, J.; Lidstrom, M.E.; Yang, S. Comprehensive molecular characterization of Methylobacterium extorquens AM1 adapted for 1-butanol tolerance. Biotechnol. Biofuels 2016, 9, 84. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Zhao, C.; Zhang, T.; Zhu, H.; Lin, Z.; Tao, W.; Zhang, Y.; Li, Y. A systematically chromosomally engineered Escherichia coli efficiently produces butanol. Metab. Eng. 2017, 44, 284–292. [Google Scholar] [CrossRef]
- Spano, G.; Capozzi, V.; Vernile, A.; Massa, S. Cloning, molecular characterization and expression analysis of two small heat shock genes isolated from wine Lactobacillus plantarum. J. Appl. Microbiol. 2004, 97, 774–782. [Google Scholar] [CrossRef] [PubMed]
- Petrova, P.; Emanuilova, M.; Petrov, K. Amylolytic Lactobacillus strains from Bulgarian fermented beverage boza. Z. Naturforsch. C 2010, 65, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Petrova, P.; Tsvetanova, F.; Petrov, K. Low cell surface hydrophobicity is one of the key factors for high butanol tolerance of Lactic acid bacteria. Eng. Life Sci. 2019, 19, 133–142. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.; Gobbetti, M. Environmental stress responses in Lactobacillus: A review. Proteomics 2004, 4, 106–122. [Google Scholar] [CrossRef]
- Spano, G.; Massa, S. Environmental stress response in wine lactic acid bacteria: Beyond Bacillus subtilis. Crit. Rev. Microbiol. 2006, 32, 77–86. [Google Scholar] [CrossRef]
- Li, J.; Zhao, J.B.; Zhao, M.; Yang, Y.L.; Jiang, W.H.; Yang, S. Screening and characterization of butanol-tolerant micro-organisms. Lett. Appl. Microbiol. 2010, 50, 373–379. [Google Scholar] [CrossRef]
- Arsov, A.; Petrov, K.; Petrova, P. How to outwit nature: Omics insight into butanol tolerance. Biotechnol. Adv. 2021, 46, 107658. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, Z. A functional and genetic overview of exopolysaccharides produced by Lactobacillus plantarum. J. Funct. Foods 2018, 47, 229–240. [Google Scholar] [CrossRef]
- Van Bokhorst-van de Veen, H.; Abee, T.; Tempelaars, M.; Bron, P.A.; Kleerebezem, M.; Marco, M.L. Short- and long-term adaptation to ethanol stress and its cross-protective consequences in Lactobacillus plantarum. Appl. Environ. Microbiol. 2011, 77, 5247–5256. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Liu, Y.; Ma, F.; Sun, M.; Mu, G.; Tuo, Y. Global transcriptomic and proteomics analysis of Lactobacillus plantarum Y44 response to 2,2-azobis(2-methylpropionamidine) dihydrochloride (AAPH) stress. J. Proteom. 2020, 226, 103903. [Google Scholar] [CrossRef]
- Siezen, R.; Boekhorst, J.; Muscariello, L.; Molenaar, D.; Renckens, B.; Kleerebezem, M. Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific gram-positive bacteria. BMC Genom. 2006, 7, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galloway-Peña, J.R.; Liang, X.; Singh, K.V.; Yadav, P.; Chang, C.; La Rosa, S.L.; Shelburne, S.; Ton-That, H.; Höök, M.; Murray, B.E. The identification and functional characterization of WxL proteins from Enterococcus faecium reveal surface proteins involved in extracellular matrix interactions. J. Bacteriol. 2015, 197, 882–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duong, T.; Barrangou, R.; Russell, W.M.; Klaenhammer, T.R. Characterization of the tre locus and analysis of trehalose cryoprotection in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 2006, 72, 1218–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, J.L.; Lindsay, E.L.; Faustoferri, R.C.; To, T.T.; Hendrickson, E.L.; He, X.; Shi, W.; McLean, J.S.; Quivey, R.G. Characterization of the trehalose utilization operon in Streptococcus mutans reveals that the TreR transcriptional regulator is involved in stress response pathways and toxin production. J. Bacteriol. 2018, 200, e00057-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basler, G.; Thompson, M.; Tullman-Ercek, D.; Keasling, J. A Pseudomonas putida efux pump acts on short-chain alcohols. Biotechnol. Biofuels 2018, 11, 136. [Google Scholar] [CrossRef] [Green Version]
- Dehoux, P.; Marvaud, J.C.; Abouelleil, A.; Earl, A.M.; Lambert, T.; Dauga, C. Comparative genomics of Clostridium bolteae and Clostridium clostridioforme reveals species-specific genomic properties and numerous putative antibiotic resistance determinants. BMC Genomics 2016, 17, 819. [Google Scholar] [CrossRef]
- Jakob, R.P.; Koch, J.R.; Burmann, B.M.; Schmidpeter, P.A.; Hunkeler, M.; Hiller, S.; Schmid, F.X.; Maier, T. Dimeric structure of the bacterial extracellular foldase PrsA. J. Biol. Chem. 2015, 290, 3278–3292. [Google Scholar] [CrossRef] [Green Version]
- Savijoki, K.; Nyman, T.A.; Kainulainen, V.; Miettinen, I.; Siljamäki, P.; Fallarero, A.; Sandholm, J.; Satokari, R.; Varmanen, P. Growth mode and carbon source impact the surfaceome dynamics of Lactobacillus rhamnosus GG. Front. Microbiol. 2019, 10, 1272. [Google Scholar] [CrossRef] [Green Version]
- Bignucolo, A.; Appanna, V.P.; Thomas, S.C.; Auger, C.; Han, S.; Omri, A.; Appanna, V.D. Hydrogen peroxide stress provokes a metabolic reprogramming in Pseudomonas fluorescens: Enhanced production of pyruvate. J. Biotechnol. 2013, 167, 309–315. [Google Scholar] [CrossRef]
- Wu, C.; Zhang, J.; Du, G.; Chen, J. Aspartate protects Lactobacillus casei against acid stress. Appl. Microbiol. Biotechnol. 2013, 97, 4083–4093. [Google Scholar] [CrossRef]
- Doi, Y. Glycerol metabolism and its regulation in lactic acid bacteria. Appl. Microbiol. Biotechnol. 2019, 103, 5079–5093. [Google Scholar] [CrossRef] [PubMed]
- Kryachko, Y.; Batbayar, B.; Tanaka, T.; Nickerson, M.T.; Korber, D.R. Production of glycerol by Lactobacillus plantarum NRRL B-4496 and formation of hexamine during fermentation of pea protein enriched flour. J. Biotechnol. 2020, 323, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Del Sol Cuenca, M.; Roca, A.; Molina-Santiago, C.; Duque, E.; Armengaud, J.; Gómez-Garcia, M.R.; Ramos, J.L. Understanding butanol tolerance and assimilation in Pseudomonas putida BIRD-1: An integrated omics approach. Microb. Biotechnol. 2016, 9, 100–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Chen, L.; Tian, X.; Gao, L.; Niu, X.; Shi, M.; Zhang, W. Global metabolomic and network analysis of Escherichia coli responses to exogenous biofuels. J. Proteome Res. 2013, 12, 5302–5312. [Google Scholar] [CrossRef]
- Winkler, J.; Kao, K.C. Transcriptional analysis of Lactobacillus brevis to n-butanol and ferulic acid stress responses. PLoS ONE 2011, 6, 8. [Google Scholar] [CrossRef] [Green Version]
- Si, H.M.; Zhang, F.; Wu, A.N.; Han, R.Z.; Xu, G.C.; Ni, Y. DNA microarray of global transcription factor mutant reveals membrane-related proteins involved in n-butanol tolerance in Escherichia coli. Biotechnol. Biofuels 2016, 9, 114. [Google Scholar] [CrossRef] [Green Version]
- Latousakis, D.; MacKenzie, D.A.; Telatin, A.; Juge, N. Serine-rich repeat proteins from gut microbes. Gut Microbes 2020, 11, 102–117. [Google Scholar] [CrossRef]
- Hevia, A.; Martínez, N.; Ladero, V.; Alvarez, M.A.; Margolles, A.; Sánchez, B. An extracellular Serine/Threonine-rich protein from Lactobacillus plantarum NCIMB 8826 is a novel aggregation-promoting factor with affinity to mucin. Appl. Environ. Microbiol. 2013, 79, 6059–6066. [Google Scholar] [CrossRef] [Green Version]
- Dijkstra, A.R.; Setyawati, M.C.; Bayjanov, J.R.; Alkema, W.; van Hijum, S.A.; Bron, P.A.; Hugenholtz, J. Diversity in robustness of Lactococcus lactis strains during heat stress, oxidative stress, and spray drying stress. Appl. Environ. Microbiol. 2014, 80, 603–611. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Zhu, Y.; Tang, H.; Yan, J.; Luo, L. Identification of a GntR family regulator BusRTha and its regulatory mechanism in the glycine betaine ABC transport system of Tetragenococcus halophilus. Extremophiles 2019, 23, 451–460. [Google Scholar] [CrossRef]
- Sun, D.; Setlow, P. Cloning and nucleotide sequence of the Bacillus subtilis ansR gene, which encodes a repressor of the ans operon coding for L-asparaginase and L-aspartase. J. Bacteriol. 1993, 175, 2501–2506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rojas-Tapias, D.F.; Helmann, J.D. Roles and regulation of Spx family transcription factors in Bacillus subtilis and related species. Adv. Microb. Physiol. 2019, 75, 279–323. [Google Scholar] [PubMed]
- Petrov, K.; Popova, L.; Petrova, P. High lactic acid and fructose production via Mn2+ mediated conversion of inulin by Lactobacillus paracasei. Appl. Microbiol. Biotechnol. 2017, 101, 4433–4445. [Google Scholar] [CrossRef] [PubMed]
- Velikova, P.; Petrov, K.; Petrova, P. The cell wall anchored β-fructosidases of Lactobacillus paracasei: Overproduction, purification, and gene expression control. Process Biochem. 2017, 52, 53–62. [Google Scholar] [CrossRef]
- Aziz, R.K.; Breitbart, M.; Edwards, R.A. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 2010, 38, 4207–4217. [Google Scholar] [CrossRef] [Green Version]
- Vandecraen, J.; Chandler, M.; Aertsen, A.; Van Houdt, R. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 2017, 43, 709–730. [Google Scholar] [CrossRef]
- Hall, B.G. Transposable elements as activators of cryptic genes in E. coli. Genetica 1999, 107, 181–187. [Google Scholar] [CrossRef]
- Pedersen, M.B.; Gaudu, P.; Lechardeur, D.; Petit, M.A.; Gruss, A. Aerobic respiration metabolism in lactic acid bacteria and uses in biotechnology. Annu. Rev. Food Sci. Technol. 2012, 3, 37–58. [Google Scholar] [CrossRef]
- Brooijmans, R.J.; de Vos, W.M.; Hugenholtz, J. Lactobacillus plantarum WCFS1 electron transport chains. Appl. Environ. Microbiol. 2009, 75, 3580–3585. [Google Scholar] [CrossRef] [Green Version]
- Tielens, A.G.; Van Hellemond, J.J. The electron transport chain in anaerobically functioning eukaryotes. Biochim. Biophys. Acta 1998, 1365, 71–78. [Google Scholar] [CrossRef] [Green Version]









| Gene Expression | Gene | Protein | FC */Ym1 | FC */8-1 |
|---|---|---|---|---|
| Ym1—upregulated 8-1—downregulated | lp_0230 | PTS mannitol transporter subunit IICBA | +6.48 | −3.22 |
| lp_1107, citD | Citrate lyase acyl carrier protein | +3.30 | −2.96 | |
| lp_1109, citF | Citrate (pro-3S)-lyase subunit α | +3.23 | −2.12 | |
| lp_1108, citE | Citrate (pro-3S)-lyase subunit β | +3.24 | −2.19 | |
| lp_0435 | GntR family transcriptional regulator | +2.83 | −7.82 | |
| lp_3313, pflB | Formate C-acetyltransferase | +2.45 | −7.41 | |
| lp_3054 | NAD(P)-dependent alcohol DH | +2.25 | −2.09 | |
| lp_3316 | Metal-sulfur cluster assembly factor | +2.25 | −2.29 | |
| lp_3487, galM3 | Galactose mutarotase | +2.21 | −2.19 | |
| Ym1—downregulated 8-1—upregulated | lp_2037, aroF | Chorismate synthase | −2.04 | +2.61 |
| lp_2035, aroE | 3-P-shikimate 1-carboxyvinyltransferase | −2.27 | +2.52 | |
| lp_2113 | Iron-sulfur cluster biosynthesis protein | −2.36 | +2.17 | |
| lp_1992 | Hypothetical protein | −2.02 | +2.09 | |
| lp_2038 | MFS transporter | −2.17 | +2.42 | |
| lp_1955 | ABC transporter permease subunit | −3.19 | +5.81 |
| Gene | Name | FC * in Ym1 | FC * in 8-1 |
|---|---|---|---|
| lp_0230, pts2CB | PTS mannitol transporter subunit IICBA | +6.48 | −3.22 |
| lp_2097, fruA | PTS transporter subunit EIIA | +4.35 | +2.48 |
| lp_3654, pts38C | PTS sorbitol transporter subunit IIC | +2.29 | NC |
| lp_0286, pts6C | PTS cellobiose transporter subunit IIC | +2.55 | NC |
| lp_2531, pts18CBA | PTS transporter subunit EIIC | NC | +3.22 |
| lp_0886, pts11BC | PTS transporter subunit EIIC | NC | −3.91 |
| lp_3240, pts28ABC | PTS transporter subunit EIIC | NC | −2.19 |
| lp_0436, pts7C | PTS sugar transporter subunit IIC | NC | −17.13 |
| lp_3507, pts29C | PTS sugar transporter subunit IIC | NC | −6.13 |
| lp_3010, pts23C | PTS sugar transporter subunit IIC | NC | −3.72 |
| lp_2954 | PTS sugar transporter subunit IIC | NC | +2.29 |
| lp_2781, pts20B | PTS sugar transporter subunit IIB | NC | −5.44 |
| lp_3653, pts38BC | PTS glucitol/sorbitol transporter subunit IIB | NC | −5.28 |
| lp_0232, pts2A | PTS sugar transporter subunit IIA | NC | −11.82 |
| lp_2780, pts20A | PTS lactose/cellobiose transporter subunit IIA | NC | −11.81 |
| lp_3652, pts38A | PTS glucitol/sorbitol transporter subunit IIA | NC | −3.11 |
| lp_0884, pts11A | PTS glucose transporter subunit IIA | NC | −2.30 |
| lp_0575, pts9AB | PTS mannose transporter subunit IIAB | NC | −6.36 |
| lp_0185, pts1BCA | PTS β-glucoside transporter subunit IIBCA | NC | −4.96 |
| lp_2649, pts19C | PTS N-acetylgalactosamine transporter IIC | −12.45 | −2.88 |
| lp_3009, pts23B | PTS sugar transporter subunit IIB | −2.19 | −3.02 |
| lp_0265, pts5ABC | PTS transporter subunit EIIC | −18.57 | −14.69 |
| lp_0264, pts4ABC | PTS transporter subunit EIIC | −38.68 | −25.45 |
| lp_0170, dak3 | PTS-dependent dihydroxyacetone kinase phosphotransferase subunit DhaM | −4.86 | −164.91 |
| Gene | Protein | FC * in Ym1 | FC * in 8-1 |
|---|---|---|---|
| lp_2033, aroI | Shikimate kinase | −4.43 | NC |
| lp_2037, aroF | Chorismate synthase | −2.04 | +2.61 |
| lp_1652, trpE | Anthranilate synthase component I | −2.31 | NC |
| lp_1653, trpG | Anthranilate synthase component II | −3.25 | −2.07 |
| lp_1654, trpD | Anthranilate phosphoribosyltransferase | −2.49 | NC |
| lp_1656, trpF | Phosphoribosylanthranilate isomerase | −2.33 | NC |
| lp_1658, trpB | Tryptophan synthase subunit α | −2.26 | NC |
| lp_1657, trpA | Tryptophan synthase subunit β | −2.11 | NC |
| Gene | Protein | FC * in Ym1 | FC * in 8-1 |
|---|---|---|---|
| lp_0435 | GntR family transcriptional regulator | +2.82 | −7.82 |
| lp_2782 | GntR family transcriptional regulator | −3.65 | NC |
| lp_3506 | GntR family transcriptional regulator | −3.53 | NC |
| lp_0917 | Helix-turn-helix transcriptional regulator | NC | +9.96 |
| lp_3290 | Helix-turn-helix transcriptional regulator | NC | +2.97 |
| lp_1557 | Helix-turn-helix transcriptional regulator | NC | +2.40 |
| lp_0921 | Helix-turn-helix transcriptional regulator | NC | +2.06 |
| lp_2782 | Helix-turn-helix transcriptional regulator | NC | −3.65 |
| lp_0949 | Helix-turn-helix transcriptional regulator | NC | −2.04 |
| lp_3415 | Helix-turn-helix transcriptional regulator | +2.88 | +2.32 |
| lp_3143 | Helix-turn-helix transcriptional regulator | +2.79 | NC |
| lp_2228, spx3 | Transcriptional regulator Spx | NC | +2.35 |
| lp_3579, spx5 | Transcriptional regulator Spx | −3.80 | −20.62 |
| lp_3479, galR | LacI family transcriptional regulator | +2.51 | NC |
| lp_0173 | LacI family transcriptional regulator | NC | −4.76 |
| lp_0172 | LacI family transcriptional regulator | NC | −3.73 |
| lp_0166, dak1A | DhaKLM operon coactivator DhaQ | NC | −3.11 |
| lp_3216 | PadR family transcriptional regulator | NC | +4.99 |
| lp_0154 | PadR family transcriptional regulator | NC | −2.86 |
| lp_2964 | HxlR family transcriptional regulator | NC | +2.90 |
| lp_2804 | LysR family transcriptional regulator | +2.62 | +2.32 |
| lp_1685 | LysR family transcriptional regulator | NC | −3.15 |
| lp_1000 | LytR family transcriptional regulator | NC | −2.09 |
| lp_1565 | LytTR family transcriptional regulator | −2.39 | NC |
| lp_0274 | TetR/AcrR family transcriptional regulator | +2.67 | +2.19 |
| lp_1442 | Crp/Fnr family transcriptional regulator | +2.05 | +2.58 |
| lp_3444 | Crp/Fnr family transcriptional regulator | NC | +2.54 |
| lp_2095, fruR | DeoR/GlpR transcriptional regulator | +4.02 | +3.47 |
| lp_1092, ttdR | AraC family transcriptional regulator | −2.60 | −2.39 |
| lp_1782, pyrR2 | PyrR transcriptional regulator | −2.10 | NC |
| lp_2704, pyrR1 | PyrR transcriptional regulator | −7.10 | −3.27 |
| lp_2967 | MarR family transcriptional regulator | −2.27 | NC |
| lp_0128 | MarR family transcriptional regulator | NC | +2.13 |
| lp_2800b | MarR family transcriptional regulator | NC | −2.35 |
| lp_3113 | MerR family transcriptional regulator | +3.06 | +3.16 |
| lp_0984 | MerR family transcriptional regulator | +2.80 | +4.88 |
| lp_2567 | MerR family transcriptional regulator | +2.20 | +2.31 |
| lp_2854 | MerR family transcriptional regulator | NC | +4.14 |
| lp_0281 | MerR family transcriptional regulator | NC | +2.50 |
| lp_3013 | MerR family transcriptional regulator | NC | +2.43 |
| lp_0992 | MerR family transcriptional regulator | NC | +2.07 |
| lp_2063, lexA | Transcriptional repressor LexA | −2.31 | NC |
| lp_2708, pucR | PucR family transcriptional regulator | −2.43 | NC |
| lp_3508 | Response regulator transcription factor | NC | −5.63 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrov, K.; Arsov, A.; Petrova, P. Butanol Tolerance of Lactiplantibacillus plantarum: A Transcriptome Study. Genes 2021, 12, 181. https://doi.org/10.3390/genes12020181
Petrov K, Arsov A, Petrova P. Butanol Tolerance of Lactiplantibacillus plantarum: A Transcriptome Study. Genes. 2021; 12(2):181. https://doi.org/10.3390/genes12020181
Chicago/Turabian StylePetrov, Kaloyan, Alexander Arsov, and Penka Petrova. 2021. "Butanol Tolerance of Lactiplantibacillus plantarum: A Transcriptome Study" Genes 12, no. 2: 181. https://doi.org/10.3390/genes12020181







