Post-Translational Mechanisms of Plant Circadian Regulation
Abstract
:1. Introduction
2. Post-Translational Mechanisms: Protein Modifications
2.1. Ubiquitination
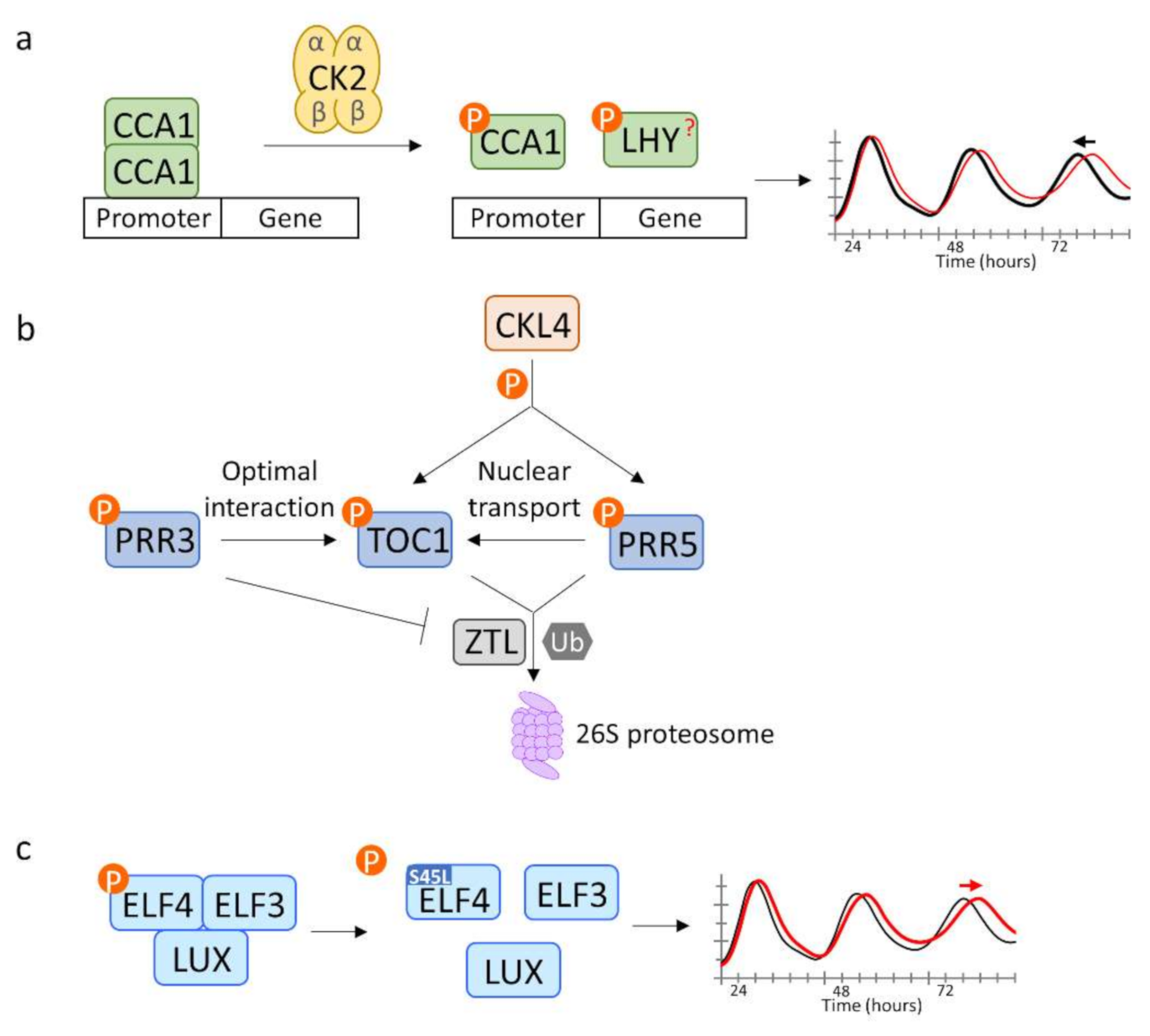
2.2. Phosphorylation
2.3. O-Glycosylation
2.4. SUMOylation
2.5. Protein Methylation
2.6. Phosphatidic Acid
3. Post-Translational Mechanisms: Protein Partitioning and Movement
3.1. Nucleocytoplasmic Partitioning
3.2. Tissue-Specific Clocks and Intercellular/Interorgan Coupling
4. Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef] [Green Version]
- Baker, C.L.; Loros, J.J.; Dunlap, J.C. The circadian clock of Neurospora crassa. FEMS Microbiol. Rev. 2012, 36, 95–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harmer, S.L.; Hogenesch, J.B.; Straume, M.; Chang, H.S.; Han, B.; Zhu, T.; Wang, X.; Kreps, J.A.; Kay, S.A. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 2000, 290, 2110–2113. [Google Scholar] [CrossRef] [PubMed]
- Seaton, D.D.; Graf, A.; Baerenfaller, K.; Stitt, M.; Millar, A.J.; Gruissem, W. Photoperiodic control of the Arabidopsis proteome reveals a translational coincidence mechanism. Mol. Syst. Biol. 2018, 14, e7962. [Google Scholar] [CrossRef]
- Graf, A.; Coman, D.; Uhrig, R.G.; Walsh, S.; Flis, A.; Stitt, M.; Gruissem, W. Parallel analysis of Arabidopsis circadian clock mutants reveals different scales of transcriptome and proteome regulation. Open Biol. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Farré, E.M.; Kay, S.A. PRR7 protein levels are regulated by light and the circadian clock in Arabidopsis. Plant J. 2007, 52, 548–560. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.-Y.; Geng, R.; Somers, D.E. Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc. Natl. Acad. Sci. USA 2003, 100, 4933–4938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Fujiwara, S.; Somers, D.E. PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 2010, 29, 1903–1915. [Google Scholar] [CrossRef] [Green Version]
- Más, P.; Kim, W.-Y.; Somers, D.E.; Kay, S.A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 2003, 426, 567–570. [Google Scholar] [CrossRef]
- Jo, H.H.; Kim, Y.J.; Kim, J.K.; Foo, M.; Somers, D.E.; Kim, P.J. Waveforms of molecular oscillations reveal circadian timekeeping mechanisms. Commun. Biol. 2018, 1, 207. [Google Scholar] [CrossRef]
- Syed, S.; Saez, L.; Young, M.W. Kinetics of doubletime kinase-dependent degradation of the Drosophila period protein. J. Biol. Chem. 2011, 286, 27654–27662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, R.; Linka, R.M.; Reinke, H. HSP90 affects the stability of BMAL1 and circadian gene expression. J. Biol. Rhythms 2014, 29, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Kiba, T.; Henriques, R.; Sakakibara, H.; Chua, N.-H. Targeted Degradation of PSEUDO-RESPONSE REGULATOR5 by an SCF ZTL Complex Regulates Clock Function and Photomorphogenesis in Arabidopsis thaliana. Plant Cell 2007, 19, 2516–2530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baudry, A.; Ito, S.; Song, Y.H.; Strait, A.A.; Kiba, T.; Lu, S.; Henriques, R.; Pruneda-Paz, J.L.; Chua, N.-H.; Tobin, E.M.; et al. F-Box Proteins FKF1 and LKP2 Act in Concert with ZEITLUPE to Control Arabidopsis Clock Progression. Plant Cell 2010, 22, 606–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srikanta, S.B.; Cermakian, N. To Ub or not to Ub: Regulation of circadian clocks by ubiquitination and deubiquitination. J. Neurochem. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, H.; Estelle, M. Plant Development: Regulation by Protein Degradation. Science 2002, 297, 793–797. [Google Scholar] [CrossRef] [Green Version]
- Vierstra, R. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 2003, 8, 135–142. [Google Scholar] [CrossRef]
- Chen, L.; Hellmann, H. Plant E3 ligases: Flexible enzymes in a sessile world. Mol. Plant 2013, 6, 1388–1404. [Google Scholar] [CrossRef] [Green Version]
- Nelson, D.C.; Lasswell, J.; Rogg, L.E.; Cohen, M.A.; Bartel, B. FKF1, a Clock-Controlled Gene that Regulates the Transition to Flowering in Arabidopsis. Cell 2000, 101, 331–340. [Google Scholar] [CrossRef] [Green Version]
- Schultz, T.F.; Kiyosue, T.; Yanovsky, M.; Wada, M.; Kay, S.A. A Role for LKP2 in the Circadian Clock of Arabidopsis. Plant Cell 2001, 13, 2659–2670. [Google Scholar] [CrossRef] [Green Version]
- Somers, D.E.; Schultz, T.F.; Milnamow, M.; Kay, S.A. ZEITLUPE Encodes a Novel Clock-Associated PAS Protein from Arabidopsis. Cell 2000, 101, 319–329. [Google Scholar] [CrossRef] [Green Version]
- Somers, D.E. Clock-associated genes in Arabidopsis: A family affair. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 1745–1753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pudasaini, A.; Shim, J.S.; Song, Y.H.; Shi, H.; Kiba, T.; Somers, D.E.; Imaizumi, T.; Zoltowski, B.D. Kinetics of the LOV domain of ZEITLUPE determine its circadian function in Arabidopsis. Elife 2017, 6. [Google Scholar] [CrossRef]
- Zoltowski, B.D.; Imaizumi, T. Structure and Function of the ZTL/FKF1/LKP2 Group Proteins in Arabidopsis. Enzymes 2014, 35, 213–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, L.; Mason, M.; Risseeuw, E.P.; Crosby, W.L.; Somers, D.E. Formation of an SCFZTL complex is required for proper regulation of circadian timing. Plant J. 2004, 40, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Harmon, F.; Imaizumi, T.; Gray, W.M. CUL1 regulates TOC1 protein stability in the Arabidopsis circadian clock. Plant J. 2008, 55, 568–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.-M.; Feke, A.; Li, M.-W.; Adamchek, C.; Webb, K.; Pruneda-Paz, J.; Bennett, E.J.; Kay, S.A.; Gendron, J.M. Decoys Untangle Complicated Redundancy and Reveal Targets of Circadian Clock F-Box Proteins. Plant Physiol. 2018, 177, 1170–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasuhara, M.; Mitsui, S.; Hirano, H.; Takanabe, R.; Tokioka, Y.; Ihara, N.; Komatsu, A.; Seki, M.; Shinozaki, K.; Kiyosue, T. Identification of ASK and clock-associated proteins as molecular partners of LKP2 (LOV kelch protein 2) in Arabidopsis. J. Exp. Bot. 2004, 55, 2015–2027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujiwara, S.; Wang, L.; Han, L.; Suh, S.S.; Salome, P.A.; McClung, C.R.; Somers, D.E. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 2008, 283, 23073–23083. [Google Scholar] [CrossRef] [Green Version]
- Para, A.; Farré, E.M.; Imaizumi, T.; Pruneda-Paz, J.L.; Harmon, F.G.; Kay, S.A. PRR3 Is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 2007, 19, 3462–3473. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.-Y.; Fujiwara, S.; Suh, S.-S.; Kim, J.; Kim, Y.; Han, L.; David, K.; Putterill, J.; Nam, H.G.; Somers, D.E. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 2007, 449, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-s.; Kim, W.Y.; Fujiwara, S.; Kim, J.; Cha, J.-Y.; Park, J.H.; Lee, S.Y.; Somers, D.E. HSP90 functions in the circadian clock through stabilization of the client F-box protein ZEITLUPE. Proc. Natl. Acad. Sci. USA 2011, 108, 16843–16848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, J.-Y.; Kim, J.; Kim, T.-S.; Zeng, Q.; Wang, L.; Lee, S.Y.; Kim, W.-Y.; Somers, D.E. GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat. Commun. 2017, 8, 3. [Google Scholar] [CrossRef] [Green Version]
- Krahmer, J.; Goralogia, G.S.; Kubota, A.; Zardilis, A.; Johnson, R.S.; Song, Y.H.; MacCoss, M.J.; Le Bihan, T.; Halliday, K.J.; Imaizumi, T.; et al. Time-resolved interaction proteomics of the GIGANTEA protein under diurnal cycles in Arabidopsis. FEBS Lett. 2019, 593, 319–338. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Somers, D.E. An HSP90 co-chaperone controls circadian proteostasis. Cell Cycle 2017, 16, 1483–1484. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-M.; Li, M.-W.; Feke, A.; Liu, W.; Saffer, A.M.; Gendron, J.M. GIGANTEA recruits the UBP12 and UBP13 deubiquitylases to regulate accumulation of the ZTL photoreceptor complex. Nat. Commun. 2019, 10, 3750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayama, R.; Yang, P.; Valverde, F.; Mizoguchi, T.; Furutani-Hayama, I.; Vierstra, R.D.; Coupland, G. Ubiquitin carboxyl-terminal hydrolases are required for period maintenance of the circadian clock at high temperature in Arabidopsis. Sci. Rep. 2019, 9, 17030. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.H.; Estrada, D.A.; Johnson, R.S.; Kim, S.K.; Lee, S.Y.; MacCoss, M.J.; Imaizumi, T. Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. USA 2014, 111, 17672–17677. [Google Scholar] [CrossRef] [Green Version]
- Feke, A.; Liu, W.; Hong, J.; Li, M.-W.; Lee, C.-M.; Zhou, E.K.; Gendron, J.M. Decoys provide a scalable platform for the identification of plant E3 ubiquitin ligases that regulate circadian function. eLife 2019, 8, e44558. [Google Scholar] [CrossRef]
- Jang, K.; Gil Lee, H.; Jung, S.-J.; Paek, N.-C.; Joon Seo, P. The E3 Ubiquitin Ligase COP1 Regulates Thermosensory Flowering by Triggering GI Degradation in Arabidopsis. Sci. Rep. 2015, 5, 12071. [Google Scholar] [CrossRef]
- Yu, J.-W.; Rubio, V.; Lee, N.-Y.; Bai, S.; Lee, S.-Y.; Kim, S.-S.; Liu, L.; Zhang, Y.; Irigoyen, M.L.; Sullivan, J.A.; et al. COP1 and ELF3 Control Circadian Function and Photoperiodic Flowering by Regulating GI Stability. Mol. Cell 2008, 32, 617–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saijo, Y.; Sullivan, J.A.; Wang, H.; Yang, J.; Shen, Y.; Rubio, V.; Ma, L.; Hoecker, U.; Deng, X.W. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003, 17, 2642–2647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.Q.; Sarmast, M.K.; Jiang, J.; Dehesh, K. The Transcriptional Regulator BBX19 Promotes Hypocotyl Growth by Facilitating COP1-Mediated EARLY FLOWERING3 Degradation in Arabidopsis. Plant Cell 2015, 27, 1128–1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, L.; Wang, S.; Song, Z.T.; Jiang, Y.; Han, J.J.; Lu, S.J.; Li, L.; Liu, J.X. Two B-Box Domain Proteins, BBX18 and BBX23, Interact with ELF3 and Regulate Thermomorphogenesis in Arabidopsis. Cell Rep. 2018, 25, 1718–1728.e1714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, O.S.; Huang, X.; Charron, J.-B.; Lee, J.-H.; Li, G.; Deng, X.W. Interaction of Arabidopsis DET1 with CCA1 and LHY in Mediating Transcriptional Repression in the Plant Circadian Clock. Mol. Cell 2011, 43, 703–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Wang, X.; Mo, X.; Tang, C.; Zhong, S.; Deng, X.W. Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc. Natl. Acad. Sci. USA 2015, 112, 3817–3822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, B.S.; Eo, H.J.; Jang, I.-C.; Kang, H.-G.; Song, J.T.; Seo, H.S. Ubiquitination of LHY by SINAT5 regulates flowering time and is inhibited by DET1. Biochem. Biophys. Res. Commun. 2010, 398, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Song, H.-R.; Carré, I.A. DET1 regulates the proteasomal degradation of LHY, a component of the Arabidopsis circadian clock. Plant Mol. Biol. 2005, 57, 761–771. [Google Scholar] [CrossRef]
- David, K.M.; Armbruster, U.; Tama, N.; Putterill, J. Arabidopsis GIGANTEA protein is post-transcriptionally regulated by light and dark. FEBS Lett. 2006, 580, 1193–1197. [Google Scholar] [CrossRef] [Green Version]
- Ito, S.; Nakamichi, N.; Kiba, T.; Yamashino, T.; Mizuno, T. Rhythmic and Light-Inducible Appearance of Clock-Associated Pseudo-Response Regulator Protein PRR9 Through Programmed Degradation in the Dark in Arabidopsis thaliana. Plant Cell Physiol. 2007, 48, 1644–1651. [Google Scholar] [CrossRef] [Green Version]
- Kangisser, S.; Yakir, E.; Green, R.M. Proteasomal regulation of CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) stability is part of the complex control of CCA1. Plant Signal. Behav. 2013, 8, e23206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perales, M.; Portolés, S.; Más, P. The proteasome-dependent degradation of CKB4 is regulated by the Arabidopsis biological clock. Plant J. 2006, 46, 849–860. [Google Scholar] [CrossRef]
- Edery, I.; Zwiebel, L.J.; Dembinska, M.E.; Rosbash, M. Temporal phosphorylation of the Drosophila period protein. Proc. Natl. Acad. Sci. USA 1994, 91, 2260–2264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Loros, J.; Dunlap, J.C. Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc. Natl. Acad. Sci. USA 2000, 97, 234–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toh, K.L.; Jones, C.R.; He, Y.; Eide, E.J.; Hinz, W.A.; Virshup, D.M.; Ptácek, L.J.; Fu, Y.H. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 2001, 291, 1040–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusakina, J.; Dodd, A.N. Phosphorylation in the plant circadian system. Trends Plant Sci. 2012, 17, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.M.; Kilman, V.L.; Keegan, K.; Paddock, B.; Emery-Le, M.; Rosbash, M.; Allada, R. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature 2002, 420, 816–820. [Google Scholar] [CrossRef]
- Akten, B.; Jauch, E.; Genova, G.K.; Kim, E.Y.; Edery, I.; Raabe, T.; Jackson, F.R. A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosci. 2003, 6, 251–257. [Google Scholar] [CrossRef]
- Yang, Y.; Cheng, P.; Liu, Y. Regulation of the Neurospora circadian clock by casein kinase II. Genes Dev. 2002, 16, 994–1006. [Google Scholar] [CrossRef] [Green Version]
- Tamaru, T.; Hirayama, J.; Isojima, Y.; Nagai, K.; Norioka, S.; Takamatsu, K.; Sassone-Corsi, P. CK2alpha phosphorylates BMAL1 to regulate the mammalian clock. Nat. Struct. Mol. Biol. 2009, 16, 446–448. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Akashi, M.; Matsuda, M.; Goto, K.; Miyata, Y.; Node, K.; Nishida, E. Involvement of the protein kinase CK2 in the regulation of mammalian circadian rhythms. Sci. Signal. 2009, 2, ra26. [Google Scholar] [CrossRef]
- Meissner, R.A.; Kilman, V.L.; Lin, J.M.; Allada, R. TIMELESS is an important mediator of CK2 effects on circadian clock function in vivo. J. Neurosci. 2008, 28, 9732–9740. [Google Scholar] [CrossRef] [PubMed]
- Diernfellner, A.C.R.; Brunner, M. Phosphorylation Timers in the Neurospora crassa Circadian Clock. J. Mol. Biol. 2020, 432, 3449–3465. [Google Scholar] [CrossRef] [PubMed]
- Sugano, S.; Andronis, C.; Green, R.M.; Wang, Z.Y.; Tobin, E.M. Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc. Natl. Acad. Sci. USA 1998, 95, 11020–11025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugano, S.; Andronis, C.; Ong, M.S.; Green, R.M.; Tobin, E.M. The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proc. Natl. Acad. Sci. USA 1999, 96, 12362–12366. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.X.; Liu, H.; Knowles, S.M.; Li, J.; Ma, L.; Tobin, E.M.; Lin, C. A role for protein kinase casein kinase2 α-subunits in the Arabidopsis circadian clock. Plant Physiol. 2011, 157, 1537–1545. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Somers, D.E. Rapid assessment of gene function in the circadian clock using artificial microRNA in Arabidopsis mesophyll protoplasts. Plant Physiol. 2010, 154, 611–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portolés, S.; Más, P. The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 2010, 6, e1001201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choudhary, M.K.; Nomura, Y.; Wang, L.; Nakagami, H.; Somers, D.E. Quantitative Circadian Phosphoproteomic Analysis of Arabidopsis Reveals Extensive Clock Control of Key Components in Physiological, Metabolic, and Signaling Pathways. Mol. Cell Proteom. 2015, 14, 2243–2260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolmos, E.; Nowak, M.; Werner, M.; Fischer, K.; Schwarz, G.; Mathews, S.; Schoof, H.; Nagy, F.; Bujnicki, J.M.; Davis, S.J. Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J. 2009, 3, 350–366. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Nusinow, D.A. Into the Evening: Complex Interactions in the Arabidopsis Circadian Clock. Trends Genet. 2016, 32, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Xu, D.; Tian, T.; Kong, F.; Lin, K.; Gan, S.; Zhang, H.; Li, G. Molecular and functional dissection of EARLY-FLOWERING 3 (ELF3) and ELF4 in Arabidopsis. Plant Sci. 2021, 303, 110786. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Barbosa, A.D.; Hutin, S.; Kumita, J.R.; Gao, M.; Derwort, D.; Silva, C.S.; Lai, X.; Pierre, E.; Geng, F.; et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 2020, 585, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Covington, M.F.; Maloof, J.N.; Straume, M.; Kay, S.A.; Harmer, S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008, 9, R130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazen, S.P.; Naef, F.; Quisel, T.; Gendron, J.M.; Chen, H.; Ecker, J.R.; Borevitz, J.O.; Kay, S.A. Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol. 2009, 10, R17. [Google Scholar] [CrossRef] [Green Version]
- Ono, A.; Sato, A.; Fujimoto, K.J.; Matsuo, H.; Yanai, T.; Kinoshita, T.; Nakamichi, N. 3,4-Dibromo-7-Azaindole Modulates Arabidopsis Circadian Clock by Inhibiting Casein Kinase 1 Activity. Plant Cell Physiol. 2019, 60, 2360–2368. [Google Scholar] [CrossRef] [Green Version]
- Uehara, T.N.; Mizutani, Y.; Kuwata, K.; Hirota, T.; Sato, A.; Mizoi, J.; Takao, S.; Matsuo, H.; Suzuki, T.; Ito, S.; et al. Casein kinase 1 family regulates PRR5 and TOC1 in the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 2019, 116, 11528–11536. [Google Scholar] [CrossRef] [Green Version]
- Montagnoli, A.; Valsasina, B.; Croci, V.; Menichincheri, M.; Rainoldi, S.; Marchesi, V.; Tibolla, M.; Tenca, P.; Brotherton, D.; Albanese, C.; et al. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat. Chem Biol. 2008, 4, 357–365. [Google Scholar] [CrossRef]
- Krahmer, J.; Hindle, M.; Perby, L.K.; Nielsen, T.H.; VanOoijen, G.; Halliday, K.J.; Bihan, T.L.; Millar, A.J. Circadian protein regulation in the green lineage II. The clock gene circuit controls a phospho-dawn in Arabidopsis thaliana. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Frank, A.; Matiolli, C.C.; Viana, A.J.C.; Hearn, T.J.; Kusakina, J.; Belbin, F.E.; Wells Newman, D.; Yochikawa, A.; Cano-Ramirez, D.L.; Chembath, A.; et al. Circadian Entrainment in Arabidopsis by the Sugar-Responsive Transcription Factor bZIP63. Curr. Biol. 2018, 28, 2597–2606.e6. [Google Scholar] [CrossRef] [Green Version]
- Uhrig, R.G.; Schläpfer, P.; Roschitzki, B.; Hirsch-Hoffmann, M.; Gruissem, W. Diurnal changes in concerted plant protein phosphorylation and acetylation in Arabidopsis organs and seedlings. Plant J. 2019, 99, 176–194. [Google Scholar] [CrossRef] [PubMed]
- Uhrig, R.G.; Echevarría-Zomeño, S.; Schlapfer, P.; Grossmann, J.; Roschitzki, B.; Koerber, N.; Fiorani, F.; Gruissem, W. Diurnal Dynamics of the Arabidopsis Rosette Proteome and Phosphoproteome. Plant Cell Environ. 2020. [Google Scholar] [CrossRef]
- Kim, E.Y.; Jeong, E.H.; Park, S.; Jeong, H.J.; Edery, I.; Cho, J.W. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 2012, 26, 490–502. [Google Scholar] [CrossRef] [Green Version]
- Li, M.D.; Ruan, H.B.; Hughes, M.E.; Lee, J.S.; Singh, J.P.; Jones, S.P.; Nitabach, M.N.; Yang, X. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 2013, 17, 303–310. [Google Scholar] [CrossRef] [Green Version]
- Zentella, R.; Sui, N.; Barnhill, B.; Hsieh, W.P.; Hu, J.; Shabanowitz, J.; Boyce, M.; Olszewski, N.E.; Zhou, P.; Hunt, D.F.; et al. The Arabidopsis O-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA. Nat. Chem. Biol. 2017, 13, 479–485. [Google Scholar] [CrossRef] [Green Version]
- Zentella, R.; Hu, J.; Hsieh, W.P.; Matsumoto, P.A.; Dawdy, A.; Barnhill, B.; Oldenhof, H.; Hartweck, L.M.; Maitra, S.; Thomas, S.G.; et al. O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis. Genes Dev. 2016, 30, 164–176. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; He, Y.; Su, C.; Zentella, R.; Sun, T.P.; Wang, L. Nuclear Localized O-Fucosyltransferase SPY Facilitates PRR5 Proteolysis to Fine-Tune the Pace of Arabidopsis Circadian Clock. Mol. Plant 2020, 13, 446–458. [Google Scholar] [CrossRef]
- Tseng, T.S.; Salomé, P.A.; McClung, C.R.; Olszewski, N.E. SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 2004, 16, 1550–1563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, P.; Shimoji, S.; Hart, G.W. Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett. 2010, 584, 2526–2538. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Xiao, J.; Yin, F.; Guo, X.; Xing, L.; Xu, Y.; Chong, K. The Protein Modifications of O-GlcNAcylation and Phosphorylation Mediate Vernalization Response for Flowering in Winter Wheat. Plant Physiol. 2019, 180, 1436–1449. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Xu, S.; Li, C.; Xu, Y.; Xing, L.; Niu, Y.; Huan, Q.; Tang, Y.; Zhao, C.; Wagner, D.; et al. O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat. Commun. 2014, 5, 4572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benlloch, R.; Lois, L.M. Sumoylation in plants: Mechanistic insights and its role in drought stress. J. Exp. Bot. 2018, 69, 4539–4554. [Google Scholar] [CrossRef] [PubMed]
- Ghimire, S.; Tang, X.; Zhang, N.; Liu, W.; Si, H. SUMO and SUMOylation in plant abiotic stress. Plant Growth Regul. 2020, 91, 317–325. [Google Scholar] [CrossRef]
- Gill, G. Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr. Opin. Genet. Dev. 2003, 13, 108–113. [Google Scholar] [CrossRef]
- Cardone, L.; Hirayama, J.; Giordano, F.; Tamaru, T.; Palvimo, J.J.; Sassone-Corsi, P. Circadian clock control by SUMOylation of BMAL1. Science 2005, 309, 1390–1394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, L.L.; van den Burg, H.A.; van Ooijen, G. Sumoylation Contributes to Timekeeping and Temperature Compensation of the Plant Circadian Clock. J. Biol. Rhythms 2017, 32, 560–569. [Google Scholar] [CrossRef] [Green Version]
- Hansen, L.L.; Imrie, L.; Le Bihan, T.; van den Burg, H.A.; van Ooijen, G. Sumoylation of the Plant Clock Transcription Factor CCA1 Suppresses DNA Binding. J. Biol. Rhythms 2017, 32, 570–582. [Google Scholar] [CrossRef] [Green Version]
- Nukarinen, E.; Tomanov, K.; Ziba, I.; Weckwerth, W.; Bachmair, A. Protein sumoylation and phosphorylation intersect in Arabidopsis signaling. Plant J. 2017, 91, 505–517. [Google Scholar] [CrossRef] [Green Version]
- Bedford, M.T.; Clarke, S.G. Protein arginine methylation in mammals: Who, what, and why. Mol. Cell 2009, 33, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Ren, J.; Wang, Y.; Liang, Y.; Zhang, Y.; Bao, S.; Xu, Z. Methylation of ribosomal protein S10 by protein-arginine methyltransferase 5 regulates ribosome biogenesis. J. Biol. Chem. 2010, 285, 12695–12705. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Lu, F.; Cui, X.; Cao, X. Histone methylation in higher plants. Annu. Rev. Plant Biol. 2010, 61, 395–420. [Google Scholar] [CrossRef] [PubMed]
- Gonsalvez, G.B.; Rajendra, T.K.; Tian, L.; Matera, A.G. The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr. Biol. 2006, 16, 1077–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anne, J.; Ollo, R.; Ephrussi, A.; Mechler, B.M. Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development 2007, 134, 137–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedford, M.T.; Richard, S. Arginine methylation an emerging regulator of protein function. Mol. Cell 2005, 18, 263–272. [Google Scholar] [CrossRef]
- Pahlich, S.; Zakaryan, R.P.; Gehring, H. Protein arginine methylation: Cellular functions and methods of analysis. Biochim. Biophys. Acta 2006, 1764, 1890–1903. [Google Scholar] [CrossRef]
- Pei, Y.; Niu, L.; Lu, F.; Liu, C.; Zhai, J.; Kong, X.; Cao, X. Mutations in the Type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol. 2007, 144, 1913–1923. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, R.J.; Sung, S.; Amasino, R.M. Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2008, 105, 411–416. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, Y.; Ma, Q.; Zhang, Z.; Xue, Y.; Bao, S.; Chong, K. SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 2007, 26, 1934–1941. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Gu, L.; Liu, C.; Lu, T.; Lu, F.; Lu, Z.; Cui, P.; Pei, Y.; Wang, B.; Hu, S.; et al. Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 2010, 107, 19114–19119. [Google Scholar] [CrossRef] [Green Version]
- Hong, S.; Song, H.R.; Lutz, K.; Kerstetter, R.A.; Michael, T.P.; McClung, C.R. Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 21211–21216. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, S.E.; Petrillo, E.; Beckwith, E.J.; Zhang, X.; Rugnone, M.L.; Hernando, C.E.; Cuevas, J.C.; Godoy Herz, M.A.; Depetris-Chauvin, A.; Simpson, C.G.; et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 2010, 468, 112–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrillo, E.; Sanchez, S.E.; Kornblihtt, A.R.; Yanovsky, M.J. Alternative splicing adds a new loop to the circadian clock. Commun. Integr. Biol. 2011, 4, 284–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Wan, Y.; Huang, G.; Wang, D.; Yu, X.; Huang, G.; Guo, J. The exosome controls alternative splicing by mediating the gene expression and assembly of the spliceosome complex. Sci. Rep. 2015, 5, 13403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, M.A.; Covington, M.F.; DiTacchio, L.; Vollmers, C.; Panda, S.; Harmer, S.L. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc. Natl. Acad. Sci. USA 2010, 107, 21623–21628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Devaiah, S.P.; Zhang, W.; Welti, R. Signaling functions of phosphatidic acid. Prog. Lipid Res. 2006, 45, 250–278. [Google Scholar] [CrossRef] [PubMed]
- Testerink, C.; Munnik, T. Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J. Exp. Bot. 2011, 62, 2349–2361. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-C.; Nusinow, D.A.; Sorkin, M.L.; Pruneda-Paz, J.; Wang, X. Interaction and Regulation Between Lipid Mediator Phosphatidic Acid and Circadian Clock Regulators. Plant Cell 2019, 31, 399–416. [Google Scholar] [CrossRef] [Green Version]
- Hirano, A.; Fu, Y.-H.; Ptáček, L.J. The intricate dance of post-translational modifications in the rhythm of life. Nat. Struct. Mol. Biol. 2016, 23, 1053–1060. [Google Scholar] [CrossRef]
- Virshup, D.M.; Eide, E.J.; Forger, D.B.; Gallego, M.; Harnish, E.V. Reversible protein phosphorylation regulates circadian rhythms. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 413–420. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Shen, Y.; Francey, L.J.; Ramanathan, C.; Sehgal, A.; Liu, A.C.; Hogenesch, J.B. The NRON complex controls circadian clock function through regulated PER and CRY nuclear translocation. Sci. Rep. 2019, 9, 11883. [Google Scholar] [CrossRef]
- Korge, S.; Maier, B.; Brüning, F.; Ehrhardt, L.; Korte, T.; Mann, M.; Herrmann, A.; Robles, M.S.; Kramer, A. The non-classical nuclear import carrier Transportin 1 modulates circadian rhythms through its effect on PER1 nuclear localization. PLoS Genet. 2018, 14, e1007189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Zhao, X.; Zhang, Y.; Tan, H.; Qiu, B.; Ma, T.; Zeng, J.; Tao, D.; Liu, Y.; Lu, Y.; et al. RAE1 promotes BMAL1 shuttling and regulates degradation and activity of CLOCK: BMAL1 heterodimer. Cell Death Dis. 2019, 10, 62. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jang, A.R.; Francey, L.J.; Sehgal, A.; Hogenesch, J.B. KPNB1 mediates PER/CRY nuclear translocation and circadian clock function. Elife 2015, 4, e08647. [Google Scholar] [CrossRef] [PubMed]
- Meier, I.; Somers, D.E. Regulation of nucleocytoplasmic trafficking in plants. Curr. Opin. Plant Biol. 2011, 14, 538–546. [Google Scholar] [CrossRef]
- Wang, W.M.; Liu, P.Q.; Xu, Y.J.; Xiao, S. Protein trafficking during plant innate immunity. J. Integr. Plant Biol. 2016, 58, 284–298. [Google Scholar] [CrossRef] [PubMed]
- Huq, E.; Tepperman, J.M.; Quail, P.H. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2000, 97, 9789–9794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khanna, R.; Kikis, E.A.; Quail, P.H. EARLY FLOWERING 4 Functions in Phytochrome B-Regulated Seedling De-Etiolation. Plant Physiol. 2003, 133, 1530–1538. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.L. ELF3 Encodes a Circadian Clock-Regulated Nuclear Protein That Functions in an Arabidopsis PHYB Signal Transduction Pathway. Plant Cell Online 2001, 13, 1293–1304. [Google Scholar] [CrossRef]
- Lu, S.X.; Knowles, S.M.; Andronis, C.; Ong, M.S.; Tobin, E.M. CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL Function Synergistically in the Circadian Clock of Arabidopsis. Plant Physiol. 2009, 150, 834–843. [Google Scholar] [CrossRef] [Green Version]
- Onai, K.; Ishiura, M. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 2005, 10, 963–972. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Kenigsbuch, D.; Sun, L.; Harel, E.; Ong, M.S.; Tobin, E.M. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 1997, 9, 491–507. [Google Scholar] [CrossRef]
- Yakir, E.; Hilman, D.; Kron, I.; Hassidim, M.; Melamed-Book, N.; Green, R.M. Posttranslational Regulation of CIRCADIAN CLOCK ASSOCIATED1 in the Circadian Oscillator of Arabidopsis. Plant Physiol. 2009, 150, 844–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClung, C.R. The Plant Circadian Oscillator. Biology 2019, 8, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrero, E.; Kolmos, E.; Bujdoso, N.; Yuan, Y.; Wang, M.; Berns, M.C.; Uhlworm, H.; Coupland, G.; Saini, R.; Jaskolski, M.; et al. EARLY FLOWERING4 Recruitment of EARLY FLOWERING3 in the Nucleus Sustains the Arabidopsis Circadian Clock. Plant Cell 2012, 24, 428–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anwer, M.U.; Boikoglou, E.; Herrero, E.; Hallstein, M.; Davis, A.M.; Velikkakam James, G.; Nagy, F.; Davis, S.J. Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock. eLife 2014, 3, e02206. [Google Scholar] [CrossRef] [Green Version]
- Kolmos, E.; Herrero, E.; Bujdoso, N.; Millar, A.J.; Tóth, R.; Gyula, P.; Nagy, F.; Davis, S.J. A Reduced-Function Allele Reveals That EARLY FLOWERING3 Repressive Action on the Circadian Clock Is Modulated by Phytochrome Signals in Arabidopsis. Plant Cell 2011, 23, 3230–3246. [Google Scholar] [CrossRef] [Green Version]
- Silva, C.S.; Nayak, A.; Lai, X.; Hutin, S.; Hugouvieux, V.; Jung, J.-H.; López-Vidriero, I.; Franco-Zorrilla, J.M.; Panigrahi, K.C.S.; Nanao, M.H.; et al. Molecular mechanisms of Evening Complex activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 6901–6909. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Han, S.; Yeom, M.; Kim, H.; Lim, J.; Cha, J.-Y.; Kim, W.-Y.; Somers, D.E.; Putterill, J.; Nam, H.G.; et al. Balanced Nucleocytosolic Partitioning Defines a Spatial Network to Coordinate Circadian Physiology in Plants. Dev. Cell 2013, 26, 73–85. [Google Scholar] [CrossRef] [Green Version]
- Nohales, M.A.; Liu, W.; Duffy, T.; Nozue, K.; Sawa, M.; Pruneda-Paz, J.L.; Maloof, J.N.; Jacobsen, S.E.; Kay, S.A. Multi-level Modulation of Light Signaling by GIGANTEA Regulates Both the Output and Pace of the Circadian Clock. Dev. Cell 2019, 49, 840–851.e8. [Google Scholar] [CrossRef]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Lim, J.; Yeom, M.; Kim, H.; Kim, J.; Wang, L.; Kim, W.Y.; Somers, D.E.; Nam, H.G. ELF4 Regulates GIGANTEA Chromatin Access through Subnuclear Sequestration. Cell Rep. 2013, 3, 671–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Geng, R.; Gallenstein, R.A.; Somers, D.E. The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 2013, 140, 4060–4069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Park, S.J.; Kim, Y.; Nam, H.G. Subcellular Localization of GIGANTEA Regulates the Timing of Leaf Senescence and Flowering in Arabidopsis. Front. Plant Sci. 2020, 11, 1814. [Google Scholar] [CrossRef]
- Kim, T.-S.; Wang, L.; Kim, Y.J.; Somers, D.E. Compensatory Mutations in GI and ZTL May Modulate Temperature Compensation in the Circadian Clock. Plant Physiol. 2020, 182, 1130–1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrero, E.; Davis, S.J. Time for a Nuclear Meeting: Protein Trafficking and Chromatin Dynamics Intersect in the Plant Circadian System. Mol. Plant 2012, 5, 554–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical Nuclear Localization Signals: Definition, Function, and Interaction with Importin α. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef] [Green Version]
- Hennessey, T.L.; Field, C.B. Evidence of multiple circadian oscillators in bean plants. J. Biol. Rhythms 1992, 7, 105–113. [Google Scholar] [CrossRef]
- Thain, S.C.; Hall, A.; Millar, A.J. Functional independence of circadian clocks that regulate plant gene expression. Curr. Biol. 2000, 10, 951–956. [Google Scholar] [CrossRef] [Green Version]
- Thain, S.C.; Murtas, G.; Lynn, J.R.; McGrath, R.B.; Millar, A.J. The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol. 2002, 130, 102–110. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Kim, Y.; Yeom, M.; Lim, J.; Nam, H.G. Age-associated circadian period changes in Arabidopsis leaves. J. Exp. Bot. 2016, 67, 2665–2673. [Google Scholar] [CrossRef] [Green Version]
- Endo, M.; Shimizu, H.; Nohales, M.A.; Araki, T.; Kay, S.A. Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 2014, 515, 419–422. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, H.; Katayama, K.; Koto, T.; Torii, K.; Araki, T.; Endo, M. Decentralized circadian clocks process thermal and photoperiodic cues in specific tissues. Nat. Plants 2015, 1, 15163. [Google Scholar] [CrossRef] [Green Version]
- Bordage, S.; Sullivan, S.; Laird, J.; Millar, A.J.; Nimmo, H.G. Organ specificity in the plant circadian system is explained by different light inputs to the shoot and root clocks. New Phytol. 2016, 212, 136–149. [Google Scholar] [CrossRef]
- Gould, P.D.; Domijan, M.; Greenwood, M.; Tokuda, I.T.; Rees, H.; Kozma-Bognar, L.; Hall, A.J.; Locke, J.C. Coordination of robust single cell rhythms in the Arabidopsis circadian clock via spatial waves of gene expression. Elife 2018, 7, e31700. [Google Scholar] [CrossRef]
- James, A.B.; Monreal, J.A.; Nimmo, G.A.; Kelly, C.L.; Herzyk, P.; Jenkins, G.I.; Nimmo, H.G. The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science 2008, 322, 1832–1835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, K.; Araki, T.; Endo, M. Oscillator networks with tissue-specific circadian clocks in plants. Semin Cell Dev. Biol. 2018, 83, 78–85. [Google Scholar] [CrossRef]
- Takahashi, N.; Hirata, Y.; Aihara, K.; Mas, P. A hierarchical multi-oscillator network orchestrates the Arabidopsis circadian system. Cell 2015, 163, 148–159. [Google Scholar] [CrossRef] [Green Version]
- Barclay, J.L.; Tsang, A.H.; Oster, H. Interaction of central and peripheral clocks in physiological regulation. Prog. Brain Res. 2012, 199, 163–181. [Google Scholar] [CrossRef]
- Mohawk, J.A.; Green, C.B.; Takahashi, J.S. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012, 35, 445–462. [Google Scholar] [CrossRef] [Green Version]
- Greenwood, M.; Domijan, M.; Gould, P.D.; Hall, A.J.W.; Locke, J.C.W. Coordinated circadian timing through the integration of local inputs in Arabidopsis thaliana. PLoS Biol. 2019, 17, e3000407. [Google Scholar] [CrossRef] [Green Version]
- Nimmo, H.G. Entrainment of Arabidopsis roots to the light:dark cycle by light piping. Plant Cell Environ. 2018, 41, 1742–1748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endo, M. Tissue-specific circadian clocks in plants. Curr. Opin. Plant Biol. 2016, 29, 44–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez, S.E.; Kay, S.A. The Plant Circadian Clock: From a Simple Timekeeper to a Complex Developmental Manager. Cold Spring Harb. Perspect. Biol. 2016, 8, a027748. [Google Scholar] [CrossRef] [PubMed]
- Haydon, M.J.; Mielczarek, O.; Frank, A.; Román, Á.; Webb, A.A.R. Sucrose and Ethylene Signaling Interact to Modulate the Circadian Clock. Plant Physiol. 2017, 175, 947–958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.W.; Takahashi, N.; Hirata, Y.; Ronald, J.; Porco, S.; Davis, S.J.; Nusinow, D.A.; Kay, S.A.; Mas, P. A mobile ELF4 delivers circadian temperature information from shoots to roots. Nat. Plants 2020, 6, 416–426. [Google Scholar] [CrossRef]
- O’Neill, J.S.; Maywood, E.S.; Hastings, M.H. Cellular mechanisms of circadian pacemaking: Beyond transcriptional loops. Handb. Exp. Pharmacol. 2013, 67–103. [Google Scholar] [CrossRef]
- Hastings, M.H.; Maywood, E.S.; O’Neill, J.S. Cellular circadian pacemaking and the role of cytosolic rhythms. Curr. Biol. 2008, 18, R805–R815. [Google Scholar] [CrossRef] [Green Version]
- Reddy, A.B.; Rey, G. Metabolic and nontranscriptional circadian clocks: Eukaryotes. Annu. Rev. Biochem. 2014, 83, 165–189. [Google Scholar] [CrossRef] [Green Version]
- Martí Ruiz, M.C.; Hubbard, K.E.; Gardner, M.J.; Jung, H.J.; Aubry, S.; Hotta, C.T.; Mohd-Noh, N.I.; Robertson, F.C.; Hearn, T.J.; Tsai, Y.C.; et al. Circadian oscillations of cytosolic free calcium regulate the Arabidopsis circadian clock. Nat. Plants 2018, 4, 690–698. [Google Scholar] [CrossRef]
- Johnson, C.H.; Stewart, P.L.; Egli, M. The cyanobacterial circadian system: From biophysics to bioevolution. Annu. Rev. Biophys. 2011, 40, 143–167. [Google Scholar] [CrossRef] [Green Version]
- Ode, K.L.; Ueda, H.R. Design Principles of Phosphorylation-Dependent Timekeeping in Eukaryotic Circadian Clocks. Cold Spring Harb. Perspect. Biol. 2018, 10, a028357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorkin, M.L.; Nusinow, D.A. Time Will Tell: Intercellular Communication in the Plant Clock. Trends Plant Sci. 2021. [Google Scholar] [CrossRef] [PubMed]
- Labib, M.; Kelley, S.O. Single-cell analysis targeting the proteome. Nat. Rev. Chem. 2020, 4, 143–158. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, J.; Kim, Y.J.; Somers, D.E. Post-Translational Mechanisms of Plant Circadian Regulation. Genes 2021, 12, 325. https://doi.org/10.3390/genes12030325
Yan J, Kim YJ, Somers DE. Post-Translational Mechanisms of Plant Circadian Regulation. Genes. 2021; 12(3):325. https://doi.org/10.3390/genes12030325
Chicago/Turabian StyleYan, Jiapei, Yeon Jeong Kim, and David E. Somers. 2021. "Post-Translational Mechanisms of Plant Circadian Regulation" Genes 12, no. 3: 325. https://doi.org/10.3390/genes12030325
APA StyleYan, J., Kim, Y. J., & Somers, D. E. (2021). Post-Translational Mechanisms of Plant Circadian Regulation. Genes, 12(3), 325. https://doi.org/10.3390/genes12030325






