Comparative Analysis, Characterization and Evolutionary Study of Dirigent Gene Family in Cucurbitaceae and Expression of Novel Dirigent Peptide against Powdery Mildew Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Disease Infection
2.2. Sequences Assembly of DIRs in Cucurbitaceae
2.3. Physical Location of DIR Genes on the Chromosome and Synteny Analysis
2.4. Predicted Protein Structure, Property Analysis and Subcellular Localization
2.5. Phylogenetic Analysis, Motif, and Gene Structures
2.6. Gene Ontology and Cis-Element in Promoter Region
2.7. Quantitative Real-Time PCR
2.8. Expression Analysis of DIR Genes in Various Tissue under PM Stress
2.9. Statistical Analysis
3. Results
3.1. Genome-Wide Analysis Reveals Diversification of DIR in Cucurbitaceae
3.2. Physiochemical Properties of DIR Proteins and Prediction of Tertiary Structures
3.3. Sequence Alignment and Evolutionary Relationship of DIR Genes
3.3.1. DIR-a
3.3.2. DIR-b/d
3.3.3. DIR-c
3.3.4. DIR-e
3.3.5. DIR-f
3.3.6. DIR-g
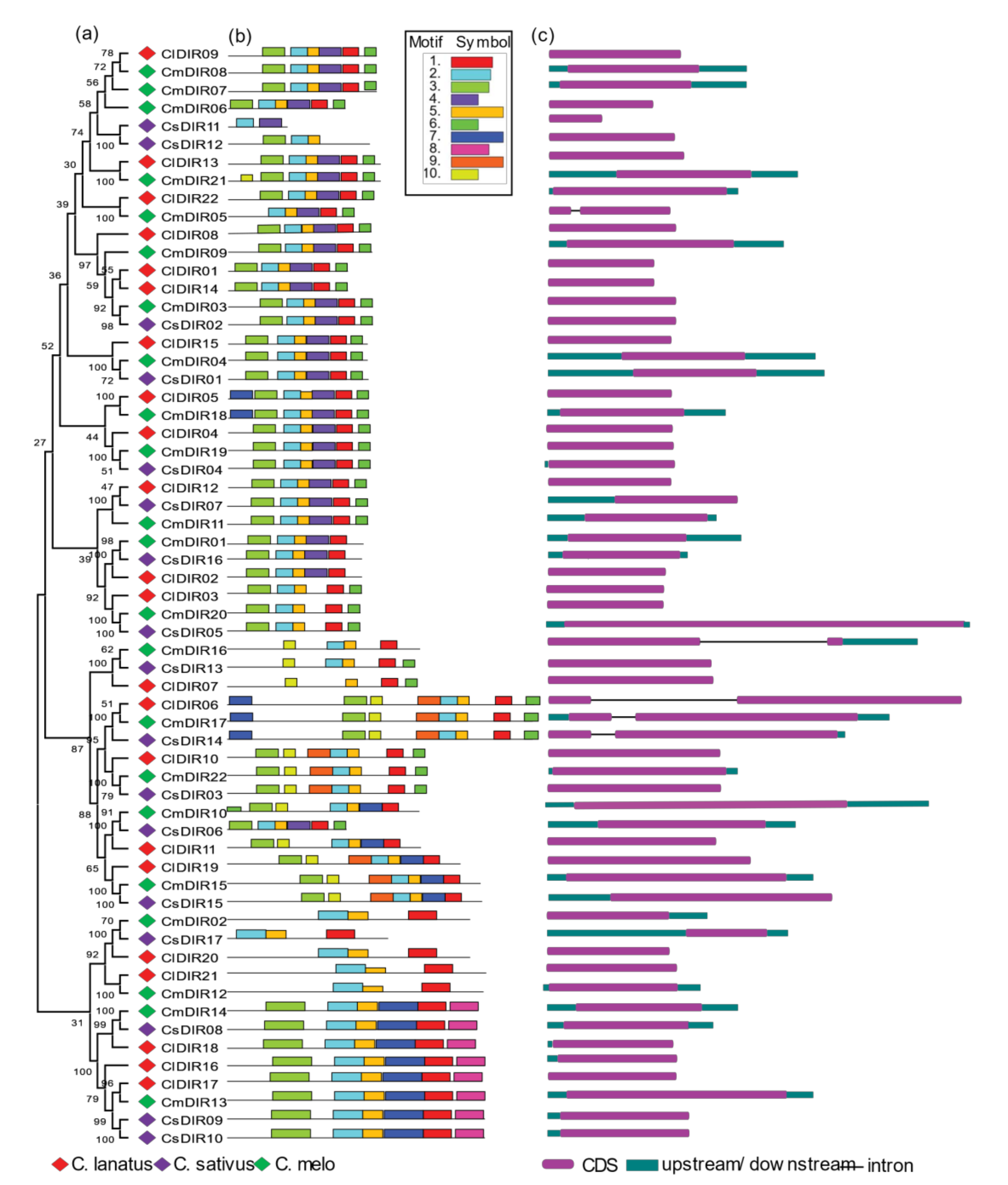
3.4. Systematic Evolutionary Relationship, Motif Analysis and Structural Gene Diversity
3.4.1. Gene Structure Diversity
3.4.2. Conserved Motif Distribution
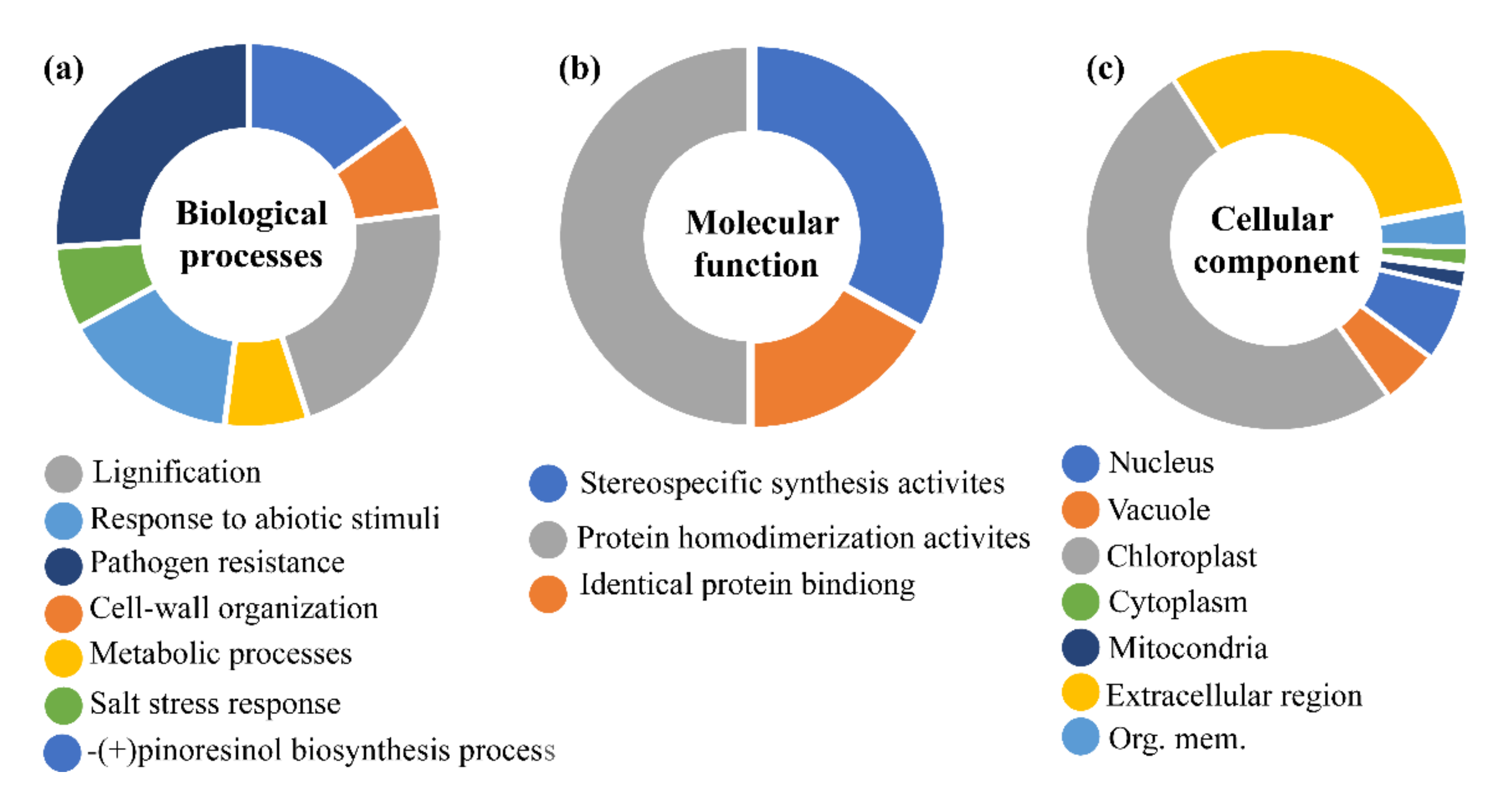
3.5. Prediction of Cis-Element and Gene Ontology (GO)
3.5.1. Phytohormones and Stress-Responsive Cis-Elements in Promoter Region
3.5.2. Role in Plant Stress Stimulus and (+)-Pinoresional Biosynthesis Processes.
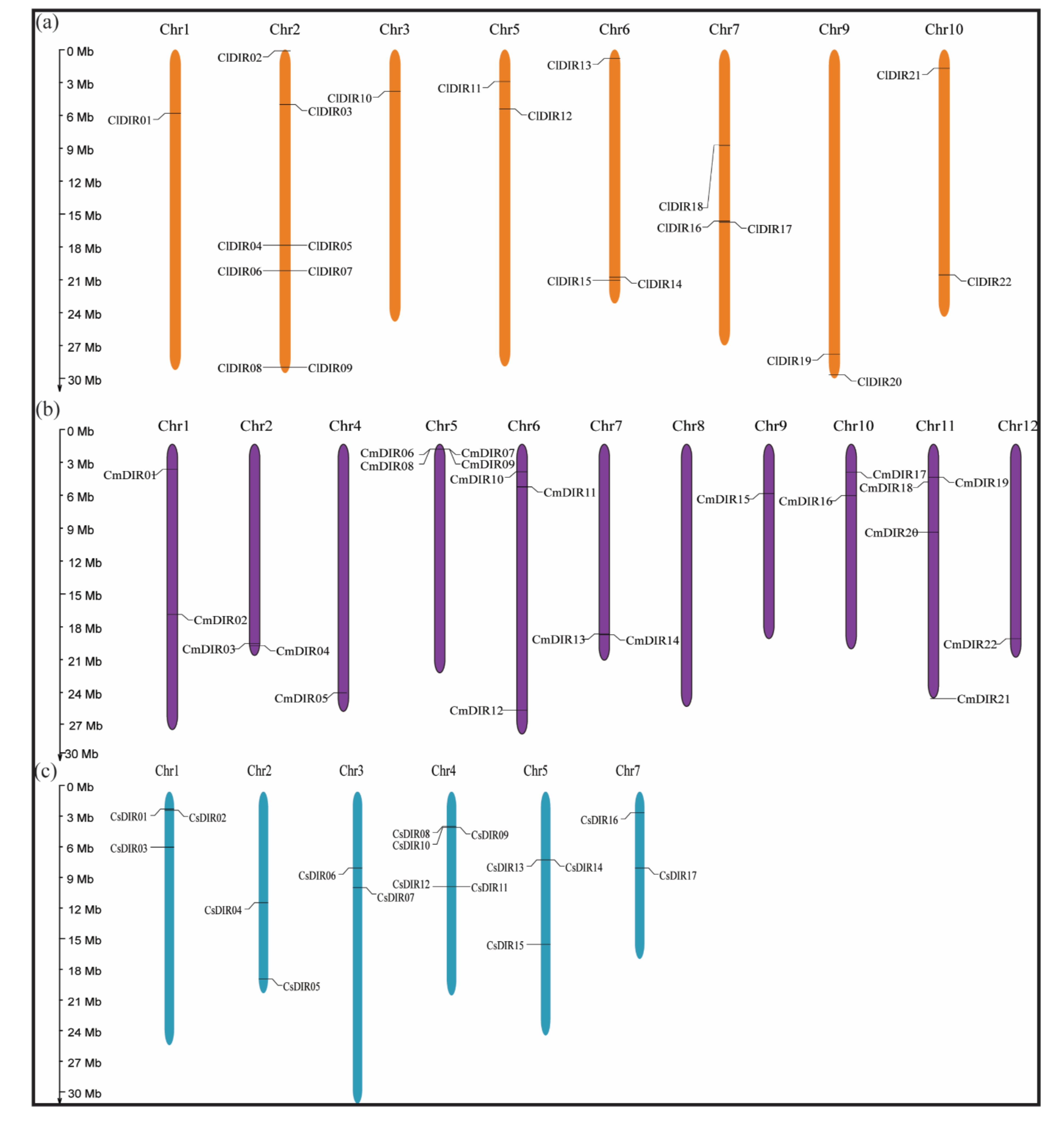
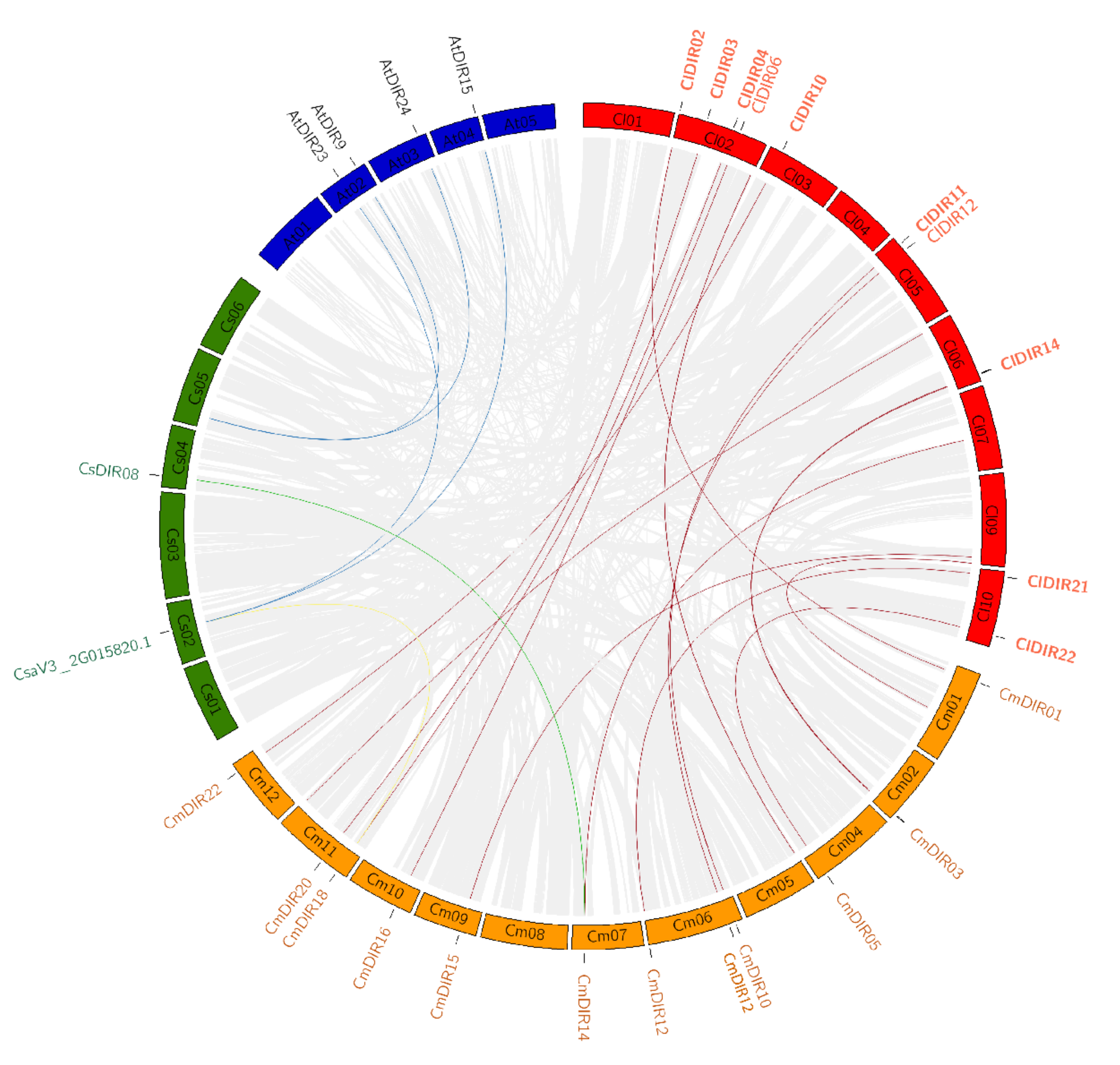
3.6. Chromosome Mapping and Synteny Analysis
3.6.1. Uneven and Wide Distribution of ClDIR, CmDIR and CsDIR Genes on Different Chromosomes
3.6.2. Duplication and Synteny Relationship Analysis
3.7. Preferential Expression of CsDIR, CmDIR and ClDIR in Root Tissue
3.7.1. qRT-PCR Expression Analysis of ClDIR Genes in Different Plant Tissues
3.7.2. Meta-Analysis of Public Expression Data for CsDIR and CmDIR in Various Tissues
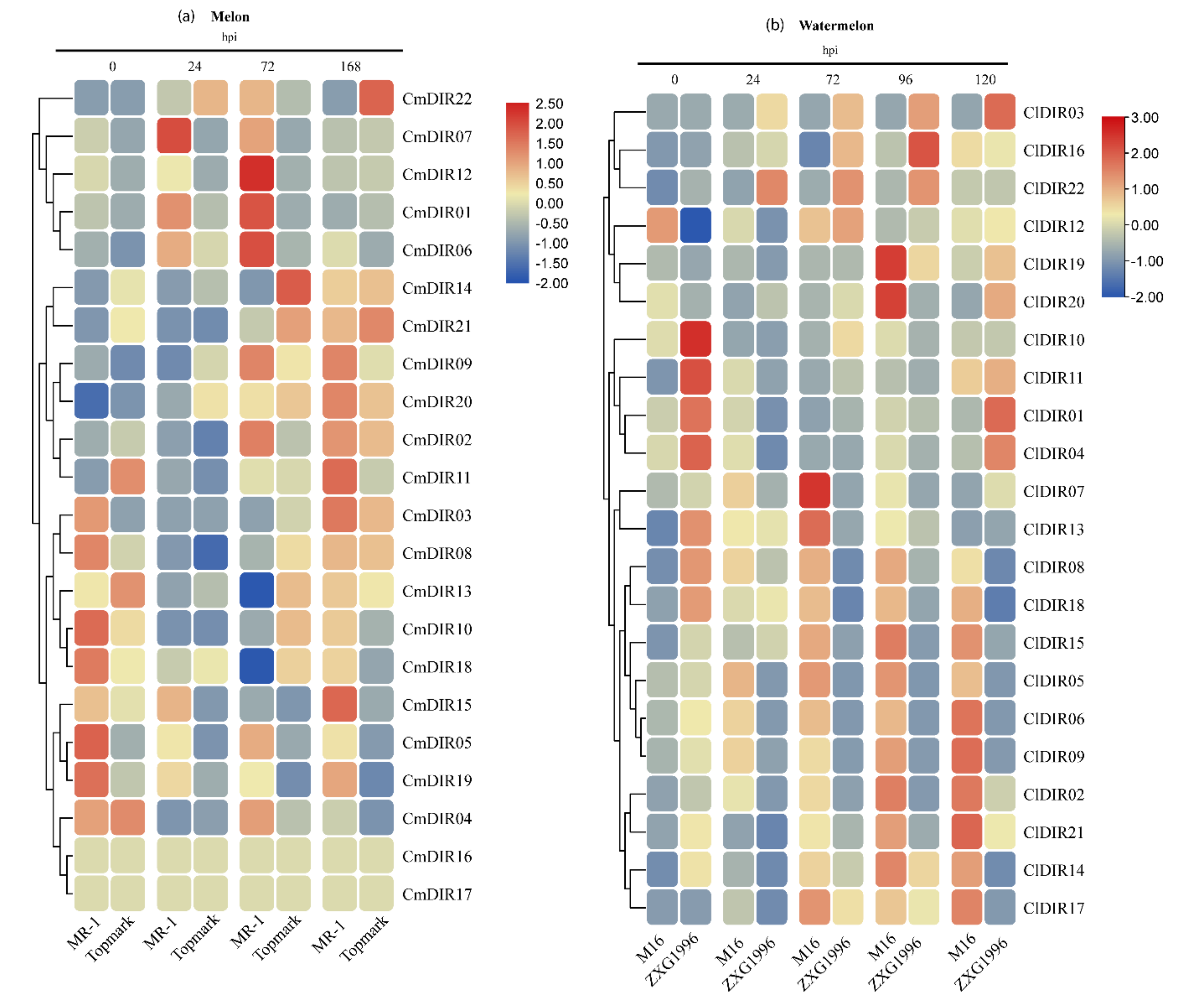
3.8. Gene Expression Profiling Provides Key Insight into a Potential Role in Disease Resistance
3.8.1. Digital Gene Expression Analysis of CmDIR Genes in Resistant and Sensitive Cultivars in Response to Powdery Mildew Infection
3.8.2. Gene Expression Profiling of Resistant and Sensitive Cultivar against Powdery Mildew Infection
4. Discussion
4.1. Functional Diversification among Subgroups from DIR-a to DIR-f
4.2. Gain in Introns during DIR Diversification
4.3. Wide Distribution of DIR on Different Chromosomes and the Role of Duplication in Gene Evolution
4.4. Transcript Accumulation Patterns in Various Plant Tissues
4.5. Cultivar Specific Inducible Expression Pattern of ClDIR and CmDIR Response to PM
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vanholme, R.; Morreel, K.; Darrah, C.; Oyarce, P.; Grabber, J.H.; Ralph, J.; Boerjan, W. Metabolic engineering of novel lignin in biomass crops. New Phytol. 2012, 196, 978–1000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barros, J.; Serk, H.; Granlund, I.; Pesquet, E. The cell biology of lignification in higher plants. Ann. Bot. 2015, 115, 1053–1074. [Google Scholar] [CrossRef] [Green Version]
- Paniagua, C.; Bilkova, A.; Jackson, P.; Dabravolski, S.; Riber, W.; Didi, V.; Houser, J.; Gigli-Bisceglia, N.; Wimmerova, M.; Budínská, E.; et al. Dirigent proteins in plants: Modulating cell wall metabolism during abiotic and biotic stress exposure. J. Exp. Bot. 2017, 68, 3287–3301. [Google Scholar] [CrossRef] [Green Version]
- Gasper, R.; Effenberger, I.; Kolesinski, P.; Terlecka, B.; Hofmann, E.; Schaller, A. Dirigent Protein Mode of Action Revealed by the Crystal Structure of AtDIR6. Plant Physiol. 2016, 172, 2165–2175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burlat, V.; Kwon, M.; Davin, L.B.; Lewis, N.G. Dirigent proteins and dirigent sites in lignifying tissues. Phytochemisty 2001, 57, 883–897. [Google Scholar] [CrossRef]
- Gang, D.R.; Costa, M.A.; Fujita, M.; Dinkova-Kostova, A.T.; Wang, H.-B.; Burlat, V.; Martin, W.; Sarkanen, S.; Davin, L.B.; Lewis, N.G. Regiochemical control of monolignol radical coupling: A new paradigm for lignin and lignan biosynthesis. Chem. Biol. 1999, 6, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Dalisay, D.S.; Kim, K.W.; Lee, C.; Yang, H.; Rübel, O.; Bowen, B.P.; Davin, L.B.; Lewis, N.G. Dirigent Protein-Mediated Lignan and Cyanogenic Glucoside Formation in Flax Seed: Integrated Omics and MALDI Mass Spectrometry Imaging. J. Nat. Prod. 2015, 78, 1231–1242. [Google Scholar] [CrossRef] [PubMed]
- Davin, L.B.; Jourdes, M.; Patten, A.M.; Kim, K.-W.; Vassão, D.G.; Lewis, N.G. Dissection of lignin macromolecular configuration and assembly: Comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis. J. Nat. Product Rep. 2008, 25, 1015–1090. [Google Scholar] [CrossRef]
- Bagniewska-Zadworna, A.; Barakat, A.; Łakomy, P.; Smoliński, D.J.; Zadworny, M. Lignin and lignans in plant defence: Insight from expression profiling of cinnamyl alcohol dehydrogenase genes during development and following fungal infection in Populus. Plant Sci. 2014, 229, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Jeon, H.S.; Kim, S.H.; Chung, J.H.; Roppolo, D.; Lee, H.; Cho, H.J.; Tobimatsu, Y.; Ralph, J.; Park, O.K. Lignin-based barrier restricts pathogens to the infection site and confers resistance in plants. EMBO J. 2019, 38, e101948. [Google Scholar] [CrossRef]
- Yadav, V.; Wang, Z.; Wei, C.; Amo, A.; Ahmed, B.; Yang, X.; Zhang, X. Phenylpropanoid Pathway Engineering: An Emerging Approach towards Plant Defense. Pathogens 2020, 9, 312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallego-Giraldo, L.; Jikumaru, Y.; Kamiya, Y.; Tang, Y.; Dixon, R.A. Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol. 2011, 190, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Emiedes, E.; Evanholme, R.; Eboerjan, W.; Emolina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef] [Green Version]
- Reboledo, G.; Del Campo, R.; Alvarez, A.; Montesano, M.; Mara, H.; De León, I.P. Physcomitrella patens Activates Defense Responses against the Pathogen Colletotrichum gloeosporioides. Int. J. Mol. Sci. 2015, 16, 22280–22298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez, A.; Montesano, M.; Schmelz, E.; De León, I.P. Activation of Shikimate, Phenylpropanoid, Oxylipins, and Auxin Pathways in Pectobacterium carotovorum Elicitors-Treated Moss. Front. Plant Sci. 2016, 7, 328. [Google Scholar] [CrossRef] [Green Version]
- De León, I.P.; Schmelz, E.A.; Gaggero, C.; Castro, A.; Álvarez, A.; Montesano, M. Physcomitrella patens activates reinforcement of the cell wall, programmed cell death and accumulation of evolutionary conserved defence signals, such as salicylic acid and 12-oxo-phytodienoic acid, but not jasmonic acid, upon Botrytis cinerea infection. Mol. Plant Pathol. 2012, 13, 960–974. [Google Scholar] [CrossRef] [PubMed]
- Arasan, S.K.T.; Park, J.-I.; Ahmed, N.U.; Jung, H.-J.; Hur, Y.; Kang, K.-K.; Lim, Y.-P.; Nou, I.-S. Characterization and expression analysis of dirigent family genes related to stresses in Brassica. Plant Physiol. Biochem. 2013, 67, 144–153. [Google Scholar] [CrossRef]
- Weidenbach, D.; Esch, L.; Möller, C.; Hensel, G.; Kumlehn, J.; Höfle, C.; Hückelhoven, R.; Schaffrath, U. Polarized Defense Against Fungal Pathogens Is Mediated by the Jacalin-Related Lectin Domain of Modular Poaceae -Specific Proteins. Mol. Plant 2016, 9, 514–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbin, C.; Drouet, S.; Markulin, L.; Auguin, D.; Lainé, É.; Davin, L.B.; Cort, J.R.; Lewis, N.G.; Hano, C. A genome-wide analysis of the flax (Linum usitatissimum L.) dirigent protein family: From gene identification and evolution to differential regulation. Plant Mol. Biol. 2018, 97, 73–101. [Google Scholar] [CrossRef]
- Ralph, S.; Park, J.-Y.; Bohlmann, J.; Mansfield, S.D. Dirigent Proteins in Conifer Defense: Gene Discovery, Phylogeny, and Differential Wound- and Insect-induced Expression of a Family of DIR and DIR-like Genes in Spruce (Picea spp.). Plant Mol. Biol. 2006, 60, 21–40. [Google Scholar] [CrossRef]
- Pérez-García, A.; Romero, D.; Fernández-Ortuño, D.; López-Ruiz, F.; De Vicente, A.; Torés, J.A. The powdery mildew fungusPodosphaera fusca(synonymPodosphaera xanthii), a constant threat to cucurbits. Mol. Plant Pathol. 2009, 10, 153–160. [Google Scholar] [CrossRef]
- Keinath, A.P.; DuBose, V.B. Evaluation of fungicides for prevention and management of powdery mildew on watermelon. Crop. Prot. 2004, 23, 35–42. [Google Scholar] [CrossRef]
- McGrath, M.T. Fungicide Sensitivity ofSphaerotheca fuligineaPopulations in the United States. Plant Dis. 1996, 80, 697–703. [Google Scholar] [CrossRef]
- Yadav, V.; Wang, Z.; Lu, G.; Sikdar, A.; Yang, X.; Zhang, X. Evaluation of watermelon germplasm and advance breeding lines against powdery mildew race ‘2F’. Pak. J. Agric. Sci. 2021, 58, 321–330. [Google Scholar] [CrossRef]
- Zhu, Q.; Gao, P.; Wan, Y.; Cui, H.; Fan, C.; Liu, S.; Luan, F. Comparative transcriptome profiling of genes and pathways related to resistance against powdery mildew in two contrasting melon genotypes. Sci. Hortic. 2018, 227, 169–180. [Google Scholar] [CrossRef]
- Mandal, M.K.; Suren, H.; Kousik, C.J.S.R. Elucidation of resistance signaling and identification of powdery mildew resistant mapping loci (ClaPMR2) during watermelon-Podosphaera xanthii interaction using RNA-Seq and whole-genome resequencing approach. Sci. Rep. 2020, 10, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.K.; Suren, H.; Ward, B.; Boroujerdi, A.; Kousik, C. Differential roles of melatonin in plant-host resistance and pathogen suppression in cucurbits. J. Pineal Res. 2018, 65, e12505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Khan, A.; Li, R.-J.; Sun, J.-T.; Ma, F.; Zhang, H.-X.; Jin, J.-H.; Ali, M.; Haq, S.U.; Wang, J.-E.; Gong, Z.-H. Genome-wide analysis of dirigent gene family in pepper (Capsicum annuum L.) and characterization of CaDIR7 in biotic and abiotic stresses. Sci. Rep. 2018, 8, 5500. [Google Scholar] [CrossRef]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L.J.N.A.R. The Pfam protein families database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, C.; Zhang, R.; Yang, X.; Zhu, C.; Li, H.; Zhang, Y.; Ma, J.; Yang, J.; Zhang, X. Comparative Analysis of Calcium-Dependent Protein Kinase in Cucurbitaceae and Expression Studies in Watermelon. Int. J. Mol. Sci. 2019, 20, 2527. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Li, H.; Yang, Y.; Wang, Y.; Mo, Y.; Zhang, R.; Zhang, Y.; Ma, J.; Wei, C.; Zhang, X. Identification and expression analyses of WRKY genes reveal their involvement in growth and abiotic stress response in watermelon (Citrullus lanatus). PLoS ONE 2018, 13, e0191308. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Wei, C.; Yang, X.; Chen, H.; Yang, Y.; Mo, Y.; Li, H.; Zhang, Y.; Ma, J.; Yang, J.; et al. Genome-wide identification and expression analysis of calcium-dependent protein kinase and its related kinase gene families in melon (Cucumis melo L.). PLoS ONE 2017, 12, e0176352. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Liu, S.; Jiang, Y.; Hu, C.; Zhang, X.; Cao, X.; Xu, Z.; Gao, X.; Li, L.; Zhu, J.; et al. Genome-wide analysis and environmental response profiling of dirigent family genes in rice (Oryza sativa). Genes Genom. 2017, 39, 47–62. [Google Scholar] [CrossRef]
- Vision, T.J.; Brown, D.G.; Tanksley, S.D. The Origins of Genomic Duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar] [CrossRef] [Green Version]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef]
- Brāzma, A.; Jonassen, I.; Vilo, J.; Ukkonen, E. Predicting Gene Regulatory Elements in Silico on a Genomic Scale. Genome Res. 1998, 8, 1202–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dabravolski, S.A. The Resurgence of Dirigent Story: Time for a Bacterial Chapter. Curr. Microbiol. 2019, 77, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Su, X.; Muhammad, A.; Li, M.; Zhang, J.; Sun, Y.; Li, G.; Jin, Q.; Cai, Y.; Lin, Y. Molecular Characterization, Evolution, and Expression Profiling of the Dirigent (DIR) Family Genes in Chinese White Pear (Pyrus bretschneideri). Front. Genet. 2018, 9, 136. [Google Scholar] [CrossRef] [PubMed]
- Davin, L.B.; Lewis, N.G. Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiol 2000, 123, 453–461. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Chen, J.; Xiao, Y.; Di, P.; Zhang, L.; Chen, W. The dirigent multigene family in Isatis indigotica: Gene discovery and differential transcript abundance. BMC Genom. 2014, 15, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Ralph, S.G.; Jancsik, S.; Bohlmann, J. Dirigent proteins in conifer defense II: Extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (Picea spp.)☆. Phytochemisty 2007, 68, 1975–1991. [Google Scholar] [CrossRef]
- Kim, M.K.; Jeon, J.-H.; Davin, L.B.; Lewis, N.G. Monolignol radical–radical coupling networks in western red cedar and Arabidopsis and their evolutionary implications. Phytochemisty 2002, 61, 311–322. [Google Scholar] [CrossRef]
- Corbin, C.; Decourtil, C.; Marosevic, D.; Bailly, M.; Lopez, T.; Renouard, S.; Doussot, J.; Dutilleul, C.; Auguin, D.; Giglioli-Guivarc’H, N.; et al. Role of protein farnesylation events in the ABA-mediated regulation of the Pinoresinol–Lariciresinol Reductase 1 (LuPLR1) gene expression and lignan biosynthesis in flax (Linum usitatissimum L.). Plant Physiol. Biochem. 2013, 72, 96–111. [Google Scholar] [CrossRef]
- Uchida, K.; Akashi, T.; Aoki, T. The Missing Link in Leguminous Pterocarpan Biosynthesis is a Dirigent Domain-Containing Protein with Isoflavanol Dehydratase Activity. Plant Cell Physiol. 2017, 58, 398–408. [Google Scholar] [CrossRef]
- Jordan, I.K.; Rogozin, I.B.; Wolf, Y.I.; Koonin, E.V. Essential Genes Are More Evolutionarily Conserved Than Are Nonessential Genes in Bacteria. Genome Res. 2002, 12, 962–968. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Liu, Z.; Zhu, L.; Zhang, C.; Chen, Y.; Zhou, Y.; Li, F.; Li, X. Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. Acta Biochim. Biophys. Sin. 2012, 44, 555–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Zhao, M.; Liu, T.; Dong, L.; Cheng, Q.; Wu, J.; Wang, L.; Chen, X.; Zhang, C.; Lu, W.; et al. A Novel Soybean Dirigent Gene GmDIR22 Contributes to Promotion of Lignan Biosynthesis and Enhances Resistance to Phytophthora sojae. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Song, M.; Peng, X. Genome-Wide Identification and Characterization of DIR Genes in Medicago truncatula. Biochem. Genet. 2019, 57, 487–506. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.N. Alternative Splicing of Pre-Messenger RNAs in Plants in the Genomic Era. Annu. Rev. Plant Biol. 2007, 58, 267–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Addendum: Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2009, 41, 762. [Google Scholar] [CrossRef]
- Bishop, E.H.; Kumar, R.; Luo, F.; Saski, C.; Sekhon, R.S. Genome-wide identification, expression profiling, and network analysis of AT-hook gene family in maize. Genomics 2020, 112, 1233–1244. [Google Scholar] [CrossRef] [PubMed]
- Picart-Picolo, A.; Grob, S.; Picault, N.; Franek, M.; Llauro, C.; Halter, T.; Maier, T.R.; Jobet, E.; Descombin, J.; Zhang, P.; et al. Large tandem duplications affect gene expression, 3D organization, and plant–pathogen response. Genome Res. 2020, 30, 1583–1592. [Google Scholar] [CrossRef] [PubMed]
- Hermansen, R.A.; Hvidsten, T.R.; Sandve, S.R.; Liberles, D.A. Extracting functional trends from whole genome duplication events using comparative genomics. Biol. Proced. Online 2016, 18, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Konrad, A.; Teufel, A.I.; Grahnen, J.A.; Liberles, D.A. Toward a General Model for the Evolutionary Dynamics of Gene Duplicates. Genome Biol. Evol. 2011, 3, 1197–1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hückelhoven, R.; Panstruga, R. Cell biology of the plant–powdery mildew interaction. Curr. Opin. Plant Biol. 2011, 14, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Fristensky, B.; Riggleman, R.C.; Wagoner, W.; Hadwiger, L. Gene expression in susceptible and disease resistant interactions of peas induced with Fusarium solani pathogens and chitosan. Physiol. Plant Pathol. 1985, 27, 15–28. [Google Scholar] [CrossRef]
- Sánchez-Elordi, E.; Contreras, R.; De Armas, R.; Benito, M.C.; Alarcón, B.; De Oliveira, E.; Del Mazo, C.; Díaz-Peña, E.M.; Santiago, R.; Vicente, C.; et al. Differential expression of SofDIR16 and SofCAD genes in smut resistant and susceptible sugarcane cultivars in response to Sporisorium scitamineum. J. Plant Physiol. 2018, 226, 103–113. [Google Scholar] [CrossRef]
- Borges, A.F.; Ferreira, R.B.; Monteiro, S. Transcriptomic changes following the compatible interaction Vitis vinifera–Erysiphe necator. Paving the way towards an enantioselective role in plant defence modulation. Plant Physiol. Biochem. 2013, 68, 71–80. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Zhao, F.; Fang, W.; Xie, D.; Hou, J.; Yang, X.; Zhao, Y.; Tang, Z.; Nie, L.; Lv, S. Identification of early salt stress responsive proteins in seedling roots of upland cotton (Gossypium hirsutum L.) employing iTRAQ-based proteomic technique. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Seneviratne, H.K.; Dalisay, D.S.; Kim, K.-W.; Moinuddin, S.G.; Yang, H.; Hartshorn, C.M.; Davin, L.B.; Lewis, N.G. Non-host disease resistance response in pea (Pisum sativum) pods: Biochemical function of DRR206 and phytoalexin pathway localization. Phytochemistry 2015, 113, 140–148. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Zhang, X.; Tu, L.; Zeng, F.; Nie, Y.; Guo, X. Isolation and characterization of two novel dirigent-like genes high-ly induced in cotton (Gossypium barbadense and G-hirsutum) after infection by Verticillium dahliae. J. Plant Pathol. 2007, 89, 41–45. [Google Scholar]
- Hano, C.; Addi, M.; Bensaddek, L.; Cronier, D.; Baltora-Rosset, S.; Doussot, J.; Maury, S.; Mesnard, F.; Chabbert, B.; Hawkins, S.; et al. Differential accumulation of monolignol-derived compounds in elicited flax (Linum usitatissimum) cell suspension cultures. Planta 2005, 223, 975–989. [Google Scholar] [CrossRef]
- Hano, C.; Addi, M.; Fliniaux, O.; Bensaddek, L.; Duverger, E.; Mesnard, F.; Lamblin, F.; Lainé, E. Molecular characterization of cell death induced by a compatible interaction between Fusarium oxysporum f. sp. linii and flax (Linum usitatissimum) cells. Plant Physiol. Biochem. 2008, 46, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Martín, J.; Heald, J.; Kingston-Smith, A.; Winters, A.; Rubiales, D.; Sanz, M.; Mur, L.A.J.; Prats, E. A metabolomic study in oats (Avena sativa) highlights a drought tolerance mechanism based upon salicylate signalling pathways and the modulation of carbon, antioxidant and photo-oxidative metabolism. Plant Cell Environ. 2015, 38, 1434–1452. [Google Scholar] [CrossRef] [Green Version]



| Plant Species | Gene ID | CDS (bp) | No of AA | MW (kDa) | PI | Instability Index | Aliphatic Index | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| C. lanatus | ClDIR01 | 474 | 157 | 17.273 | 7.10 | 27.66 | 83.25 | −0.067 | chlo |
| ClDIR02 | 525 | 174 | 18.864 | 6.55 | 24.12 | 98.62 | 0.329 | extr. sp | |
| ClDIR03 | 525 | 174 | 18.441 | 9.64 | 9.42 | 99.77 | 0.272 | extr. sp | |
| ClDIR04 | 564 | 187 | 20.495 | 8.59 | 30.7 | 88.61 | −0.079 | extr | |
| ClDIR05 | 555 | 184 | 19.795 | 9.19 | 30.59 | 92.66 | 0.147 | extr | |
| ClDIR06 | 1191 | 396 | 41.460 | 4.36 | 43.18 | 79.34 | −0.14 | extr | |
| ClDIR07 | 738 | 245 | 27.495 | 9.63 | 33.34 | 68.86 | −0.329 | extr | |
| ClDIR08 | 567 | 188 | 20.582 | 10.04 | 28.82 | 100.64 | 0.176 | extr | |
| ClDIR09 | 588 | 195 | 21.721 | 9.85 | 31.9 | 83.54 | −0.05 | chlo | |
| ClDIR10 | 768 | 255 | 27.451 | 5.33 | 44.6 | 77.22 | −0.292 | chlo | |
| ClDIR11 | 753 | 250 | 25.619 | 5 | 38.14 | 88.6 | 0.221 | extr: | |
| ClDIR12 | 549 | 182 | 20.060 | 5.41 | 38.82 | 93.63 | 0.116 | vacu | |
| ClDIR13 | 603 | 200 | 22.527 | 6.51 | 42.14 | 89.8 | −0.007 | chlo | |
| ClDIR14 | 474 | 157 | 17.482 | 6.58 | 44.91 | 75.1 | −0.207 | extr | |
| ClDIR15 | 552 | 183 | 20.038 | 7.89 | 36.89 | 92.19 | 0.236 | extr | |
| ClDIR16 | 579 | 192 | 21.159 | 7.74 | 44.41 | 86.35 | 0.169 | extr | |
| ClDIR17 | 573 | 190 | 21.150 | 8.38 | 22.75 | 83.74 | 0.127 | extr | |
| ClDIR18 | 558 | 185 | 20.586 | 8.95 | 32.29 | 84.43 | 0.039 | chlo | |
| ClDIR19 | 906 | 301 | 30.69 | 4.92 | 35.85 | 77.74 | −0.052 | nucl | |
| ClDIR20 | 546 | 181 | 20.235 | 9.69 | 38.17 | 85.75 | −0.223 | chlo | |
| ClDIR21 | 582 | 193 | 21.146 | 7.34 | 27.1 | 94.87 | 0.172 | extr | |
| ClDIR22 | 579 | 191 | 20.730 | 9.74 | 52.29 | 93.93 | 0.231 | org. mem. | |
| C. melo | CmDIR01 | 531 | 176 | 19.135 | 7.78 | 28.23 | 93.69 | 0.207 | chlo |
| CmDIR02 | 546 | 191 | 20.924 | 9.01 | 24.49 | 92.77 | 0.138 | vacu | |
| CmDIR03 | 573 | 190 | 20.663 | 9.12 | 38.02 | 81.05 | −0.091 | extr | |
| CmDIR04 | 552 | 183 | 19.983 | 6.06 | 34.77 | 93.17 | 0.245 | chlo | |
| CmDIR05 | 501 | 166 | 17.805 | 9.3 | 48.42 | 76.93 | 0.172 | chlo | |
| CmDIR06 | 465 | 154 | 17.041 | 10.04 | 29.77 | 74.16 | −0.212 | extr | |
| CmDIR07 | 588 | 195 | 21.555 | 9.48 | 34.81 | 85.54 | 0.004 | chlo | |
| CmDIR08 | 588 | 195 | 21.646 | 9.52 | 29.96 | 84.51 | −0.007 | chlo | |
| CmDIR09 | 570 | 189 | 21.189 | 9.49 | 28.92 | 93.39 | 0.105 | nucl | |
| CmDIR10 | 747 | 248 | 25.332 | 5.16 | 31.55 | 90.48 | 0.234 | extr | |
| CmDIR11 | 549 | 182 | 19.864 | 4.99 | 33.04 | 89.34 | 0.149 | chlo | |
| CmDIR12 | 576 | 191 | 20.924 | 9.01 | 24.49 | 92.77 | 0.138 | nucl | |
| CmDIR13 | 579 | 192 | 21.353 | 7.72 | 26.75 | 86.41 | 0.189 | chlo | |
| CmDIR14 | 564 | 187 | 20.963 | 8.48 | 40.91 | 76.15 | 0.01 | extr | |
| CmDIR15 | 984 | 327 | 33.789 | 5.08 | 37.1 | 82.57 | −0.003 | extr | |
| CmDIR16 | 750 | 244 | 27.605 | 9.67 | 42.98 | 65.94 | −0.396 | chlo | |
| CmDIR17 | 1185 | 394 | 41.230 | 4.29 | 47.64 | 75.33 | −0.166 | chlo | |
| CmDIR18 | 555 | 184 | 19.947 | 9.16 | 30.57 | 86.9 | 0.055 | chlo | |
| CmDIR19 | 564 | 187 | 20.493 | 9.32 | 39.66 | 89.63 | −0.028 | chlo | |
| CmDIR20 | 519 | 172 | 18.21 | 9.57 | 12.26 | 95.29 | 0.197 | chlo | |
| CmDIR21 | 603 | 200 | 22.350 | 5.94 | 38 | 100 | 0.084 | chlo | |
| CmDIR22 | 777 | 258 | 27.890 | 5.96 | 41.65 | 76.32 | −0.288 | chlo | |
| C. sativus | CsDIR01 | 552 | 183 | 19.943 | 6.06 | 32.24 | 90 | 0.255 | nucl |
| CsDIR02 | 573 | 190 | 20.685 | 9.39 | 45.56 | 85.16 | −0.055 | chlo | |
| CsDIR03 | 774 | 257 | 27.810 | 5.69 | 46.08 | 75.53 | −0.284 | chlo | |
| CsDIR04 | 564 | 187 | 20.450 | 8.93 | 36.84 | 88.07 | −0.004 | chlo | |
| CsDIR05 | 519 | 172 | 18.340 | 9.75 | 10.99 | 91.28 | 0.119 | cyto | |
| CsDIR06 | 750 | 249 | 25.447 | 5.35 | 32.07 | 90.88 | 0.248 | vacu | |
| CsDIR07 | 549 | 182 | 20.146 | 5.14 | 26.75 | 86.1 | 0.056 | chlo | |
| CsDIR08 | 561 | 186 | 20.951 | 8.6 | 30.79 | 87.04 | 0.111 | chlo | |
| CsDIR09 | 576 | 191 | 21.309 | 8.86 | 28.57 | 87.8 | 0.169 | chlo | |
| CsDIR10 | 576 | 191 | 21.309 | 8.86 | 28.57 | 87.8 | 0.169 | chlo | |
| CsDIR11 | 237 | 78 | 8.696 | 9.85 | 36.49 | 70 | −0.338 | chlo | |
| CsDIR12 | 561 | 186 | 21.484 | 9.62 | 31.97 | 80.22 | −0.294 | ogr. mem | |
| CsDIR13 | 729 | 242 | 27.432 | 9.67 | 40.72 | 64.05 | −0.413 | chlo | |
| CsDIR14 | 1185 | 394 | 41.42 | 4.31 | 47.97 | 77.54 | −0.177 | chlo | |
| CsDIR15 | 990 | 329 | 34.014 | 5.08 | 33.9 | 79.12 | −0.011 | chlo | |
| CsDIR16 | 525 | 174 | 18.969 | 5.4 | 28.27 | 97.53 | 0.274 | extr | |
| CsDIR17 | 363 | 120 | 13.329 | 6.91 | 47.2 | 73.08 | −0.203 | mito |
| Motif | E Value | Site | Width | Consensus Sequence | Motif Logo |
|---|---|---|---|---|---|
| 1 | 5.8e-629 | 59 | 23 | RELPVVGGTGKFRFARGYATAKT |  |
| 2 | 2.6e-524 | 60 | 22 | FGTVFVIDBPLTEGPELGSKLV | 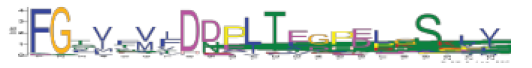 |
| 3 | 7.6e-417 | 51 | 21 | TNLVFYMHDILSGKNPTAIAV |  |
| 4 | 2.4e-328 | 60 | 15 | GRAQGFYVSSSQDGF |  |
| 5 | 2.6e-357 | 30 | 29 | LLMAMNFAFTSGKYNGSSJTILGRNPVME |  |
| 6 | 2.9e-187 | 40 | 15 | DTGDAVVEYNVYVLH |  |
| 7 | 2.0e-124 | 19 | 29 | WLAFSFMFNSTEYKGSJNFVGADPJMVKT |  |
| 8 | 1.9e-098 | 8 | 21 | DAFEGEVYFRLRVDIKFYECW |  |
| 9 | 1.9e-080 | 9 | 29 | GGLNGNEDNNQPFVTAGQLPSGVTLQQLM |  |
| 10 | 7.5e-066 | 20 | 15 | SSQLPFSKPNKFFPP |  |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, V.; Wang, Z.; Yang, X.; Wei, C.; Changqing, X.; Zhang, X. Comparative Analysis, Characterization and Evolutionary Study of Dirigent Gene Family in Cucurbitaceae and Expression of Novel Dirigent Peptide against Powdery Mildew Stress. Genes 2021, 12, 326. https://doi.org/10.3390/genes12030326
Yadav V, Wang Z, Yang X, Wei C, Changqing X, Zhang X. Comparative Analysis, Characterization and Evolutionary Study of Dirigent Gene Family in Cucurbitaceae and Expression of Novel Dirigent Peptide against Powdery Mildew Stress. Genes. 2021; 12(3):326. https://doi.org/10.3390/genes12030326
Chicago/Turabian StyleYadav, Vivek, Zhongyuan Wang, Xiaozhen Yang, Chunhua Wei, Xuan Changqing, and Xian Zhang. 2021. "Comparative Analysis, Characterization and Evolutionary Study of Dirigent Gene Family in Cucurbitaceae and Expression of Novel Dirigent Peptide against Powdery Mildew Stress" Genes 12, no. 3: 326. https://doi.org/10.3390/genes12030326
APA StyleYadav, V., Wang, Z., Yang, X., Wei, C., Changqing, X., & Zhang, X. (2021). Comparative Analysis, Characterization and Evolutionary Study of Dirigent Gene Family in Cucurbitaceae and Expression of Novel Dirigent Peptide against Powdery Mildew Stress. Genes, 12(3), 326. https://doi.org/10.3390/genes12030326







