Circadian Clock Components Offer Targets for Crop Domestication and Improvement
Abstract
:1. Introduction
1.1. Plant Domestication
1.2. The Plant Circadian Clock
1.2.1. The Plant Circadian Clock Consists of Multiple Interlocked Feedback Loops
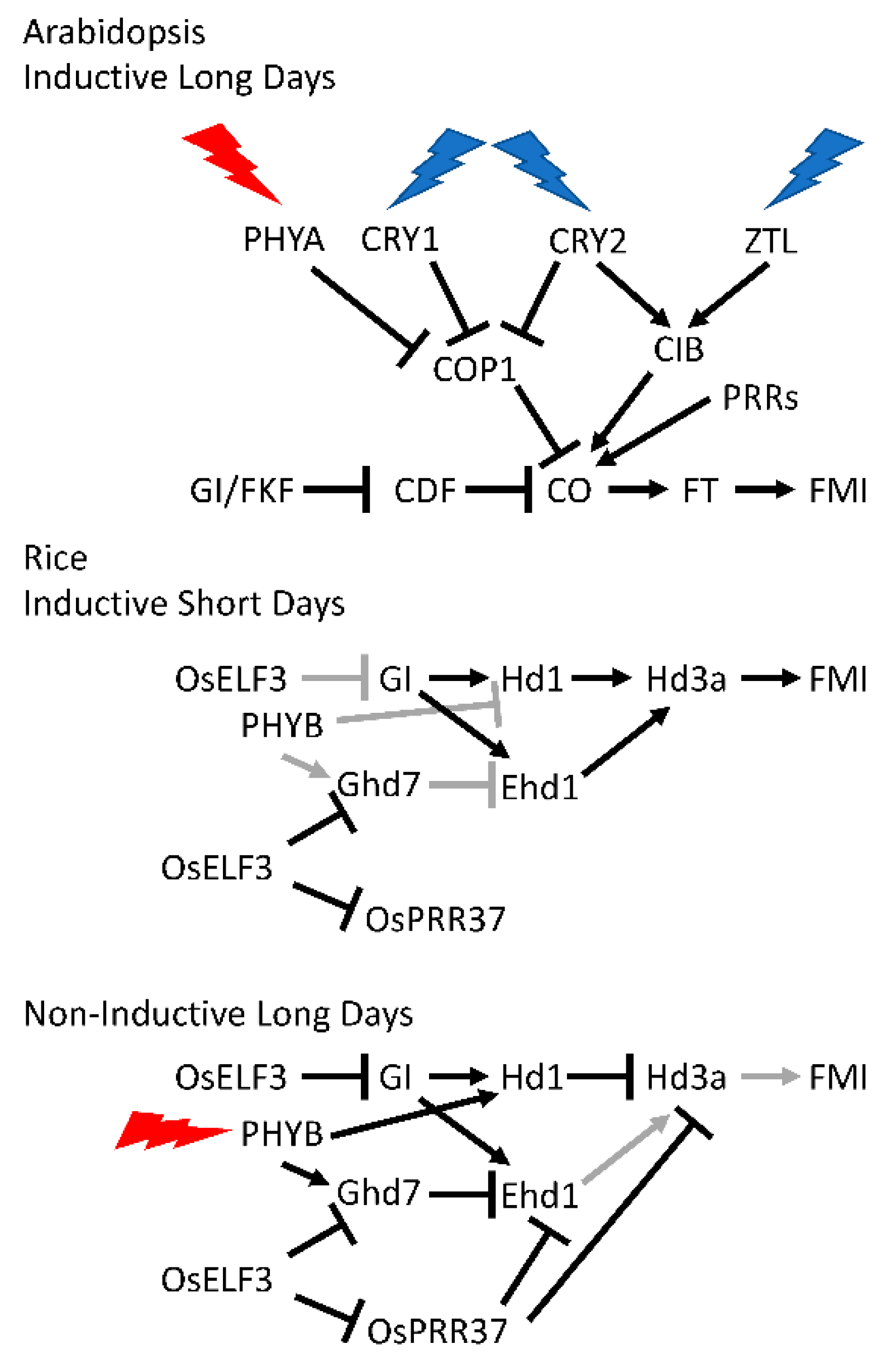
1.2.2. Photoperiodic Induction of Flowering
2. Circadian Clock Components as Domestication and Crop Improvement Loci
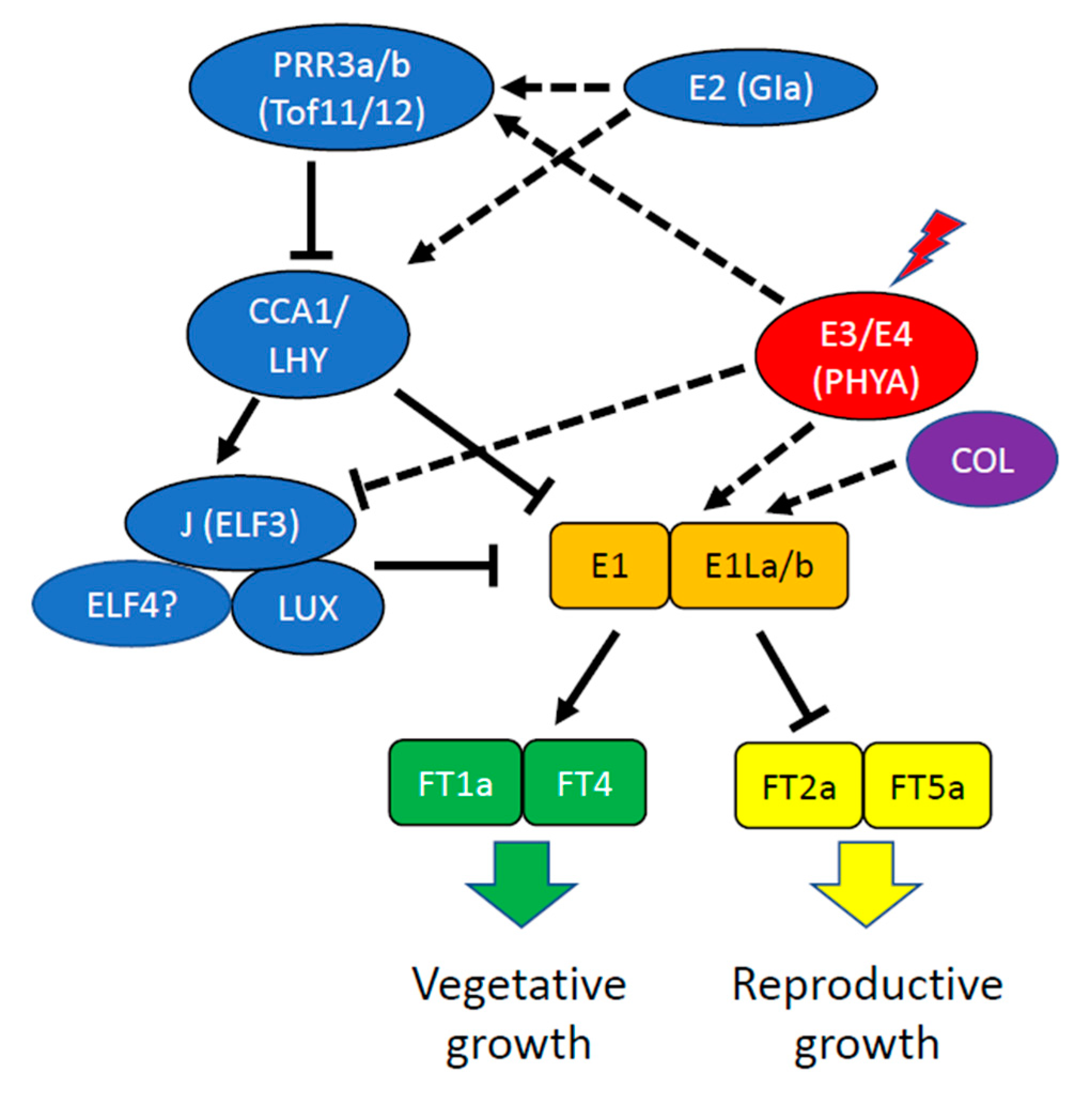
2.1. Soybean
2.2. Tomato
2.3. Sugar Beet
2.4. Monocot Clocks
2.5. Rice
2.6. Barley
3. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The molecular genetics of crop domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina, J.; Sikora, M.; Garud, N.; Flowers, J.M.; Rubinstein, S.; Reynolds, A.; Huang, P.; Jackson, S.; Schaal, B.A.; Bustamante, C.D.; et al. Molecular evidence for a single evolutionary origin of domesticated rice. Proc. Natl. Acad. Sci. USA 2011, 108, 8351–8356. [Google Scholar] [CrossRef] [Green Version]
- Matsuoka, Y.; Vigouroux, Y.; Goodman, M.M.; Sanchez, G.J.; Buckler, E.; Doebley, J. A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA 2002, 99, 6080–6084. [Google Scholar] [CrossRef] [Green Version]
- Purugganan, M.D.; Fuller, D.Q. The nature of selection during plant domestication. Nature 2009, 457, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Purugganan, M.D. Evolutionary insights into the nature of plant domestication. Curr. Biol. 2019, 29, R705–R714. [Google Scholar] [CrossRef] [Green Version]
- Hammer, K. Das Domestikationssyndrom. Kulturpflanze 1984, 32, 11–34. [Google Scholar] [CrossRef]
- Eyre-Walker, A.; Gaut, R.L.; Hilton, H.; Feldman, D.L.; Gaut, B.S. Investigation of the bottleneck leading to the domestication of maize. Proc. Natl. Acad. Sci. USA 1998, 95, 4441–4446. [Google Scholar] [CrossRef] [Green Version]
- Wright, S.I.; Bi, I.V.; Schroeder, S.G.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S. The effects of artificial selection on the maize genome. Science 2005, 308, 1310–1314. [Google Scholar] [CrossRef]
- Meyer, R.S.; Purugganan, M.D. Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 2013, 14, 840–852. [Google Scholar] [CrossRef] [PubMed]
- Bendix, C.; Marshall, C.M.; Harmon, F.G. Circadian clock genes universally control key agricultural traits. Mol. Plant 2015, 8, 1135–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yerushalmi, S.; Green, R.M. Evidence for the adaptive significance of circadian rhythms. Ecol. Lett. 2009, 12, 970–981. [Google Scholar] [CrossRef]
- Greenham, K.; McClung, C.R. Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 2015, 16, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.J. The intracellular dynamics of circadian clocks reach for the light of ecology and evolution. Annu. Rev. Plant Biol. 2016, 67, 595–618. [Google Scholar] [CrossRef]
- Edgar, R.S.; Green, E.W.; Zhao, Y.; van Ooijen, G.; Olmedo, M.; Qin, X.; Xu, Y.; Pan, M.; Valekunja, U.K.; Feeney, K.A.; et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature 2012, 485, 459–464. [Google Scholar] [CrossRef] [Green Version]
- Young, M.W.; Kay, S.A. Time zones: A comparative genetics of circadian clocks. Nat. Rev. Genet. 2001, 2, 702–715. [Google Scholar] [CrossRef]
- Lou, P.; Wu, J.; Cheng, F.; Cressman, L.G.; Wang, X.; McClung, C.R. Preferential retention of circadian clock genes during diploidization following whole genome triplication in Brassica rapa. Plant Cell 2012, 24, 2415–2426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClung, C.R. The plant circadian oscillator. Biology 2019, 8, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamichi, N. The transcriptional network in the Arabidopsis circadian clock system. Genes 2020, 11, 1284. [Google Scholar] [CrossRef]
- James, A.B.; Syed, N.H.; Bordage, S.; Marshall, J.; Nimmo, G.A.; Jenkins, G.I.; Herzy, P.; Brown, J.W.S.; Nimmo, H.G. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 2012, 24, 961–981. [Google Scholar] [CrossRef] [Green Version]
- Filichkin, S.A.; Priest, H.D.; Givan, S.A.; Shen, R.; Bryant, D.W.; Fox, S.E.; Wong, W.-K.; Mockler, T.C. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010, 20, 45–58. [Google Scholar] [CrossRef] [Green Version]
- Romanowski, A.; Schlaen, R.G.; Perez-Santangelo, S.; Mancini, E.; Yanovsky, M.J. Global transcriptome analysis reveals circadian control of splicing events in Arabidopsis thaliana. Plant J. 2020, 103, 889–902. [Google Scholar] [CrossRef]
- Sanchez, S.E.; Petrillo, E.; Beckwith, E.J.; Zhang, X.; Rugnone, M.L.; Hernando, C.E.; Cuevas, J.C.; Godoy Herz, M.A.; Depetris-Chauvin, A.; Simpson, C.G.; et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 2010, 468, 112–116. [Google Scholar] [CrossRef] [Green Version]
- Hong, S.; Song, H.-R.; Lutz, K.; Kerstetter, R.A.; Michael, T.P.; McClung, C.R. Type II Protein Arginine Methyltransferase PRMT5 is required for circadian period determination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 21211–21216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, M.A.; Williams, B.A.; McNicol, J.; Simpson, C.G.; Brown, J.W.S.; Harmer, S.L. Mutation of Arabidopsis SPLICEOSOMAL TIMEKEEPER LOCUS1 causes circadian clock defects. Plant Cell 2012, 24, 4066–4082. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wu, F.; Xie, Q.; Wang, H.; Wang, Y.; Yue, Y.; Gahura, O.; Ma, S.; Liu, L.; Cao, Y.; et al. SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 2012, 24, 3278–3295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, P.J.; Mas, P. Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell 2014, 26, 79–87. [Google Scholar] [CrossRef] [Green Version]
- Mateos, J.L.; de Leone, M.J.; Torchio, J.; Reichel, M.; Staiger, D. Beyond transcription—Fine tuning of circadian timekeeping by posttranscriptional regulation. Genes 2018, 9, 616. [Google Scholar] [CrossRef] [Green Version]
- Más, P.; Kim, W.-Y.; Somers, D.E.; Kay, S.A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 2003, 426, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Wang, L.; Han, L.; Suh, S.S.; Salomé, P.A.; McClung, C.R.; Somers, D.E. Post-translational regulation of the circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 2008, 283, 23073–23083. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Fujiwara, S.; Somers, D.E. PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 2010, 29, 1903–1915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.-s.; Kim, W.Y.; Fujiwara, S.; Kim, J.; Cha, J.-Y.; Park, J.H.; Lee, S.Y.; Somers, D.E. HSP90 functions in the circadian clock through stabilization of the client F-box protein ZEITLUPE. Proc. Natl. Acad. Sci. USA 2011, 108, 16843–16848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Geng, R.; Gallenstein, R.A.; Somers, D.E. The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 2013, 140, 4060–4069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.; Han, S.; Yeom, M.; Kim, H.; Lim, J.; Cha, J.-Y.; Kim, W.-Y.; Somers, D.E.; Putterill, J.; Nam, H.G.; et al. Balanced nucleocytosolic partitioning defines a spatial network to coordinate circadian physiology in plants. Dev. Cell 2013, 26, 73–85. [Google Scholar] [CrossRef] [Green Version]
- Cha, J.-Y.; Kim, J.; Kim, T.-S.; Zeng, Q.; Wang, L.; Lee, S.Y.; Kim, W.-Y.; Somers, D.E. GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat. Commun. 2017, 8, 3. [Google Scholar] [CrossRef] [Green Version]
- Romanowski, A.; Yanovsky, M.J. Circadian rhythms and post-transcriptional regulation in higher plants. Front. Plant Sci. 2015, 6, 437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Ma, D.; Lu, S.X.; Hu, X.; Huang, R.; Liang, T.; Xu, T.; Tobin, E.M.; Liu, H. Blue light- and low temperature-regulated COR27 and COR28 play roles in the Arabidopsis circadian clock. Plant Cell 2016, 28, 2755–2769. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Cui, X.; Zhao, C.; Shi, L.; Zhang, G.; Sun, F.; Cao, X.; Yuan, L.; Xie, Q.; Xu, X. COR27 and COR28 encode nighttime repressors integrating Arabidopsis circadian clock and cold response. J. Int. Plant Biol. 2017, 59, 78–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millar, A.J.; Carré, I.A.; Strayer, C.A.; Chua, N.-H.; Kay, S.A. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 1995, 267, 1161–1163. [Google Scholar] [CrossRef] [PubMed]
- Strayer, C.; Oyama, T.; Schultz, T.F.; Raman, R.; Somers, D.E.; Más, P.; Panda, S.; Kreps, J.A.; Kay, S.A. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 2000, 289, 768–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Más, P.; Alabadí, D.; Yanovsky, M.J.; Oyama, T.; Kay, S.A. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 2003, 15, 223–236. [Google Scholar] [CrossRef] [Green Version]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef] [Green Version]
- Schaffer, R.; Ramsay, N.; Samach, A.; Corden, S.; Putterill, J.; Carré, I.A.; Coupland, G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 1998, 93, 1219–1229. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Tobin, E.M. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 1998, 93, 1207–1217. [Google Scholar] [CrossRef] [Green Version]
- Alabadí, D.; Yanovsky, M.J.; Más, P.; Harmer, S.L.; Kay, S.A. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 2002, 12, 757–761. [Google Scholar] [CrossRef] [Green Version]
- Mizoguchi, T.; Wheatley, K.; Hanzawa, Y.; Wright, L.; Mizoguchi, M.; Song, H.-R.; Carré, I.A.; Coupland, G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2002, 2, 629–641. [Google Scholar] [CrossRef] [Green Version]
- Alabadí, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Más, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef] [PubMed]
- Matsushika, A.; Makino, S.; Kojima, M.; Mizuno, T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 2000, 41, 1002–1012. [Google Scholar] [CrossRef] [Green Version]
- Nakamichi, N.; Kita, M.; Ito, S.; Sato, E.; Yamashino, T.; Mizuno, T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 686–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamichi, N.; Kiba, T.; Henriques, R.; Mizuno, T.; Chua, N.-H.; Sakakibara, H. PSEUDO-RESPONSE REGULATORS 9, 7 and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 2010, 22, 594–605. [Google Scholar] [CrossRef] [Green Version]
- Pruneda-Paz, J.L.; Breton, G.; Para, A.; Kay, S.A. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 2009, 323, 1481–1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Kim, J.; Somers, D.E. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA 2013, 110, 761–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nusinow, D.A.; Helfer, A.; Hamilton, E.E.; King, J.J.; Imaizumi, T.; Schultz, T.F.; Farre, E.M.; Kay, S.A. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 2011, 475, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Herrero, E.; Kolmos, E.; Bujdoso, N.; Yuan, Y.; Wang, M.; Berns, M.C.; Uhlworm, H.; Coupland, G.; Saini, R.; Jaskolski, M.; et al. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 2012, 24, 428–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, S.; Wei, X.; Pei, L.; Thompson, R.L.; Liu, Y.; Heard, J.E.; Ruff, T.G.; Beachy, R.N. BROTHER OF LUX ARRHYTHMO is a component of the Arabidopsis circadian clock. Plant Cell 2011, 23, 961–972. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.-F.; Tsai, H.-L.; Joanito, I.; Wu, Y.-C.; Chang, C.-W.; Li, Y.-H.; Wang, Y.; Hong, J.C.; Chu, J.-W.; Hsu, C.-P.; et al. LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat. Commun. 2016, 7, 13181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mockler, T.C.; Michael, T.P.; Priest, H.D.; Shen, R.; Sullivan, C.M.; Givan, S.A.; McEntee, C.; Kay, S.A.; Chory, J. The Diurnal Project: Diurnal and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Rawat, R.; Takahashi, N.; Hsu, P.Y.; Jones, M.A.; Schwartz, J.; Salemi, M.R.; Phinney, B.S.; Harmer, S.L. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 2011, 7, e1001350. [Google Scholar] [CrossRef]
- Hsu, P.Y.; Devisetty, U.K.; Harmer, S.L. Accurate timekeeping is controlled by a cycling activator in Arabidopsis. ELife 2013, 2, e00473. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Wang, P.; Liu, X.; Yuan, L.; Wang, L.; Zhang, C.; Li, Y.; Xing, H.; Zhi, L.; Yue, Z.; et al. LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 2014, 26, 2843–2857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- José de Leone, M.; Esteban, H.C.; Romanowski, A.; García-Hourquet, M.; Careno, D.; Casal, J.; Rugnone, M.; Mora-García, S.; Yanovsky, M.J. The LNK gene family: At the crossroad between light signaling and the circadian clock. Genes 2019, 10, 2. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Siddiqui, H.; Teng, Y.; Lin, R.; Wan, X.-y.; Li, J.; Lau, O.-S.; Ouyang, X.; Dai, M.; Wan, J.; et al. Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol. 2011, 13, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ma, M.; Li, G.; Yuan, L.; Xie, Y.; Wei, H.; Ma, X.; Li, Q.; Devlin, P.F.; Xu, X.; et al. Transcription factors FHY3 and FAR1 regulate light-induced CIRCADIAN CLOCK ASSOCIATED1 gene expression in Arabidopsis. Plant Cell 2020, 32, 1464–1478. [Google Scholar] [CrossRef]
- Zhu, W.; Zhou, H.; Lin, F.; Zhao, X.; Jiang, Y.; Xu, D.; Deng, X.W. COLD-REGULATED GENE27 integrates signals from light and the circadian clock to promote hypocotyl growth in Arabidopsis. Plant Cell 2020, 32, 3155–3169. [Google Scholar] [CrossRef] [PubMed]
- Perales, M.; Más, P. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 2007, 19, 2111–2123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.; Mas, P.; Seo, P.J. The EC-HDA9 complex rhythmically regulates histone acetylation at the TOC1 promoter in Arabidopsis. Commun. Biol. 2019, 2. [Google Scholar] [CrossRef]
- Park, H.J.; Baek, D.; Cha, J.-Y.; Liao, X.; Kang, S.-H.; McClung, C.R.; Lee, S.Y.; Yun, D.-J.; Kim, W.-Y. HOS15 interacts with the histone deactetylase HDA9 and the Evening Complex to epigenetically regulate the floral activator GIGANTEA. Plant Cell 2019, 31, 37–51. [Google Scholar] [CrossRef] [Green Version]
- Maric, A.; Mas, P. Chromatin dynamics and transcriptional control of circadian rhythms in Arabidopsis. Genes 2020, 11, 1170. [Google Scholar] [CrossRef]
- Kende, H.; Zeevart, J.A.D. Anton Lang. In Biographical Memoirs; The National Academies Press: Washington, DC, USA, 1998; Volume 74, pp. 49–61. [Google Scholar]
- Romera-Branchat, M.; Andres, F.; Coupland, G. Flowering responses to seasonal cues: What’s new? Curr. Op. Plant Biol. 2014, 21, 120–127. [Google Scholar] [CrossRef]
- Song, Y.H.; Shim, J.S.; Kinmonth-Schultz, H.A.; Imaizumi, T. Photoperiodic flowering: Time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 2015, 66, 441–464. [Google Scholar] [CrossRef] [Green Version]
- Zuo, Z.; Liu, H.; Liu, B.; Liu, X.; Lin, C. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 2011, 21, 841–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarid-Krebs, L.; Panigrahi, K.C.S.; Fornara, F.; Takahashi, Y.; Hayama, R.; Jang, S.; Tilmes, V.; Valverde, F.; Coupland, G. Phosphorylation of CONSTANS and its COP1-dependent degradation during photoperiodic flowering of Arabidopsis. Plant J. 2015, 84, 451–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayama, R.; Sarid-Krebs, L.; Richter, R.; Fernandez, V.; Jang, S.; Coupland, G. PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J. 2017, 36, 904–918. [Google Scholar] [CrossRef]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1538. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Li, X.; Li, K.; Liu, H.; Lin, C. Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 2013, 9, e1003861. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; Zhu, S.; Cui, S.; Hou, H.; Wu, H.; Hao, B.; Cai, L.; Xu, Z.; Liu, L.; Jiang, L.; et al. Transcriptional and post-transcriptional regulation of heading date in rice. New Phytol. 2021, 229. [Google Scholar] [CrossRef]
- Izawa, T.; Oikawa, T.; Sugiyama, N.; Tanisaka, T.; Yano, M.; Shimamoto, K. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 2002, 16, 2006–2020. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, R.; Aoki, M.; Kurotani, K.-i.; Yokoi, S.; Shinomura, T.; Takano, M.; Shimamoto, K. Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol. Genet. Genom. 2011, 285, 461–470. [Google Scholar] [CrossRef]
- Doi, K.; Izawa, T.; Fuse, T.; Yamanouchi, U.; Kubo, T.; Shimatani, Z.; Yano, M.; Yoshimura, A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004, 18, 926–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Itoh, H.; Nonoue, Y.; Yano, M.; Izawa, T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 2010, 42, 635–638. [Google Scholar] [CrossRef] [PubMed]
- Osugi, A.; Itoh, H.; Ikeda-Kawakatsu, K.; Takano, M.; Izawa, T. Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol. 2011, 157, 1128–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsubara, K.; Ogiso-Tanaka, E.; Hori, K.; Ebana, K.; Ando, T.; Yano, M. Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol. 2012, 53, 709–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, H.; Ogiso-Tanaka, E.; Okumoto, Y.; Yoshitake, Y.; Izumi, H.; Yokoo, T.; Matsubara, K.; Hori, K.; Yano, M.; Inoue, H.; et al. Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions. Plant Cell Physiol. 2012, 53, 717–728. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Huang, X.; Ouyang, X.; Chen, W.; Du, A.; Zhu, L.; Wang, S.; Deng, X.W.; Li, S. OsELF3-1, an ortholog of Arabidopsis EARLY FLOWERING 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS ONE 2012, 7, e43705. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Qu, X.; Zhou, Y.; Song, G.; Abiri, N.; Xiao, Y.; Liang, F.; Jiang, D.; Hu, Z.; Yang, D. OsPRR37 confers an expanded regulation of the diurnal rhythms of the transcriptome and photoperiodic flowering pathways in rice. Plant Cell Environ. 2018, 41, 630–645. [Google Scholar] [CrossRef] [PubMed]
- Hut, R.A.; Paolucci, S.; Dor, R.; Kyriacou, C.P.; Daan, S. Latitudinal clines: An evolutionary view on biological rhythms. Proc. R. Soc. B Biol. Sci. 2013, 280, 20130433. [Google Scholar] [CrossRef] [Green Version]
- Majercak, J.; Sidote, D.; Hardin, P.E.; Edery, I. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 1999, 24, 219–230. [Google Scholar] [CrossRef] [Green Version]
- Costa, R.; Kyriacou, C.P. Functional and evolutionary implications of natural variation in clock genes. Curr. Opin. Neurobiol. 1998, 8, 659–664. [Google Scholar] [CrossRef]
- Rosato, E.; Piccin, A.; Kyriacou, C.P. Circadian rhythms: From behaviour to molecules. BioEssays 1997, 19, 1075–1082. [Google Scholar] [CrossRef]
- Sawyer, L.A.; Hennessy, J.M.; Peixoto, A.A.; Rosato, E.; Parkinson, H.; Costa, R.; Kyriacou, C.P. Natural variation in a Drosophila clock gene and temperature compensation. Science 1997, 278, 2117–2120. [Google Scholar] [CrossRef]
- Johnsen, A.; Fidler, A.E.; Kuhn, S.; Carter, K.L.; Hoffmann, A.; Barr, I.R.; Biard, C.; Charmantier, A.; Eens, M.; Korsten, P.; et al. Avian Clock gene polymorphism: Evidence for a latitudinal cline in allele frequencies. Mol. Ecol. 2007, 16, 4867–4880. [Google Scholar] [CrossRef]
- O’Malley, K.G.; Banks, M.A. A latitudinal cline in the Chinook salmon (Oncorhynchus tshawytscha) Clock gene: Evidence for selection on PolyQ length variants. Proc. R. Soc. Lond. B 2008, 275, 2813–2822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michael, T.P.; Salomé, P.A.; Yu, H.J.; Spencer, T.R.; Sharp, E.L.; Alonso, J.M.; Ecker, J.R.; McClung, C.R. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 2003, 302, 1049–1053. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.D.; Lynn, J.R.; Gyula, P.; Nagy, F.; Millar, A.J. Natural allelic variation in the temperature compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics 2005, 170, 387–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenham, K.; Lou, P.; Puzey, J.R.; Kumar, G.; Arnevik, C.; Farid, H.; Willis, J.H.; McClung, C.R. Geographic variation of plant circadian clock function in natural and agricultural settings. J. Biol. Rhythms 2017, 32, 26–34. [Google Scholar] [CrossRef]
- Kim, M.Y.; Van, K.; Kang, Y.J.; Kim, K.; Lee, S.-H. Tracing soybean domestication history: From nucleotide to genome. Breed. Sci. 2012, 61, 445–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedivy, E.J.; Wu, F.; Hanzawa, Y. Soybean domestication: The origin, genetic architecture and molecular bases. New Phytol. 2017, 214, 539–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, F.; Liu, B.; Xia, Z.; Sato, S.; Kim, B.M.; Watanabe, S.; Yamada, T.; Tabata, S.; Kanazawa, A.; Harada, K.; et al. Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 2010, 154, 1220–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cober, E.R.; Morrison, M.J. Regulation of seed yield and agronomic characters by photoperiod sensitivity and growth habit genes in soybean. Theor. Appl. Genet. 2010, 120, 1005–1012. [Google Scholar] [CrossRef]
- Kim, M.Y.; Shin, J.H.; Kang, Y.J.; Shim, S.R.; Lee, S.-H. Divergence of flowering genes in soybean. J. Biosci. 2012, 37, 857–870. [Google Scholar] [CrossRef]
- Lin, X.; Liu, B.; Weller, J.L.; Abe, J.; Kong, F. Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J. Integr. Plant Biol. 2021. [Google Scholar] [CrossRef]
- Cao, D.; Takeshima, R.; Zhao, C.; Liu, B.; Abe, J.; Kong, F. Molecular mechanisms of flowering under long days and stem growth habit in soybean. J. Exp. Bot. 2017, 68, 1873–1884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samanfar, B.; Molnar, S.J.; Charette, M.; Schoenrock, A.; Dehne, F.; Golshani, A.; Belzile, F.; Cober, E.R. Mapping and identification of a potential candidate gene for a novel maturity locus, E10, in soybean. Theor. Appl. Genet. 2017, 130, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Nan, H.; Chen, L.; Fang, C.; Zhang, H.; Su, T.; Li, S.; Cheng, Q.; Dong, L.; Liu, B.; et al. A new dominant locus, E11, controls early flowering time and maturity in soybean. Mol. Breed. 2019, 39, 70. [Google Scholar] [CrossRef]
- Watanabe, S.; Xia, Z.; Hideshima, R.; Tsubokura, Y.; Sato, S.; Yamanaka, N.; Takahashi, R.; Anai, T.; Tabata, S.; Kitamura, K.; et al. A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene Is Involved in soybean maturity and flowering. Genetics 2011, 188, 395–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, S.; Hideshima, R.; Xia, Z.; Tsubokura, Y.; Sato, S.; Nakamoto, Y.; Yamanaka, N.; Takahashi, R.; shimoto, M.; Anai, T.; et al. Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 2009, 182, 1251–1262. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Kanazawa, A.; Matsumura, H.; Takahashi, R.; Harada, K.; Abe, J. Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 2008, 180, 995–1007. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Kim, M.; Kang, Y.J.; Van, K.; Lee, Y.-H.; Srinives, P.; Yuan, D.; Lee, S.-H. QTL identification of flowering time at three different latitudes reveals homeologous genomic regions that control flowering in soybean. Theor. Appl. Genet. 2011, 123, 545–553. [Google Scholar] [CrossRef]
- Jiang, B.; Nan, H.; Gao, Y.; Tang, L.; Yue, Y.; Lu, S.; Ma, L.; Cao, D.; Sun, S.; Wang, J.; et al. Allelic combinations of soybean maturity loci E1, E2, E3 and E4 result in diversity of maturity and adaptation to different latitudes. PLoS ONE 2014, 9, e106042. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Jiang, B.; Wu, C.; Lu, W.; Hou, W.; Sun, S.; Yan, H.; Han, T. Maturity group classification and maturity locus genotyping of early-maturing soybean varieties from high-latitude cold regions. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Tsubokura, Y.; Watanabe, S.; Xia, Z.; Kanamori, H.; Yamagata, H.; Kaga, A.; Katayose, Y.; Abe, J.; Ishimoto, M.; Harada, K. Natural variation in the genes responsible for maturity loci E1, E2, E3 and E4 in soybean. Ann. Bot. 2014, 113, 429–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, H.; Lu, S.; Wang, Y.; Chen, X.; Ren, H.; Yang, J.; Cheng, W.; Zong, C.; Gu, H.; Qiu, H.; et al. Allelic variations at four major maturity E genes and transcriptional abundance of the E1 gene are associated with flowering time and maturity of soybean cultivars. PLoS ONE 2014, 9, e97636. [Google Scholar] [CrossRef] [Green Version]
- Zhai, H.; Lu, S.; Wu, H.; Zhang, Y.; Zhang, X.; Yang, J.; Wang, Y.; Yang, G.; Qiu, H.; Cui, T.; et al. Diurnal expression pattern, allelic variation, and association analysis reveal functional features of the E1 gene in control of photoperiodic flowering in soybean. PLoS ONE 2015, 10, e0135909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Li, Y.; Wang, J.; Srinives, P.; Nan, H.; Cao, D.; Wang, Y.; Li, J.; Li, X.; Fang, C.; et al. QTL mapping for flowering time in different latitude in soybean. Euphytica 2015, 206, 725–736. [Google Scholar] [CrossRef]
- Kurasch, A.K.; Hahn, V.; Leiser, W.L.; Vollmann, J.; Schori, A.; Bétrix, C.A.; Mayr, B.; Winkler, J.; Mechtler, K.; Aper, J.; et al. Identification of mega-environments in Europe and effect of allelic variation at maturity E loci on adaptation of European soybean. Plant Cell Environ. 2017, 40, 765–778. [Google Scholar] [CrossRef]
- Li, J.; Wang, X.; Song, W.; Huang, X.; Zhou, J.; Zeng, H.; Sun, S.; Jia, H.; Li, W.; Zhou, X.; et al. Genetic variation of maturity groups and four E genes in the Chinese soybean mini core collection. PLoS ONE 2017, 12, e0172106. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Takeshima, R.; Zhu, J.; Xu, M.; Sato, M.; Watanabe, S.; Kanazawa, A.; Liu, B.; Kong, F.; Yamada, T.; et al. A recessive allele for delayed flowering at the soybean maturity locus E9 is a leaky allele of FT2a, a FLOWERING LOCUS T ortholog. BMC Plant Biol. 2016, 16, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Y.; Chen, L.; Liu, X.; Guo, C.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol. J. 2018, 16, 176–185. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Wang, L.; Chen, L.; Wu, T.; Liu, L.; Sun, S.; Wu, C.; Yao, W.; Jiang, B.; Yuan, S.; et al. Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol. J. 2020, 18, 298–309. [Google Scholar] [CrossRef] [Green Version]
- Zhai, H.; Lü, S.; Liang, S.; Wu, H.; Zhang, X.; Liu, B.; Kong, F.; Yuan, X.; Li, J.; Xia, Z. GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PLoS ONE 2014, 9, e89030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Zhou, Z.; Liu, Y.; Liu, T.; Li, Q.; Ji, Y.; Li, C.; Fang, C.; Wang, M.; Wu, M.; et al. Functional evolution of phosphatidylethanolamine binding proteins in soybean and Arabidopsis. Plant Cell 2015, 27, 323–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickland, D.P.; Hanzawa, Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: Functional evolution and molecular mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Jiang, B.; Ma, L.; Zhang, S.; Zhai, H.; Xu, X.; Hou, W.; Xia, Z.; Wu, C.; Sun, S.; et al. Functional diversification of Flowering Locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytol. 2018, 217, 1335–1345. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Takeshima, R.; Harigai, K.; Xu, M.; Kong, F.; Liu, B.; Kanazawa, A.; Yamada, T.; Abe, J. Loss of function of the E1-Like-b ene associates with early flowering under long-day conditions in soybean. Front. Plant Sci. 2019, 9, 1867. [Google Scholar] [CrossRef]
- Xu, M.; Yamagishi, N.; Zhao, C.; Takeshima, R.; Kasai, M.; Watanabe, S.; Kanazawa, A.; Yoshikawa, N.; Liu, B.; Yamada, T.; et al. Soybean-specific E1 family of floral repressors controls night-break responses through down-regulation of FLOWERING LOCUS T orthologs. Plant Physiol. 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeshima, R.; Hayashi, T.; Zhu, J.; Zhao, C.; Xu, M.; Yamaguchi, N.; Sayama, T.; Ishimoto, M.; Kong, L.; Shi, X.; et al. A soybean quantitative trait locus that promotes flowering under long days is identified as FT5a, a FLOWERING LOCUS T ortholog. J. Exp. Bot. 2016, 67, 5247–5258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koornneef, M.; Hanhart, C.J.; van der Veen, J.H. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 1991, 229, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Dunford, R.P.; Griffiths, S.; Christodoulou, V.; Laurie, D.A. Characterisation of a barley (Hordeum vulgare L.) homologue of the Arabidopsis flowering time regulator GIGANTEA. Theor. Appl. Genet. 2005, 110, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Y.; Liu, M.S.; Li, J.R.; Guan, C.M.; Zhang, X.S. The wheat TaGI1, involved in photoperiodic flowering, encodes an Arabidopsis GI ortholog. Plant Mol. Biol. 2005, 58, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Hecht, V.; Knowles, C.L.; Schoor, J.K.V.; Liew, L.C.; Jones, S.E.; Lambert, M.J.M.; Weller, J.L. Pea LATE BLOOMER1 Is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 2007, 144, 648–661. [Google Scholar] [CrossRef] [Green Version]
- Xie, Q.; Lou, P.; Hermand, V.; Aman, R.; Park, H.J.; Yun, D.-J.; Kim, W.Y.; Salmela, M.J.; Ewers, B.E.; Weinig, C.; et al. Allelic polymorphism of GIGANTEA is responsible for naturally occurring variation in circadian period in Brassica rapa. Proc. Natl. Acad. Sci. USA 2015, 112, 3829–3834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayama, R.; Yokoi, S.; Tamaki, S.; TYano, M.; Shimamoto, K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 2003, 422, 719–722. [Google Scholar] [CrossRef]
- Li, F.; Zhang, X.; Hu, R.; Wu, F.; Ma, J.; Meng, Y.; Fu, Y. Identification and molecular characterization of FKF1 and GI homologous genes in soybean. PLoS ONE 2013, 8, e79036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Gu, Y.; Gao, H.; Qiu, L.; Chang, R.; Chen, S.; He, C. Molecular and geographic evolutionary support for the essential role of GIGANTEAa in soybean domestication of flowering time. BMC Evol. Biol. 2016, 16, 79. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-K.; Kim, E.-S.; Kim, K.H.; Jeong, N.; Lee, J.S.; Kang, S. Genetic variance for flowering time conferring E2 gene in photoperiod-insensitive early-maturing soybean accessions and topological distribution in Korea peninsula. Mol. Breed. 2018, 38, 148. [Google Scholar] [CrossRef]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.H.; Smith, R.W.; To, B.J.; Millar, A.J.; Imaizumi, T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 2012, 336, 1045–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imaizumi, T.; Schultz, T.F.; Harmon, F.G.; Ho, L.A.; Kay, S.A. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 2005, 309, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Seo, Y.-H.; Seo, P.J.; Reyes, J.L.; Yun, J.; Chua, N.-H.; Park, C.-M. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 2007, 19, 2736–2748. [Google Scholar] [CrossRef] [Green Version]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-LIKE target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Sun, M.-Y.; Wang, X.-S.; Li, W.-B.; Li, Y.-G. Over-expression of GmGIa-regulated soybean miR172a confers early flowering in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2016, 17, 645. [Google Scholar] [CrossRef] [Green Version]
- Hartwig, E.E.; Kiihl, R.A.S. Identification and utilization of a delayed flowering character in soybeans for short-day condtions. Field Crops Res. 1979, 2, 145–151. [Google Scholar] [CrossRef]
- Destro, D.; Carpentieri-Pipolo, V.; Kiihl, R.A.S.; de Almeida, L.A. Photoperiodism and genetic control of the long juvenile period in soybean: A review. Crop Breed. Appl. Biotechnol. 2001, 1, 72–92. [Google Scholar] [CrossRef]
- Ray, J.D.; Hinson, K.; Mankono, E.B.; Malo, M.F. Genetic control of a long-juvenile trait in soybean. Crop Sci. 1995, 35, 1001–1006. [Google Scholar] [CrossRef]
- Bonato, E.R.; Vello, N.A. E6, a dominant gene conditioning early flowering and maturity in soybeans. Genet. Mol. Biol. 1999, 22, 229–232. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Fang, C.; Xu, M.; Zhang, F.; Lu, S.; Nan, H.; Su, T.; Li, S.; Zhao, X.; Kong, L.; et al. Quantitative Trait Locus mapping of soybean maturity gene E6. Crop Sci. 2017, 57, 2547–2554. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Zhao, X.; Hu, Y.; Liu, S.; Nan, H.; Li, X.; Fang, C.; Cao, D.; Shi, X.; Kong, L.; et al. Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 2017, 49, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Preuss, S.B.; Meister, R.; Xu, Q.; Urwin, C.P.; Tripodi, F.A.; Screen, S.E.; Anil, V.S.; Zhu, S.; Morrell, J.A.; Liu, G.; et al. Expression of the Arabidopsis thaliana BBX32 gene in soybean increases grain yield. PLoS ONE 2012, 7, e30717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridge, S.; Deokar, A.; Lee, R.; Daba, K.; Macknight, R.C.; Weller, J.L.; Tar’an, B. The Chickpea Early Flowering 1 (Efl1) locus Is an ortholog of Arabidopsis ELF3. Plant Physiol. 2017, 175, 802–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weller, J.L.; Liew, L.C.; Hecht, V.F.G.; Rajandran, V.; Laurie, R.E.; Ridge, S.; Wenden, B.; Vander Schoor, J.K.; Jaminon, O.; Blassiau, C.; et al. A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc. Natl. Acad. Sci. USA 2012, 109, 21158–21163. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Li, H.; Li, R.; Hu, R.; Fan, C.; Chen, F.; Wang, Z.; Liu, X.; Fu, Y.; Lin, C. Association of the circadian rhythmic expression of GmCRY1a with a latitudinal cline in photoperiodic flowering of soybean. Proc. Natl. Acad. Sci. USA 2008, 105, 21028–21033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.-W.; Liu, W.; Lam, H.-M.; Gendron, J.M. Characterization of two growth period QTLs reveals modification of PRR3 genes during soybean domestication. Plant Cell Physiol. 2019, 60, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Shen, Y.; Chang, H.-C.; Hou, Y.; Harris, A.; Ma, S.F.; McPartland, M.; Hymus, G.J.; Adam, L.; Marion, C.; et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010, 187, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yuan, L.; Su, T.; Wang, Q.; Gao, Y.; Zhang, S.; Jia, Q.; Yu, G.; Fu, Y.; Cheng, Q.; et al. Light and temperature-entrainable circadian clock in soybean development. Plant Cell Environ. 2020, 43, 637–648. [Google Scholar] [CrossRef]
- Lu, S.; Dong, L.; Fang, C.; Liu, S.; Kong, L.; Cheng, Q.; Chen, L.; Su, T.; Nan, H.; Zhang, D.; et al. Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 2020, 52, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, Y.-h.; Li, Y.; Lu, H.; Hong, H.; Tian, Y.; Li, H.; Zhao, T.; Zhou, X.; Liu, J.; et al. A domestication-associated gene GmPRR3b regulates the circadian clock and flowering time in soybean. Mol. Plant 2020, 13, 745–759. [Google Scholar] [CrossRef]
- Li, Y.; Dong, Y.; Wu, H.; Hu, B.; Zhai, H.; Yang, J.; Xia, Z. Positional cloning of the flowering time QTL qFT12-1 reveals the link between the clock related PRR homolog with photoperiodic response in soybeans. Front. Plant Sci. 2019, 10, 1303. [Google Scholar] [CrossRef] [Green Version]
- Hotta, C.T.; Gardner, M.J.; Hubbard, K.E.; Baek, S.J.; Dalchau, N.; Suhita, D.; Dodd, A.N.; Webb, A.A.R. Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 2007, 30, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; McClung, C.R.; Zhang, C. Tick Tock: Circadian regulation of plant innate immunity. Annu. Rev. Phytopathol. 2017, 55, 287–311. [Google Scholar] [CrossRef] [Green Version]
- Fowler, S.G.; Cook, D.; Thomashow, M.F. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005, 137, 961–968. [Google Scholar] [CrossRef] [Green Version]
- Wilkins, O.; Bräutigam, K.; Campbell, M.M. Time of day shapes Arabidopsis drought transcriptomes. Plant J. 2010, 63, 715–727. [Google Scholar] [CrossRef]
- Goodspeed, D.; Chehab, E.W.; Min-Venditti, A.; Braam, J.; Covington, M.F. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. USA 2012, 109, 4674–4677. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Carlsson, J.; Takeuchi, T.; Newton, L.; Farré, E.M. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 2013, 76, 101–114. [Google Scholar] [CrossRef]
- Lei, J.; Jayaprakasha, G.K.; Singh, J.; Uckoo, R.; Borrego, E.J.; Finlayson, S.; Kolomiets, M.; Patil, B.S.; Braam, J.; Zhu-Salzman, K. CIRCADIAN CLOCK-ASSOCIATED1 controls resistance to aphids by altering indole glucosinolate production. Plant Physiol. 2019, 181, 1344–1359. [Google Scholar] [CrossRef] [Green Version]
- Mody, T.; Bonnot, T.; Nagel, D.H. Interaction between the circadian clock and regulators of heat stress responses in plants. Genes 2020, 11. [Google Scholar] [CrossRef] [Green Version]
- Wilkins, O.; Waldron, L.; Nahal, H.; Provart, N.J.; Campbell, M.M. Genotype and time of day shape the Populus drought response. Plant J. 2009, 60, 703–715. [Google Scholar] [CrossRef]
- Greenham, K.; Guadagno, C.R.; Gehan, M.A.; Mockler, T.C.; Weinig, C.; Ewers, B.E.; McClung, C.R. Temporal network analysis identifies early physiological and transcriptomic indicators of mild drought in Brassica rapa. eLIFE 2017, 6, e29655. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cao, L.; Mwimba, M.; Zhou, Y.; Li, L.; Zhou, M.; Schnable, P.S.; O’Rourke, J.A.; Dong, X.; Wang, W. Comprehensive mapping of abiotic stress inputs into the soybean circadian clock. Proc. Natl. Acad. Sci. USA 2019, 116, 23840–23849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Bu, T.; Cheng, Q.; Dong, L.; Su, T.; Chen, Z.; Kong, F.; Gong, Z.; Liu, B.; Li, M. Two homologous LHY pairs negatively control soybean drought tolerance by repressing the abscisic acid responses. New Phytol. 2020. [Google Scholar] [CrossRef]
- Blanca, J.; Cañizares, J.; Cordero, L.; Pascual, L.; Diez, M.J.; Nuez, F. Variation revealed by SNP genotyping and morphology provides insight into the origin of the tomato. PLoS ONE 2012, 7, e48198. [Google Scholar] [CrossRef] [Green Version]
- Müller, N.A.; Wijnen, C.; Srinivasan, A.; Ryngajllo, M.; Ofner, I.; Lin, T.; Ranjan, A.; West, D.; Maloof, J.N.; Sinha, N.R.; et al. Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat. Genet. 2016, 48, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Dieterle, M.; Zhou, Y.-C.; Schäfer, E.; Funk, M.; Kretsch, T. EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev. 2001, 15, 939–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, N.A.; Zhang, L.; Koornneef, M.; Jiménez-Gómez, J.M. Mutations in EID1 and LNK2 caused light-conditional clock deceleration during tomato domestication. Proc. Natl. Acad. Sci. USA 2018, 115, 7135–7140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rugnone, M.L.; Faigón Soverna, A.; Sanchez, S.E.; Schlaen, R.G.; Hernando, C.E.; Seymour, D.K.; Mancini, E.; Chernomoretz, A.; Weigel, D.; Más, P.; et al. LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc. Natl. Acad. Sci. USA 2013, 110, 12120–12125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abegg, F.A. A genetic factor for the annual habit in beets and linkage relationship. J. Agric. Res. 1936, 53, 493–511. [Google Scholar]
- Büttner, B.; Abou-Elwafa, S.F.; Zhang, W.; Jung, C.; Müller, A.E. A survey of EMS-induced biennial β vulgaris mutants reveals a novel bolting locus which is unlinked to the bolting gene B. Theor. Appl. Genet. 2010, 121, 1117–1131. [Google Scholar] [CrossRef] [PubMed]
- Pin, P.A.; Zhang, W.; Vogt, S.H.; Dally, N.; Büttner, B.; Schulze-Buxloh, G.; Jelly, N.S.; Chia, T.Y.; Mutasa-Göttgens, E.S.; Dohm, J.C.; et al. The role of a pseudo-response regulator gene in life cycle adaptation and domestication of beet. Curr. Biol. 2012, 22, 1095–1101. [Google Scholar] [CrossRef] [Green Version]
- Pin, P.A.; Benlloch, R.; Bonnet, D.; Wremerth-Weich, E.; Kraft, T.; Gielen, J.J.; Nilsson, O. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 2010, 330, 1397–1400. [Google Scholar] [CrossRef] [PubMed]
- Dally, N.; Xiao, K.; Holtgräwe, D.; Jung, C. The B2 flowering time locus of beet encodes a zinc finger transcription factor. Proc. Natl. Acad. Sci. USA 2014, 111, 10365–10370. [Google Scholar] [CrossRef] [Green Version]
- Höft, N.; Dally, N.; Jung, C. Sequence variation in the bolting time regulator BTC1 changes the life cycle regime in sugar beet. Plant Breed. 2019, 137, 412–422. [Google Scholar] [CrossRef]
- Nagy, F.; Kay, S.A.; Chua, N.H. A circadian clock regulates transcription of the wheat Cab-1 gene. Genes Dev. 1988, 2, 376–382. [Google Scholar] [CrossRef] [Green Version]
- Redinbaugh, M.G.; Sabre, M.; Scandalios, J.G. Expression of the maize Cat3 catalase gene is under the influence of a circadian rhythm. Proc. Natl. Acad. Sci. USA 1990, 87, 6853–6857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takata, N.; Saito, S.; Saito, C.T.; Nanjo, T.; Shinohara, K.; Uemura, M. Molecular phylogeny and expression of poplar circadian clock genes,LHY1 and LHY2. New Phytol. 2009, 181, 808–819. [Google Scholar] [CrossRef]
- Takata, N.; Saito, S.; Saito, C.T.; Uemura, M. Phylogenetic footprint of the plant clock system in angiosperms: Evolutionary processes of Pseudo-Response Regulators BMC Evol. Biol. 2010, 10, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClung, C.R. A modern circadian clock in the common angiosperm ancestor of monocots and eudicots. BMC Biol. 2010, 8, 55. [Google Scholar] [CrossRef] [Green Version]
- Murakami, M.; Ashikari, M.; Miura, K.; Yamashino, T.; Mizuno, T. The evolutionarily conserved OsPRR quintet: Rice Pseudo-Response Regulators implicated in circadian rhythm. Plant Cell Physiol. 2003, 44, 1229–1236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, M.; Tago, Y.; Yamashino, T.; Mizuno, T. Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 2007, 48, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Miwa, K.; Serikawa, M.; Suzuki, S.; Kondo, T.; Oyama, T. Conserved expression profiles of circadian clock-related genes in two Lemna species showing long-day and short-day photoperiodic flowering responses. Plant Cell Physiol. 2006, 47, 601–612. [Google Scholar] [CrossRef] [Green Version]
- Serikawa, M.; Miwa, K.; Kondo, T.; Oyama, T. Functional conservation of clock-related genes in flowering plants: Overexpression and RNAi analyses of the circadian rhythm in the monocotyledon Lemna gibba. Plant Physiol. 2008, 146, 1952–1963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muranaka, T.; Okada, M.; Yomo, J.; Kubota, S.; Oyama, T. Characterisation of circadian rhythms of various duckweeds. Plant Biol. 2015, 17, 66–74. [Google Scholar] [CrossRef] [Green Version]
- Campoli, C.; Shtaya, M.; Davis, S.J.; von Korff, M. Expression conservation within the circadian clock of a monocot: Natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol. 2012, 12, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faure, S.; Turner, A.S.; Gruszka, D.; Christodoulou, V.; Davis, S.J.; von Korff, M.; Laurie, D.A. Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. USA 2012, 109, 8328–8333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campoli, C.; Pankin, A.; Drosse, B.; Casao, C.M.; Seth, J.; Davis, S.J.; von Korff, M. HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: Phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol. 2013, 199, 1045–1059. [Google Scholar] [CrossRef] [Green Version]
- Müller, L.M.; Mombaerts, L.; Pankin, A.; Davis, S.J.; Webb, A.A.R.; Goncalves, J.; von Korff, M. Differential effects of day/night cues and the circadian clock on the barley transcriptome. Plant Physiol. 2020, 183, 765–779. [Google Scholar] [CrossRef] [PubMed]
- Izawa, T.; Mihara, M.; Suzuki, Y.; Gupta, M.; Itoh, H.; Nagano, A.J.; Motoyama, R.; Sawada, Y.; Yano, M.; Hirai, M.Y.; et al. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell 2011, 23, 1741–1755. [Google Scholar] [CrossRef] [Green Version]
- Murakami, M.; Tago, Y.; Yamashino, T.; Mizuno, T. Characterization of the rice circadian clock-associated Pseudo-Response Regulators in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2007, 71, 1107–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, L.; Zhang, Z.; Cheng, N.; Liu, H.; Song, S.; Hu, Y.; Zhou, X.; Zhang, J.; Xing, Y. The transcriptional repressor OsPRR73 links the circadian clock and photoperiod pathway to control heading date in rice. Plant Cell Environ. 2021, 44, 842–855. [Google Scholar] [CrossRef]
- Pankin, A.; Campoli, C.; Dong, X.; Kilian, B.; Sharma, R.; Himmelbach, A.; Saini, R.; Davis, S.J.; Stein, N.; Schneeberger, K.; et al. Mapping-by-sequencing identifies HvPHYTOCHROME C as a candidate gene for the early maturity 5 locus modulating the circadian clock and photoperiodic flowering in barley. Genetics 2014, 198, 383–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakhrabekova, S.; Gough, S.P.; Braumann, I.; Müller, A.H.; Lundqvist, J.; Ahmann, K.; Dockter, C.; Matyszczak, I.; Kurowska, M.; Druka, A.; et al. Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc. Natl. Acad. Sci. USA 2012, 109, 4326–4331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, H.; Jin, M.; Zheng, X.-M.; Chen, J.; Yuan, D.; Xin, Y.; Wang, M.; Huang, D.; Zhang, Z.; Zhou, K.; et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 16337–16342. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.-M.; Feng, L.; Wang, J.; Qiao, W.; Zhang, L.; Cheng, Y.; Yang, Q. Nonfunctional alleles of long-day suppressor genes independently regulate flowering time. J. Integr. Plant Biol. 2016, 58, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Wang, J.; Feng, L.; Liu, S.; Li, J.; Qiao, W.; Song, Y.; Zhang, Z.; Cheng, Y.; Zhang, L.; et al. A combination of long-day suppressor genes contributes to the northward expansion of rice. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Gao, H.; Zheng, X.-M.; Fei, G.; Chen, J.; Jin, M.; Ren, Y.; Wu, W.; Zhou, K.; Sheng, P.; Zhou, F.; et al. Ehd4 encodes a novel and Oryza-genus-specific regulator of photoperiodic flowering in rice. PLoS Genet. 2013, 9, e1003281. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Chen, H.; Ren, D.; Tang, H.; Qiu, R.; Feng, J.; Long, Y.; Niu, B.; Chen, D.; Zhong, T.; et al. Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/ RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa). New Phytol. 2015, 208, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Turner, A.; Beales, J.; Faure, S.; Dunford, R.P.; Laurie, D.A. The Pseudo-Response Regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 2005, 310, 1031–1034. [Google Scholar] [CrossRef] [PubMed]
- Baudry, A.; Ito, S.; Song, Y.H.; Strait, A.A.; Kiba, T.; Lu, S.; Henriques, R.; Pruneda-Paz, J.L.; Chua, N.-H.; Tobin, E.M.; et al. F-Box Proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 2010, 22, 606–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Digel, B.; Tavakol, E.; Verderio, G.; Tondelli, A.; Xu, X.; Cattivelli, L.; Rossini, L.; von Korff, M. Photoperiod-H1 (Ppd-H1) controls leaf size. Plant Physiol. 2016, 172, 405–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruneda-Paz, J.L.; Kay, S.A. An expanding universe of circadian networks in higher plants. Trends Plant Sci. 2010, 15, 259–265. [Google Scholar] [CrossRef] [Green Version]
- Green, R.M.; Tingay, S.; Wang, Z.-Y.; Tobin, E.M. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 2002, 129, 576–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yerushalmi, S.; Yakir, E.; Green, R.M. Circadian clocks and adaptation in Arabidopsis. Mol. Ecol. 2011, 20, 1155–1165. [Google Scholar] [CrossRef]
- Dodd, A.N.; Salathia, N.; Hall, A.; Kevei, E.; Toth, R.; Nagy, F.; Hibberd, J.M.; Millar, A.J.; Webb, A.A.R. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005, 309, 630–633. [Google Scholar] [CrossRef] [Green Version]
- Toniutti, L.; Breitler, J.-C.; Guittin, C.; Doulbeau, S.; Etienne, H.; Campa, C.; Lambot, C.; Herrera Pinilla, J.-C.; Bertrand, B. An altered circadian clock coupled with a higher photosynthesis efficiency could explain the better agronomic performance of a new coffee clone when compared with a standard variety. Intl. J. Mol. Sci. 2019, 20, 736. [Google Scholar] [CrossRef] [Green Version]
- Ni, Z.; Kim, E.-D.; Ha, M.; Lackey, E.; Liu, J.; Zhang, Y.; Sun, Q.; Chen, Z.J. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 2009, 457, 327–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, D.W.-K.; Miller, M.; Yu, H.H.; Huang, T.-Y.; Kim, E.-D.; Lu, J.; Xie, Q.; McClung, C.R.; Chen, Z.J. A role for CHH methylation in the parent-of-origin effect on altered circadian rhythms and biomass heterosis in Arabidopsis intraspecific hybrids. Plant Cell 2014, 26, 2430–2440. [Google Scholar] [CrossRef] [Green Version]
- Ko, D.K.; Rohozinski, D.; Song, Q.; Taylor, S.H.; Juenger, T.E.; Harmon, F.G.; Chen, Z.J. Temporal shift of circadian-mediated gene expression and carbon fixation contributes to biomass heterosis in maize hybrids. PLoS Genet. 2016, 12, e1006197. [Google Scholar] [CrossRef]
- Howden, S.M.; Soussana, J.-F.; Tubiello, F.N.; Chhetri, N.; Dunlop, M.; Meinke, H. Adapting agriculture to climate change. Proc. Natl. Acad. Sci. USA 2007, 104, 19691–19696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Massel, K.; Godwin, I.D.; Gao, C. Applications and potential of genome editing in crop improvement. Genome Biol. 2018, 19, 210. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McClung, C.R. Circadian Clock Components Offer Targets for Crop Domestication and Improvement. Genes 2021, 12, 374. https://doi.org/10.3390/genes12030374
McClung CR. Circadian Clock Components Offer Targets for Crop Domestication and Improvement. Genes. 2021; 12(3):374. https://doi.org/10.3390/genes12030374
Chicago/Turabian StyleMcClung, C. Robertson. 2021. "Circadian Clock Components Offer Targets for Crop Domestication and Improvement" Genes 12, no. 3: 374. https://doi.org/10.3390/genes12030374
APA StyleMcClung, C. R. (2021). Circadian Clock Components Offer Targets for Crop Domestication and Improvement. Genes, 12(3), 374. https://doi.org/10.3390/genes12030374





