Two Archaeal Metagenome-Assembled Genomes from El Tatio Provide New Insights into the Crenarchaeota Phylum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Information and Sample Collection
2.2. DNA Extraction and Sequencing.
2.3. Metagenome-Assembled Genomes Binning
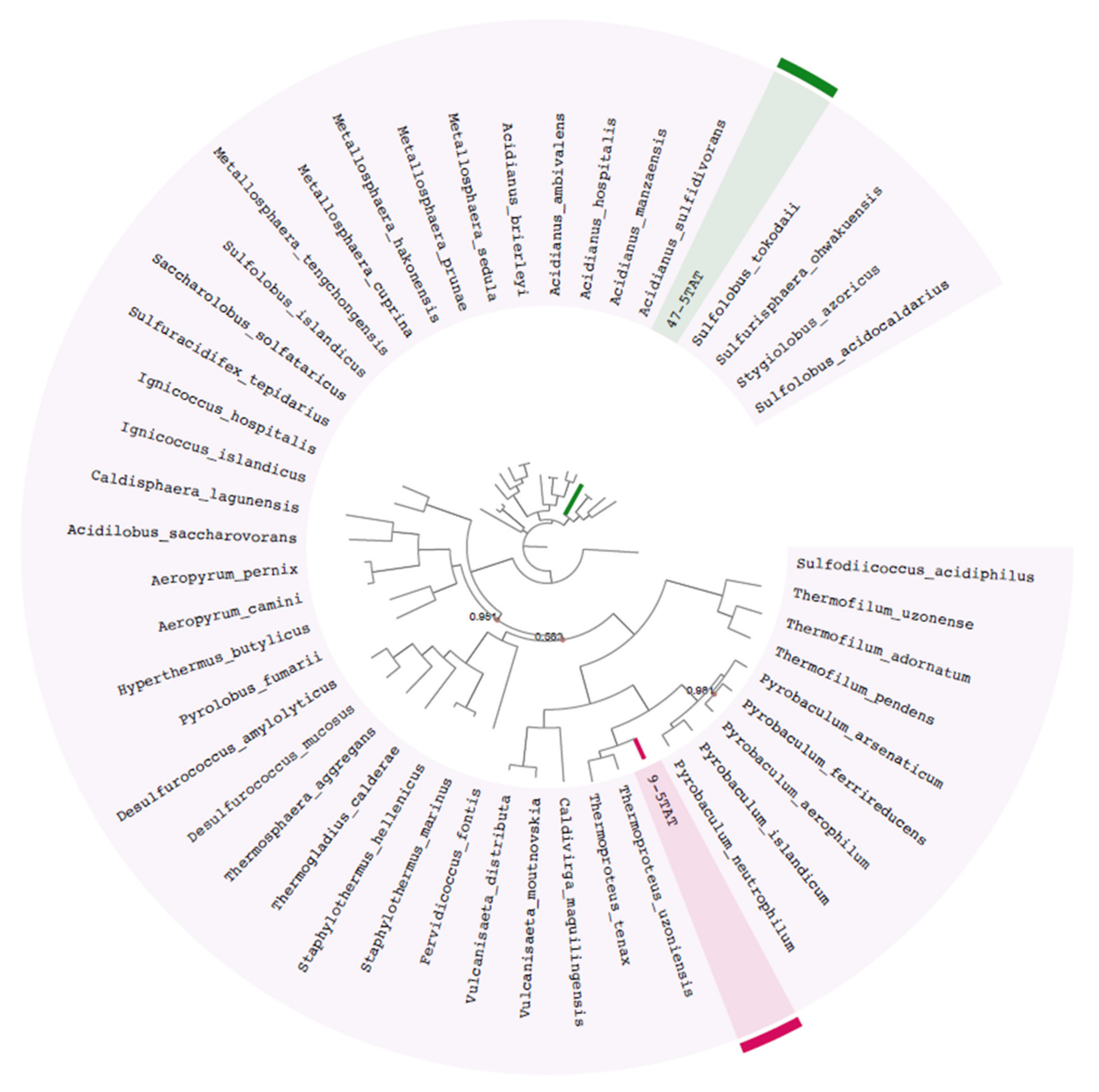
2.4. Taxonomic and Phylogenetic Inference of MAGs
2.5. Metabolic Reconstruction and Functional Annotation
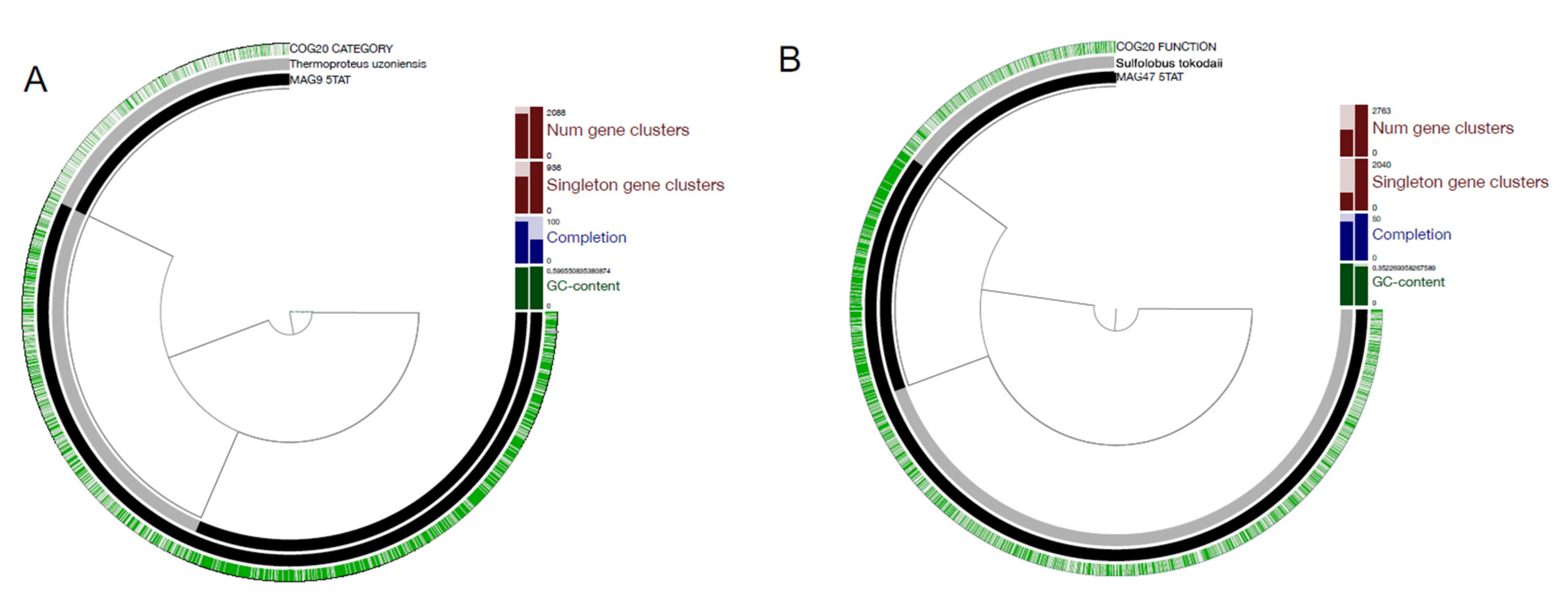
2.6. Pangenomic Comparison
3. Results and Discussion
3.1. Metagenome-Assembled Genome Binning
3.2. Phylogenetic Placement of the MAGs
3.3. Functional Annotation and Metabolic Reconstruction of the MAGs
3.4. Pangenomic Analysis of the MAGs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Landrum, J.; Bennett, P.; Engel, A.; Alsina, M.; Pastén, P.; Milliken, K. Partitioning geochemistry of arsenic and antimony, El Tatio Geyser Field, Chile. Appl. Geochem. 2009, 24, 664–676. [Google Scholar] [CrossRef]
- Fernandez-Turiel, J.; Garcia-Valles, M.; Gimeno-Torrente, D.; Saavedra-Alonso, J.; Martinez-Manent, S. The hot spring and geyser sinters of El Tatio, Northern Chile. Sediment. Geol. 2005, 180, 125–147. [Google Scholar] [CrossRef]
- Engel, A.S.; Johnson, L.R.; Porter, M.L. Arsenite oxidase gene diversity amongChloroflexiandProteobacteriafrom El Tatio Geyser Field, Chile. FEMS Microbiol. Ecol. 2013, 83, 745–756. [Google Scholar] [CrossRef] [Green Version]
- Phoenix, V.; Bennett, P.C.; Engel, A.; Tyler, S.W.; Ferris, F.G. Chilean high-altitude hot-spring sinters: A model system for UV screening mechanisms by early Precambrian cyanobacteria. Geobiology 2006, 4, 15–28. [Google Scholar] [CrossRef]
- Orellana, R.; Macaya, C.; Bravo, G.; Dorochesi, F.; Cumsille, A.; Valencia, R.; Rojas, C.; Seeger, M. Living at the Frontiers of Life: Extremophiles in Chile and Their Potential for Bioremediation. Front. Microbiol. 2018, 9, 2309. [Google Scholar] [CrossRef]
- Nelson, W.C.; Wollerman, L.; Bhaya, D.; Heidelberg, J.F. Analysis of Insertion Sequences in Thermophilic Cyanobacteria: Exploring the Mechanisms of Establishing, Maintaining, and Withstanding High Insertion Sequence Abundance. Appl. Environ. Microbiol. 2011, 77, 5458–5466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nunoura, T.; Takaki, Y.; Kakuta, J.; Nishi, S.; Sugahara, J.; Kazama, H.; Chee, G.-J.; Hattori, M.; Kanai, A.; Atomi, H.; et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2010, 39, 3204–3223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plenge, M.F.; Engel, A.S.; Omelon, C.R.; Bennett, P.C. Thermophilic Archaeal Diversity and Methanogenesis from El Tatio Geyser Field, Chile. Geomicrobiol. J. 2016, 34, 220–230. [Google Scholar] [CrossRef]
- Leigh, J.; Whitman, W. Archaeal Genetics. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 188–191. ISBN 9780080961569. [Google Scholar]
- Merino, N.; Aronson, H.S.; Bojanova, D.P.; Feyhl-Buska, J.; Wong, M.L.; Zhang, S.; Giovannelli, D. Living at the Extremes: Extremophiles and the Limits of Life in a Planetary Context. Front. Microbiol. 2019, 10, 780. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Campos, M.; Larama, G.; Acuña, J.J.; Valenzuela, B.; Solis, F.; Zamorano, P.; Araya, R.; Sadowsky, M.J.; Jorquera, M.A. Composition and predicted functions of the bacterial community in spouting pool sediments from the El Tatio Geyser field in Chile. Arch. Microbiol. 2021, 203, 389–397. [Google Scholar] [CrossRef]
- Krueger, F. Trim Galore! A Wrapper Tool around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ files. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 28 December 2020).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alneberg, J.; Bjarnason, B.S.; de Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning metagenomic contigs by coverage and composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Rinke, C.; Chuvochina, M.; Chaumeil, P.-A.; Woodcroft, B.J.; Evans, P.N.; Hugenholtz, P.; Tyson, G.W. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017, 2, 1533–1542. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.E.; Jospin, G.; Lowe, E.; Iv, F.A.M.; Bik, H.M.; Eisen, J.A. PhyloSift: Phylogenetic analysis of genomes and metagenomes. PeerJ 2014, 2, e243. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Eren, A.M.; Özcan, E.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An advanced analysis and visualization platform for ‘omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.J.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, srep08365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delmont, T.O.; Eren, E.M. Linking pangenomes and metagenomes: The Prochlorococcus metapangenome. PeerJ 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues-Oliveira, T.; Belmok, A.; Vasconcellos, D.; Schuster, B.; Kyaw, C.M. Archaeal S-Layers: Overview and Current State of the Art. Front. Microbiol. 2017, 8, 2597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madigan, M.T.; Martinko, J.M.; Stahl, D.A.; Clark, D.P. Brock Biology of Microorganisms, 13th ed.; Pearson: Benjamin Cummings: San Francisco, CA, USA, 2009; ISBN 1402472633. [Google Scholar]
- Farías, M.E.; Villafañe, P.G.; Lencina, A.I. Integral Prospection of Andean Microbial Ecosystem Project. In Microbial Ecosystems in Central Andes Extreme Environments; Springer: Chem, Switzerland, 2020. [Google Scholar]
- Dorador, C.; Vila, I.; Remonsellez, F.; Imhoff, J.F.; Witzel, K.-P. Unique clusters of Archaea in Salar de Huasco, an athalassohaline evaporitic basin of the Chilean Altiplano. FEMS Microbiol. Ecol. 2010, 73, 291–302. [Google Scholar] [CrossRef] [Green Version]
- Molina, V.; Eissler, Y.; Cornejo, M.; Galand, P.E.; Dorador, C.; Hengst, M.; Fernandez, C.; Francois, J.P. Distribution of greenhouse gases in hyper-arid and arid areas of northern Chile and the contribution of the high altitude wetland microbiome (Salar de Huasco, Chile). Antonie van Leeuwenhoek 2018, 111, 1421–1432. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Medlar, A.J.; Törönen, P.; Holm, L. AAI-profiler: Fast proteome-wide exploratory analysis reveals taxonomic identity, misclassification and contamination. Nucleic Acids Res. 2018, 46, W479–W485. [Google Scholar] [CrossRef] [Green Version]
- López-López, O.; Knapik, K.; Cerdán, M.-E.; González-Siso, M.-I. Metagenomics of an Alkaline Hot Spring in Galicia (Spain): Microbial Diversity Analysis and Screening for Novel Lipolytic Enzymes. Front. Microbiol. 2015, 6, 1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, E.-S.; Cha, I.-T.; Roh, S.W.; Nam, Y.-D.; Seo, M.-J. Haloplanus rallus sp. nov., a halophilic archaeon isolated from crude solar salt. Int. J. Syst. Evol. Microbiol. 2018, 68, 3226–3231. [Google Scholar] [CrossRef] [PubMed]
- Zillig, W.; Reysenbach, A. Thermoproteus. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–4. [Google Scholar]
- Shukla, A.K. Exploitation of Potential Extremophiles for Bioremediation of Xenobiotics Compounds: A Biotechnological Approach. Curr. Genom. 2020, 21, 161–167. [Google Scholar] [CrossRef]
- Quehenberger, J.; Shen, L.; Albers, S.-V.; Siebers, B.; Spadiut, O. Sulfolobus—A Potential Key Organism in Future Biotechnology. Front. Microbiol. 2017, 8, 2474. [Google Scholar] [CrossRef] [PubMed]
- Torregrosa-Crespo, J.; Espliego, J.M.E.; Saiz, V.B.; Pire, C.; Gates, A.J.; Richardson, D.J.; Luque, A.V.; Carrasco, M.L.C.; Bonete, M.J.; Martínez-Espinosa, R.M. Denitrification in Extreme Environments. In Extremophiles; CRC Press: Boca Raton, FL, USA, 2018; pp. 209–226. [Google Scholar]
- Giuliano, M.; Schiraldi, C.; Marotta, M.R.; Hugenholtz, J.; de Rosa, M. Expression of Sulfolobus solfataricus α-glucosidase in Lactococcus lactis. Appl. Microbiol. Biotechnol. 2004, 64, 829–832. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Iwasaki, T.; Uzawa, T.; Hara, K.; Nemoto, N.; Kon, T.; Ueki, T.; Yamagishi, A.; Oshima, T. Sulfolobus tokodaii sp. nov. (f. Sulfolobus sp. strain 7), a new member of the genus Sulfolobus isolated from Beppu Hot Springs, Japan. Extremophiles 2002, 6, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Wang, H.; Zhang, Z.; Li, K.; Zhang, X.; Mora-López, M.; Jiang, C.; Liu, C.; Wang, L.; Zhu, Y.; et al. Genome Sequencing of Sulfolobus sp. A20 from Costa Rica and Comparative Analyses of the Putative Pathways of Carbon, Nitrogen, and Sulfur Metabolism in Various Sulfolobus Strains. Front. Microbiol. 2016, 7, 1902. [Google Scholar] [CrossRef]
| MAG | Completeness (%) | Contamination (%) | GC Content | Genome Length | CDSs | COGs | KEEG Pathways |
|---|---|---|---|---|---|---|---|
| 9-5TAT | 84.9 | 2.21 | 57.5 | 1,447,267 | 2109 | 827 | 121 |
| 47-5TAT | 67.3 | 1.9 | 35.2 | 1,265,490 | 1673 | 746 | 71 |
| MAG | Closest Species | Orthologous Genes | Mean AAI | Mean ANI | Closest species GC | Closest Species Genome Length |
|---|---|---|---|---|---|---|
| 9-5TAT | T. uzoniensis | 848 | 70.35 | 76.9 | 59.6 | 2,839,046 |
| 47-5TAT | S. tokodaii | 1093 | 66.02 | 58.7 | 35.2 | 1,960,328 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, A.; Bruna, P.; Martinez-Urtaza, J.; Solís, F.; Valenzuela, B.; Zamorano, P.; Barrientos, L. Two Archaeal Metagenome-Assembled Genomes from El Tatio Provide New Insights into the Crenarchaeota Phylum. Genes 2021, 12, 391. https://doi.org/10.3390/genes12030391
Santos A, Bruna P, Martinez-Urtaza J, Solís F, Valenzuela B, Zamorano P, Barrientos L. Two Archaeal Metagenome-Assembled Genomes from El Tatio Provide New Insights into the Crenarchaeota Phylum. Genes. 2021; 12(3):391. https://doi.org/10.3390/genes12030391
Chicago/Turabian StyleSantos, Andrés, Pablo Bruna, Jaime Martinez-Urtaza, Francisco Solís, Bernardita Valenzuela, Pedro Zamorano, and Leticia Barrientos. 2021. "Two Archaeal Metagenome-Assembled Genomes from El Tatio Provide New Insights into the Crenarchaeota Phylum" Genes 12, no. 3: 391. https://doi.org/10.3390/genes12030391
APA StyleSantos, A., Bruna, P., Martinez-Urtaza, J., Solís, F., Valenzuela, B., Zamorano, P., & Barrientos, L. (2021). Two Archaeal Metagenome-Assembled Genomes from El Tatio Provide New Insights into the Crenarchaeota Phylum. Genes, 12(3), 391. https://doi.org/10.3390/genes12030391







