Abstract
Sulfide inhibits oxygenic photosynthesis by blocking electron transfer between H2O and the oxygen-evolving complex in the D1 protein of Photosystem II. The ability of cyanobacteria to counter this effect has implications for understanding the productivity of benthic microbial mats in sulfidic environments throughout Earth history. In Lake Fryxell, Antarctica, the benthic, filamentous cyanobacterium Phormidium pseudopriestleyi creates a 1–2 mm thick layer of 50 µmol L−1 O2 in otherwise sulfidic water, demonstrating that it sustains oxygenic photosynthesis in the presence of sulfide. A metagenome-assembled genome of P. pseudopriestleyi indicates a genetic capacity for oxygenic photosynthesis, including multiple copies of psbA (encoding the D1 protein of Photosystem II), and anoxygenic photosynthesis with a copy of sqr (encoding the sulfide quinone reductase protein that oxidizes sulfide). The genomic content of P. pseudopriestleyi is consistent with sulfide tolerance mechanisms including increasing psbA expression or directly oxidizing sulfide with sulfide quinone reductase. However, the ability of the organism to reduce Photosystem I via sulfide quinone reductase while Photosystem II is sulfide-inhibited, thereby performing anoxygenic photosynthesis in the presence of sulfide, has yet to be demonstrated.
1. Introduction
Cyanobacterial production of O2 from oxygenic photosynthesis (OP) oxidized the Earth’s atmosphere during the Great Oxidation Event 2.4 billion years ago, which changed various elemental cycles, including the sulfur cycle [1]. Specifically, the Great Oxidation Event increased oxidative weathering, leading to a large flux of sulfate to the ocean, allowing more microbial sulfate reduction and increased sulfide concentrations in high productivity environments [2]. The biogeochemistry of OP is influenced by the presence of sulfide, which normally inhibits OP, presenting a challenge for cyanobacteria living in the presence of sulfide in diverse environments since the Great Oxidation Event. About 750 million years ago, a global “Snowball Earth” glaciation is associated with significant sulfate reduction [3,4,5], suggesting that cyanobacteria may have sustained OP in cold, sulfidic environments during at least one global glaciation. The only cold environment where sulfide-tolerant OP has been described is Middle Island Sinkhole in Michigan, USA (8–10 °C) [6], even though cyanobacteria are the dominant primary producers in many extreme cold ecosystems [7]. Sulfide-tolerant O2 production has also been documented in springs and sink holes in warmer environments, including Frasassi springs in Italy [8,9] and Little Salt Spring sinkhole in Florida, USA [10,11].
Prior work identified a cyanobacterium, P. pseudopriestleyi in ice-covered Lake Fryxell, Antarctica [12,13], that creates a thin layer of O2 in sulfidic pore water, an “oxygen oasis” [12,13]. Compared to other environments where sulfide-tolerant OP has been observed, Lake Fryxell is colder (2.4–2.7 °C), and its high latitude leads to months of continuous winter darkness. Based on sulfide and O2 fluxes, the oxygen oasis is expected to disappear during the dark winter months when photosynthesis does not occur. The increasing availability of light in spring allows benthic mats dominated by P. pseudopriestleyi to transition from sulfidic to oxic conditions due to photosynthetic O2 production [12]. However, the mechanism by which P. pseudopriestleyi tolerates sulfide and initiates this redox change is unknown.
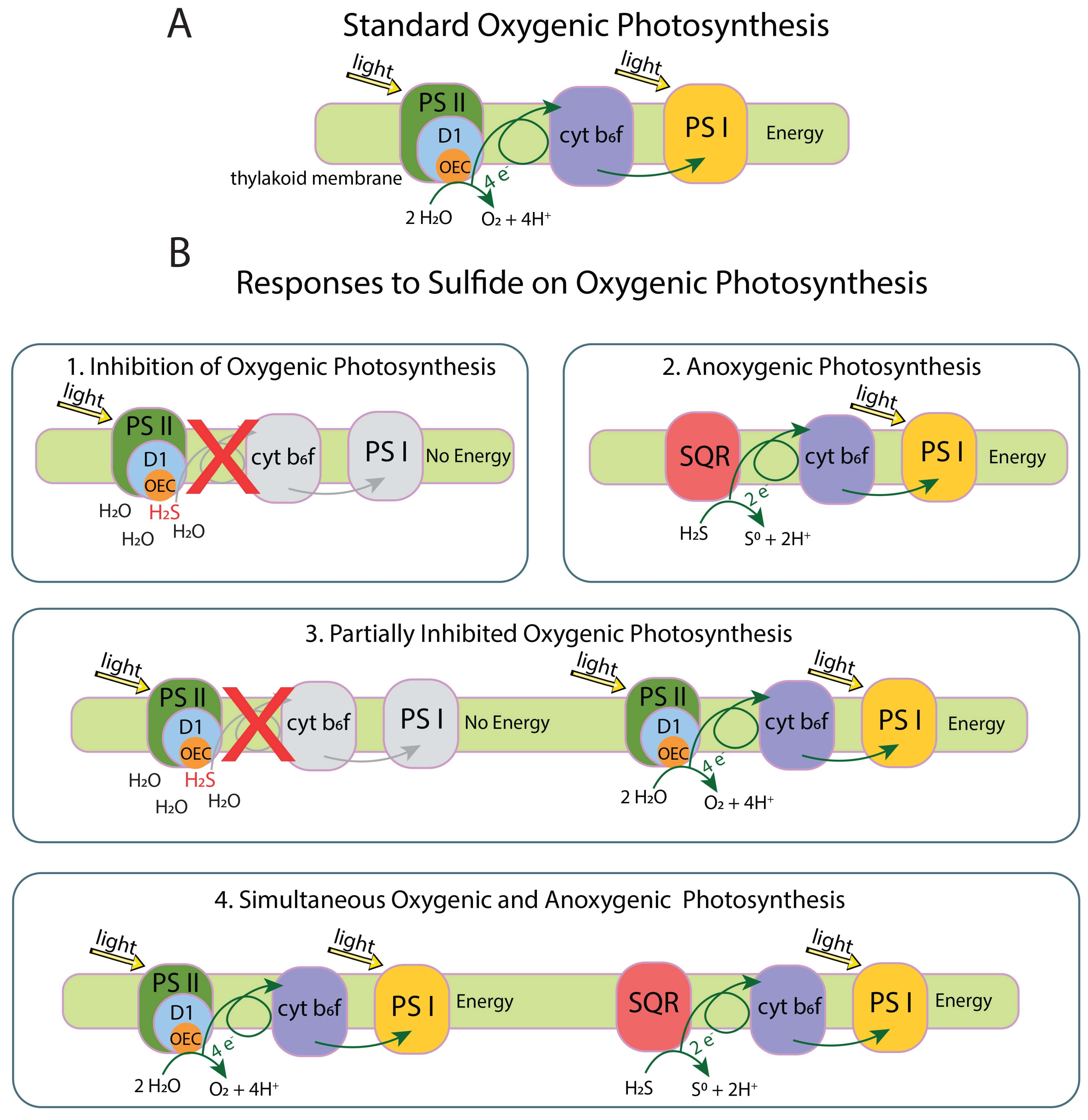
Sulfide inhibits OP by blocking electron donation interaction between H2O and the Mn4CaO5 cluster at the oxygen-evolving complex (OEC) in the D1 protein of Photosystem II (PS II). Cyanobacteria respond to sulfide in one of four ways: (1) complete inhibition of OP, (2) continued but partial inhibition of OP, (3) simultaneous OP and anoxygenic photosynthesis (AP), or (4) shutting down of OP and use of AP until enough sulfide is oxidized that OP can start again [14,15,16] (Figure 1). Response 1 results in a cessation of the electron flow by blocking the OEC in PS II with no source of electrons for Photosystem I (PS I). Response 2 consists of the sulfide quinone reductase (SQR) protein oxidizing sulfide to elemental sulfur and providing electrons to PS I to perform AP. Response 3 involves modification of the D1 protein by increasing psbA expression or switching to an alternate variant of the D1 protein allowing reduced O2 production and electron flow to and functioning of PS I. Response 4 involves the mechanism from response 3 with SQR providing additional electrons to PS I [10,17,18].
The direct oxidation of sulfide to sulfur by SQR removes the OP inhibiting chemical from the surrounding environment whether or not it provides electrons to fuel AP. Thus, its activity can eventually allow OP. Alternatively, a cyanobacterium that can maintain OP in sulfidic conditions produces O2, which can react with the sulfide in its environment through the abiotic reaction:
H2S + 2O2 → SO42− + 2H+.
SO42− does not interfere with the OEC and does not affect OP. Therefore, depending on the rate of OP and kinetics of abiotic sulfide oxidation, which may be slow at low temperatures, the production of a small amount of O2 might lead to a positive feedback loop by reducing OP inhibition.
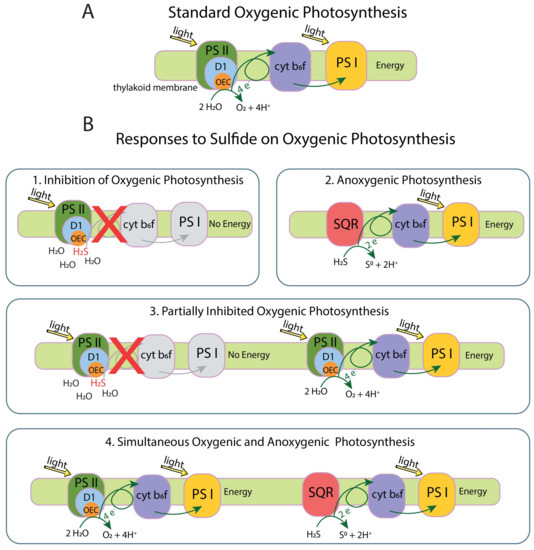
Figure 1.
(A) standard electron flow in oxygenic photosynthesis. (B1) inhibition of oxygenic photosynthesis (OP) by sulfide blocking the oxygen-evolving complex (OEC) in the D1 protein, prohibiting H2O from interacting with the OEC and from electrons to flow through the system. (B2) anoxygenic photosynthesis (AP) occurs when sulfide quinone reductase (SQR) extracts electrons from sulfide and passes them along the photosynthetic electron transport chain. Sulfide is oxidized to S0, which does not interact with the OEC. (B3) partially inhibited OP occurs when some D1 proteins are blocked by sulfide while others extract electrons from H2O and pass them along to carry out OP. Excess O2 produced from OP will oxidize sulfide to sulfate, removing sulfide from the environment. Some cyanobacteria increase psbA expression to replace the D1 protein in response to stress, which may support this response. (B4) some cyanobacteria can do OP and AP at the same time, or alternate between the two processes, until sulfide is fully depleted. Oxygenic photosynthesis protein complex image modified from the Kyoto Encyclopedia of Genes and Genomes (KEGG) [19,20].
Figure 1.
(A) standard electron flow in oxygenic photosynthesis. (B1) inhibition of oxygenic photosynthesis (OP) by sulfide blocking the oxygen-evolving complex (OEC) in the D1 protein, prohibiting H2O from interacting with the OEC and from electrons to flow through the system. (B2) anoxygenic photosynthesis (AP) occurs when sulfide quinone reductase (SQR) extracts electrons from sulfide and passes them along the photosynthetic electron transport chain. Sulfide is oxidized to S0, which does not interact with the OEC. (B3) partially inhibited OP occurs when some D1 proteins are blocked by sulfide while others extract electrons from H2O and pass them along to carry out OP. Excess O2 produced from OP will oxidize sulfide to sulfate, removing sulfide from the environment. Some cyanobacteria increase psbA expression to replace the D1 protein in response to stress, which may support this response. (B4) some cyanobacteria can do OP and AP at the same time, or alternate between the two processes, until sulfide is fully depleted. Oxygenic photosynthesis protein complex image modified from the Kyoto Encyclopedia of Genes and Genomes (KEGG) [19,20].
The D1 protein is directly affected by sulfide, and many cyanobacteria have multiple psbA genes encoding this protein. One model for reducing sulfide inhibition is altering the production of the D1 protein, which is essential for extracting electrons from H2O for OP. The effects of sulfide on D1 protein production have not been studied in cyanobacteria, although the effects of low light and 3-(3,4-dichlorophenyl)-1,1-dimehtylurea (DCMU) have [21,22,23]. DCMU blocks the electron flow from PS II to PS I, which is similar to sulfide due to preventing electron flow to PS I by blocking the extraction of electrons from H2O. Understanding how DCMU affects OP may provide insights as to how cyanobacteria deal with sulfide. In response to inhibition from light or DCMU, some cyanobacteria increase their expression of psbA to support replacement of the D1 protein [22,23,24]. Cyanobacteria can also respond to some environmental stressors by using different types of D1 proteins. The D1 proteins are divided into four groups: One standard group present in all cyanobacteria that is used under normal conditions, a version specialized for microaerobic conditions, a version for red light, and a nonfunctional version to support nitrogen fixation [25]. To allow flexible responses to environmental conditions, most cyanobacteria contain multiple copies of psbA, which allows cyanobacteria to increase expression or use the version of the protein most appropriate for the environmental conditions [22].
Instead of oxidizing sulfide with excess O2, some cyanobacteria directly oxidize sulfide to sulfur with SQR [26,27]. Cyanobacteria able to do AP pass these electrons through the quinone pool to PS I [28,29]. Some cyanobacteria can switch between OP and AP, while others can do both simultaneously, with both H2O and sulfide donating electrons [16,29]. However, some cyanobacteria, such as Aphanothece halophytica, that have sqr can survive in the presence of sulfide but not grow, suggesting that the electrons are not shuttled to an energy-producing pathway [26,28].
If P. pseudopriestleyi can create an oxygen oasis in the sulfidic benthic environment in Lake Fryxell [12], then it must be able to perform photosynthesis in the presence of sulfide or oxidize the sulfide before producing O2. To evaluate the genomic potential for tolerance mechanisms, we obtained a metagenome-assembled genome (MAG) from a natural sample and an enrichment culture of P. pseudopriestleyi. We use this MAG to evaluate the genomic potential for survival in elevated sulfide, low light, and cold temperatures and build on the phylogenetic characterization of the P. pseudopriestleyi 16S rRNA gene sequence in Jungblut et al. [13]. This study presents an investigation into the metabolic potential of this MAG to gain a better understanding of the connection between metabolic potential and environmental function with implications for primary productivity in sulfidic environments throughout Earth history.
Site Description
Lake Fryxell is a perennially ice-covered lake located at 77°36′ S 162°6′ E in the McMurdo Dry Valleys of east Antarctica. It is 5 times 1.5 km with a maximum depth of ~20 m [30]. The floor of Lake Fryxell is covered with photosynthetically active microbial mats to depths of ~10 m [12]. At the 9.8 m sampling depth, the lake floor is covered by flat prostrate mats dominated by a single diatom species and P. pseudopriestleyi.
The lake receives water from thirteen glacial meltwater streams [31]. Evaporation and ablation from the surface allow for water balance, as no streams flow out from the lake [32]. Salts remaining in the lake water and the historical balance of inflow and sublimation have led to density stratification of the lake water [13,33]. At the 9.8 m sampling depth, the salinity is approximately 4 mS cm−1 with 1.2 M NaCl [34,35,36], and sulfide (H2S + HS−) was present based on diver observations (rotten egg smell). Above 9 m, sulfide was undetectable, and below 10 m, sulfide concentration was 69.9 µM and increased to 1210 µM at the bottom of the lake [30].
Water temperature varies from 2.4 to 2.7 °C, and pH varies from 7.50 to 7.52 along a dive transect established from 8.9 m to 11.0 m in depth [13]. The water column has a sharp oxycline; dissolved oxygen is super saturated below the ice cover to a depth of 9.1 m where it decreases rapidly [33,37]. At 9.8 m depth, there is no O2 in the water column, but a microlayer of 50 μmol O2 L−1 is at least transiently present in the top ~1 mm of the mat [12].
Irradiance is highly seasonal. The lake experiences four months of darkness in the winter, followed by two months of diurnal light variations in spring, four months of continuous summer illumination, and two months of autumn diurnal light variations [31]. Even at peak illumination during the summer, only 0.5–3% of incident light penetrates the ice cover, and light is further attenuated by planktonic communities in the water column [12,38]. A daily average of 1–2 μmol photons m−2 s−1 reaches the mat at ~10 m water depth [12]. The most penetrating waveband at ~10 m in Lake Fryxell is 520–580 nm [39].
2. Materials and Methods
2.1. Sulfide Concentrations
Water samples (12 mL) for total sulfide analysis were collected on 17 January 2020 from within 10 mm of the lake floor by a diver using syringes. On return to the surface, samples were immediately injected into glass tubes and preserved for later analysis with 0.2 mL of 2 M zinc acetate. On return to New Zealand, samples were analyzed using the methylene blue method from Standard Methods for the Examination of Water and Wastewater (21st Edition) from the American Public Health Association [40].
2.2. Field Work
Samples of the microbial mat were collected in November 2012. Divers accessed the lake through a hole melted in the ice cover and sampled the microbial mat at 9.8 m depth [41]. Sampling and dissection of samples were performed using sterile technique. Briefly, divers used spatulas to cut samples of the mat and transfer them to plastic boxes underwater. In the field lab, mat samples were dissected according to morphology and pigmentation of layers. A blue-green biofilm from 9.8 m water depth was dominated by a single cyanobacterial morphotype based on field microscopy. Samples of this biofilm were peeled off the top of a prostrate microbial mat using sterile forceps [13]. Samples for metagenomic sequencing were preserved in the field within a few hours of collection with Xpedition Soil/Fecal DNA MiniPrep kit (Zymo Research, Irvine, CA, USA) and stored on ice. They were shipped to UC Davis where they were stored at −80 °C until they were processed for sequencing. One subsample for culturing was transferred to a sterile plastic vial filled with lake water that was filtered through a sterile syringe and 0.2 µm syringe filter. The cyanobacteria culture was stored at ambient indoor light at the lakeside laboratory for approximately ten days and then shipped to the Natural History Museum, London, UK where it was grown in BG11 liquid medium at 10 °C, and 24 h light at an average of 9.25 µmol photons m−2 s−1 [42].
2.3. DNA Extraction and Sequencing
DNA from the blue-green biofilm subsample was extracted from frozen samples using an Xpedition Soil/Fecal DNA MiniPrep kit (Zymo Research, Irvine, CA, USA) as per manufacturer instructions. Metagenomic sequencing on mat samples was performed at the Genome Center DNA Technologies Core at the University of California using the Illumina HiSeq 2500, PE250 platform. Illumina’s Nextera DNA Kit was used for library preparation (Oligonucleotide sequences (c) 2007–2013 Illumina, Inc., San Diego, CA, USA)
DNA was extracted from the cyanobacteria enrichment culture from Lake Fryxell using the MoBio Powerbio DNA extraction kit according to the manufacturer’s instructions. The culture was sequenced on the Illumina HiSeq platform (2000 PE 100, Illumina, Inc., San Diego, CA, USA) at the University of Michigan DNA Sequencing Core.
2.4. Bioinformatics Analysis
The sequencing of the biofilm sample from 9.8 m depth resulted in 3,911,904 reads. The biofilm sample data were quality filtered to Q20, and forward and reverse reads were joined using PEAR v0.9.6 [43]. Singletons and replicates with fewer than 10,000 reads were removed from downstream analysis. Sequencing of the culture resulted in 47,243,886 reads. For the lab culture data, trimmomatic v0.36 was used to trim sequencing adaptors with a LEADING and TRAILING parameter of 3, a SLIDINGWINDOW parameter of 4:15, and a MINLEN parameter of 25 [44]. The interleave-reads.py script from khmer v2.1.2 was used to interleave the reads [45]. The biofilm sample and lab-cultured sample were assembled separately and by coassembly with MEGAHIT v1.1.2 [46]. QUAST v4.4 was used to generate assembly statistics [47]. Mapping of both sets of reads to the coassembly was done with bwa v2.3 and samtools v1.9 [48,49]. Anvi’o v2.2.2 was used to bin and visualize the samples with the CONCOCT binning algorithm [50,51]. CheckM v1.0.7 was used to assess the quality of the bins and assign phylogeny based on marker genes of interest [52]. Taxonomy of the bins was assigned using the Genome Taxonomy Database (GTDB-Tk) on KBaseGhostKOALA v2.2 and Prokka v1.11 were used to annotate genes in the cyanobacterial bin of interest [53,54,55,56,57,58,59,60,61,62,63,64]. To refine the bin, spacegraphcats was used to extract additional content of the bin with a k size of 21 [65]. The code used for the analyses presented here is available at https://github.com/jessicalumian/fryxell-phormidium, accessed on 1 March 2021.
A custom Basic Local Alignment Search Tool (BLAST) database containing a reference amino acid sequence was constructed using the makeblastdb command in BLAST+, and D1 protein sequences from the P. pseudopriestleyi MAG were retrieved by using a blastx search with an e value of 1e-20 [66]. Subsequent analysis was performed on XSEDE Cipres Science Gateway [67]. D1 protein sequence fragments were aligned to D1 and D2 protein sequences compiled by Cardona et al. [25] with ClustalW v2.1 using standard parameters [68]. A best-fit model of evolution of LG + G4 was selected with ModelTest-ng v0.1.5 using maximum likelihood for the tree topology parameter and the discrete gamma rate categories option was selected for the candidate model’s rate heterogeneity parameter [69]. A phylogenetic tree was generated with RAxML-HPC2 v8.2.12 using a protein gamma model with an LG substitution matrix and 1000 bootstrap iterations [70].
To determine if the SQR in the P. pseudopriestleyi is type I or type II, the SQR amino acid sequence from the MAG was aligned to type I and type II references from Shahak and Hauska 2008 [71]. The sequences were aligned with ClustalW v2.1 and then trimmed with TrimAl v1.2.59 using standard parameters [68,72]. A best-fit model of evolution of WAG + G4 was selected with ModelTest-ng v0.1.5 using maximum likelihood for the tree topology parameter and the discrete gamma rate categories option was selected for the candidate model’s rate heterogeneity parameter [69]. A phylogenetic tree was generated with RAxML-HPC2 v8.2.12 using a protein gamma model, WAG substitution matrix and 1000 bootstrap iterations [70].
The average nucleotide identity (ANI) was calculated between the P. pseudopriestleyi MAG and available Antarctic cyanobacteria genomes (Leptolyngbya sp. BC1307, accession number NRTA00000000.1, Aurora vandensis, accession number JAAXLU010000000, Synechococcus sp. CS-601, accession number CP018091, and Phormidesmis priestleyi ULC007, accession number MPPI01000000) [73,74,75,76]. The ANI was also calculated between the MAG and an Arctic cyanobacterium closely related to an Antarctic strain (Phormidesmis priestleyi BC1401, accession number LXYR01000000) [77], and the closest related genome according to 16S rRNA gene sequence (Oscillatoria acuminata PCC 6304, accession number CP003607.1) [78]. Calculations were performed using the ANI calculator from the Kostas lab using the default parameters [79]. The alignment options required a 700 bp minimum length, 70% minimum identity, and 50 minimum alignments. The fragment option window size was set to 1000 bp with a step size of 200 bp.
To identify the presence P. pseudopriestleyi in other locations, the 16S rRNA gene sequence reported in Jungblut et al. [13] (accession number KT347094) was aligned with blastn to sequences from Jungblut et al. [80] (accession numbers AY541534 and AY541575) and Taton et al. [81] (accession number DQ181670). The phylogenetic tree of the 16S rRNA gene sequence of P. pseudopriestleyi and the most closely related operational taxonomic units (OTUs) in Jungblut et al. [13] was used for context.
3. Results
3.1. Sulfide Concentrations
Total sulfide ([H2S] + [HS−]) was measured to be <0.01 mg L−1 at 9.5 m and above. At 9.8 m, total sulfide was 0.091 mg L−1 and rose to 2.2 mg L−1 at 10.7 m depth. See Table 1 for all measurements.

Table 1.
Total sulfide measurements on Lake Fryxell water samples collected on 17 January 2020.
3.2. Assembly and Binning Statistics
Thirty-one bins were retrieved from the coassembly from the environmental biofilm samples and the strain P. pseudopriestleyi (see Table 2 for assembly statistics and Table 3 binning statistics), but there was only one bin belonging to the phylum Cyanobacteria based on comparison with GTDB-tk on KBASE database [13]. This bin is 5.97 MB with 42,524 reads, 678 contigs, 4738 protein coding genes, and a GC content of 62.32%. The longest contig generated was 400,416 bp. The closest relative to P. pseudopriestleyi according to 16S rRNA OTUs, Oscillatoria acuminata PCC 6304 has a genome size of 7.7 MB and contains 5,687 protein coding genes. The P. pseudopriestleyi MAG is 91.73% complete, has 1.35% contamination and 8.33% strain heterogeneity. Because the bin used for analysis is incomplete, it may not contain genes that are present in the organism’s genome. However, the MAG is sufficiently complete that it can be used to analyze the genomic potential for key metabolisms. A complete list of KEGG annotated genes is available in Supplementary Table S1.

Table 2.
Quality metrics of coassembly generated from sequencing of the mat and laboratory culture samples. All statistics are from QUAST v4.4, except for mapping statistics which were generated from samtools v1.9 using the flagstat parameter.

Table 3.
Quality metrics of the P. pseudopriestleyi metagenome-assembled genome (MAG). All statistics are from QUAST v4.4, except for completion and contamination statistics which were generated from CheckM v1.0.7 and the number of protein coding genes from Prokka v1.11.
3.3. Taxonomic Assignment of MAG
The MAG obtained from P. pseudopriestleyi was classified as family Oscillatoriaceae and genus Oscillatoria based on comparison with GTDB-tk on KBASE database [13]. The P. pseudopriestleyi MAG and O. acuminata PCC 6304 had an ANI of 88.99%, with a standard deviation of 3.25% based on 14,129 fragments. The P. pseudopriestleyi MAG and P. priestleyi BC1401 had an ANI of 75.69% with a standard deviation of 5.94% based on 75 fragments. The ANI score between the P. pseudopriestleyi MAG and P. priestleyi ULC007 was 72.03% with a standard deviation of 3.65% based on 72 fragments. Each of the ANIs between the P. pseudopriestleyi MAG and A. vandensis, Synechococcus sp. CS-601, and Leptolyngbya sp. BC1307 were less than 70%. These results suggest that the P. pseudopriestleyi MAG was most similar to O. acuminata [57,79,82]. However, additional cyanobacteria strains in Oscillatoriaceae need to be isolated and sequenced from Antarctica to allow phylogenomic interference to better resolve the relationship between Phormidium and Oscillatoria.
3.4. Photosynthetic and Electron Transport Machinery
The P. pseudopriestleyi MAG contains genes for the pigments phycoerythrocyanin, phycocyanin, and allophycocyanin, which have absorption peaks at 575, 620, and 650 nm, respectively, and lacks the gene for phycoerythrin, the light-harvesting protein that absorbs wavelengths from 495 to 560 nm [83] (Table 4). The genes for the D1/D2 protein cluster in PS II (psbA and psbD) are present along with two genes for proteins that hold the Mn4CaO5 OEC cluster in place (psbP and psbO) [25]. The MAG contains four of the eight subunits that make up the cytochrome b6f complex (petB, petD, petA, and petC). Notably, petA codes for apocytochrome f and petC codes for the iron-sulfur subunit within the b6f complex. The genes for the remaining subunits, petL, petM, petN, and petG, were not identified in the bin (Table 4). The majority of PS I genes are present in the MAG, including the core chlorophyll dimer made up of psaA and psaB (Table 4). Genes for both ferredoxin (petF) and ferredoxin-NADP+ reductase (petH) are present. The MAG does not contain petE, which codes for the electron transport protein plastocyanin that connects PS II to PS I, however cytochrome c6 encoded by petJ can perform the same function [84]. The MAG has all the genes necessary to create an F-type ATPase. Notably, the MAG has a type II sqr (Figure 2), which is necessary for sulfide oxidation in AP. The MAG also contains genes encoding arsenic resistance (arsR and acr3) that may be involved with transcriptional regulation of sqr [85].

Table 4.
Phycobilisome, photosynthesis, and respiratory machinery genes present in the P. pseudopriestleyi MAG generated with GhostKoala v2.2.
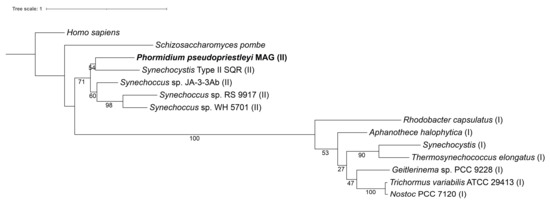
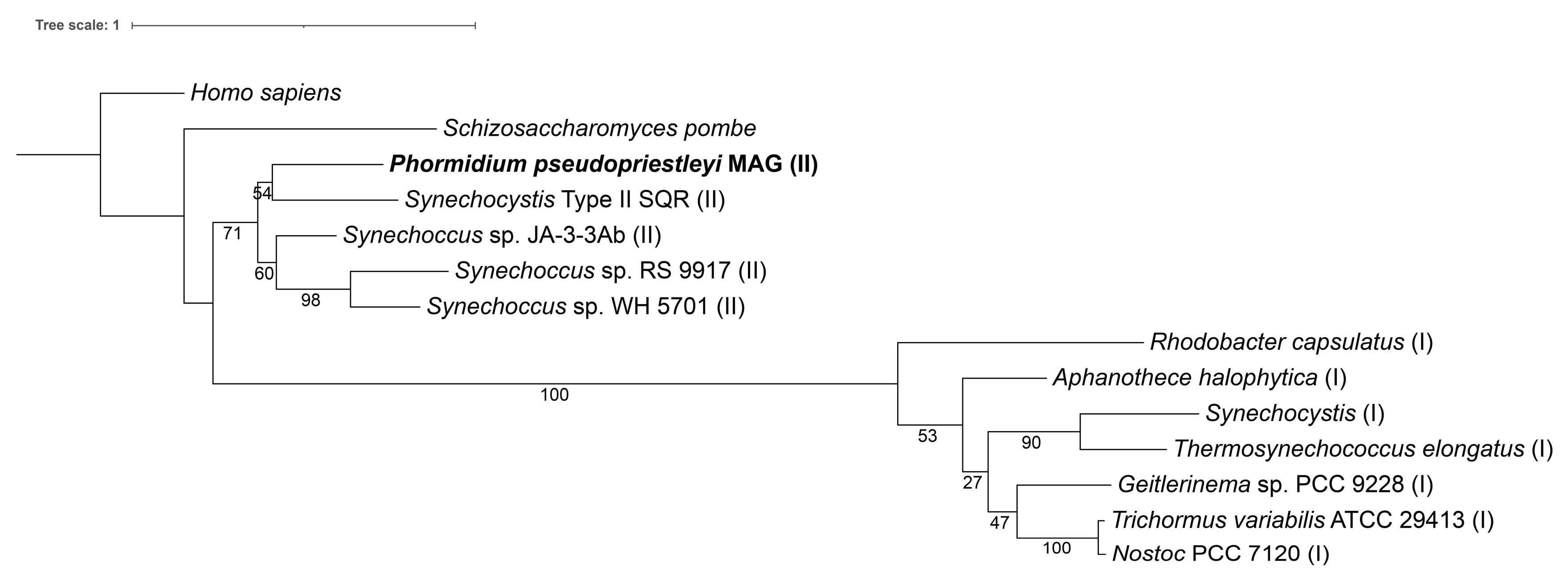
Figure 2.
A maximum likelihood tree of type I and II SQR amino acid sequences from Shahak and Hauska, 2008. Homo sapiens (accession number AAH16836), Schizosaccharomyces pombe (accession number CAA21882), Phormidium pseudopriestleyi (presented in paper), Synechocystis SQR-type II (accession number WP_010872226), Synechococcus strain sp. JA-3-3Ab (accession number ABD00861), Synechococcus sp. RS 9917 (accession number EAQ69368), Synechococcus strain WH 5701 (accession number EAQ74835), Rhodobacter capsulatus (accession number CAA66112), Aphanothece halophytica (accession number AAF72963), Synechocystis SQR-type I (accession number WP_011153573), Thermosynechococcus elongatus (accession number WP_011056143), Geitlerinema sp. PCC 9228 (formerly known as Oscillatoria limnetica, accession number AAF72962), Trichormus variabilis ATCC 29413 (formerly known as Anabaena variabilis accession number ABA22985), and Nostoc PCC 7120 (accession number WP_010998645). A type I SQR is indicated by (I) after the organism name, while SQR type II is indicated by (II).
The MAG contains all genes for a type 1 NADH dehydrogenase except for ndhN, subunit n of the complex. The succinate dehydrogenase gene sdhD, encoding a membrane anchor subunit, is absent but sdhA, shdB, and sdhC are present. The genes ctaC, ctaD, and ctaE for aa3-type cytochrome c oxidase genes are present but ctaF is not. The genes for cytochrome bd-quinol oxidase cydA and cydB are present, but cydX is not. Genes for alternative respiratory terminal oxidase and plastid terminal oxidase were not found in the MAG [86,87].
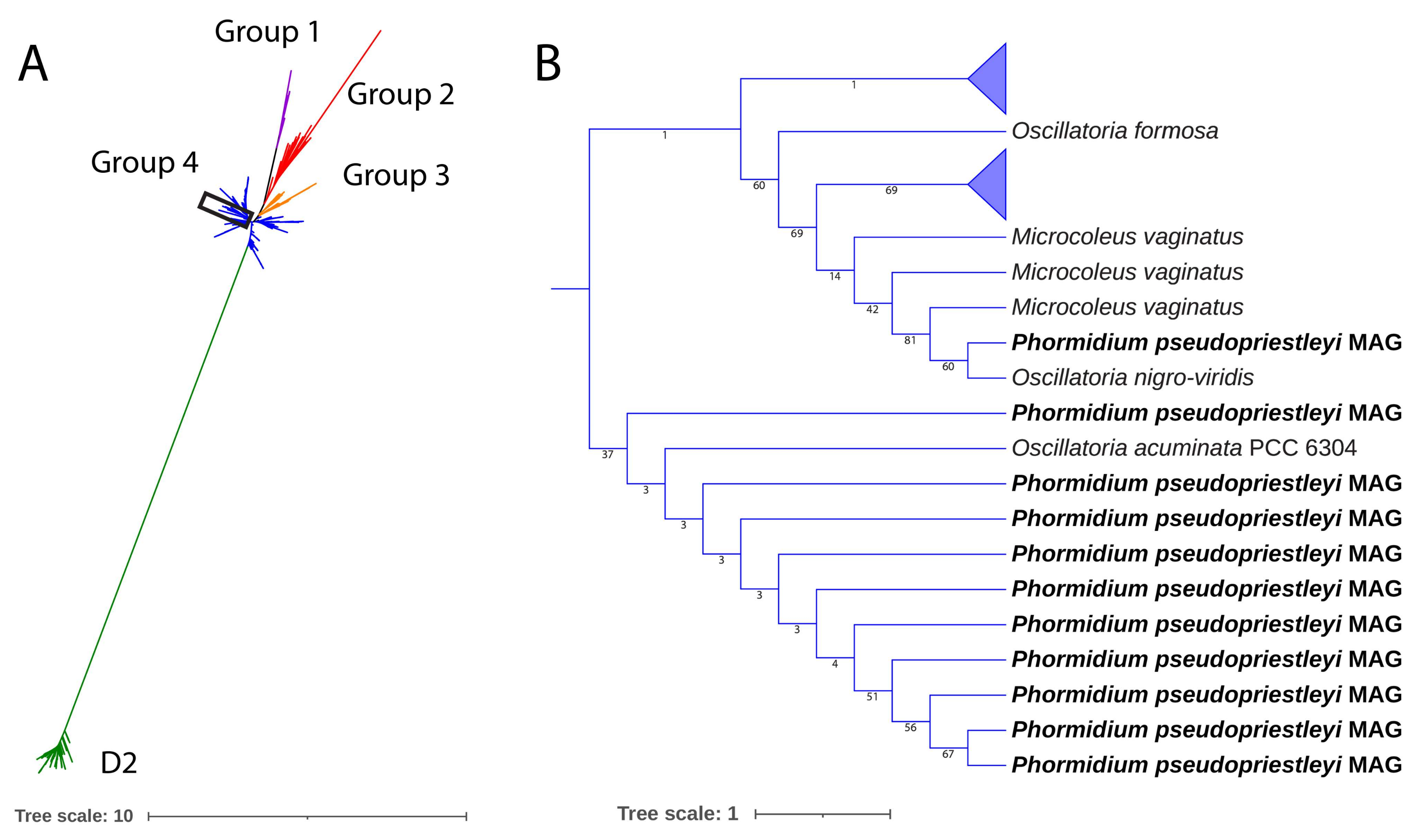
Because of the importance of the D1 protein to OP, we examined psbA sequence in more detail. Although spacegraphcats software was used to refine the MAG, a full copy of the gene for the D1 protein could not be obtained. Fragments of the D1 protein sequence are present, including the C-terminal and N-terminal portions ranging between 80 and 264 amino acids out of the full length 360 amino acids protein sequence (Alignment S1). Additionally present are all seven of the amino acids involved in ligating with the Mn4CaO5 cluster in the OEC: Asp170, Glu189, His332, Glu333, His337, Asp342, and Ala344 [88,89]. Each of these amino acids were present in fragments that contained the appropriate part of the protein sequence for psbA. A phylogenetic tree was constructed of the MAG’s D1 fragments and reference sequences from all four D1 groups, and all fragments grouped closely with group 4 D1 proteins, demonstrating that the MAG does not contain an alternative version of the D1 protein (Figure 3). Where overlapping, the sequences of the fragments are not identical, indicating the presence of at least two copies of the D1 gene in the MAG.
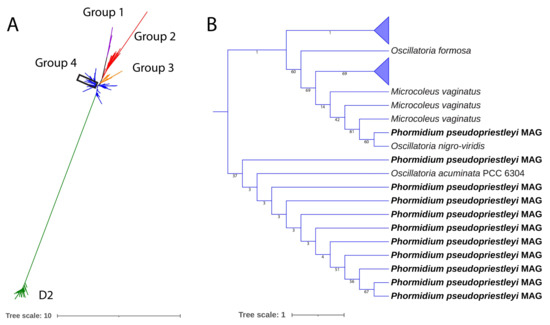
Figure 3.
(A) a maximum likelihood tree of D1 and D2 proteins of sequences presented in Cardona et al. [25] and the 11 D1 protein fragments in the P. pseudopriestleyi MAG. All P. pseudopriestleyi fragments (enclosed in the rectangle) grouped with group 4 D1 proteins. (B) region of the tree showing P. pseudopriestleyi and the most closely related D1 proteins.
3.5. Metabolic Pathways
The MAG contains genetic capacity for the Calvin cycle for carbon fixation. The MAG also contains genes for glycolysis via the Embden–Meyerhof pathway except for tpiA encoding for triosephosphate isomerase. Capacity for the tricarboxylic acid cycle is present except for genes for 2-oxoglutarate dehydrogenase, which is absent in cyanobacteria, and fumarate hydratase. The MAG contains genes for the pentose phosphate pathway and glycogen and trehalose biosynthesis pathways. It also contains full capacity for the initiation, elongation, and β-oxidation of fatty acids (Table 5).

Table 5.
Number of genes present in the P. pseudopriestleyi MAG in functional categories based on KEGG annotations. A full list of these genes is available in supplemental materials.
All genes necessary for assimilatory sulfate reduction are present in the MAG (sat, cysNC, cysH, and sir), but essential genes for dissimilatory sulfur metabolism are not (aprAB and dsrAB). Additionally, the MAG has the genes necessary for a sulfate ion transport system through a membrane (cysPUWA, sbp). The presence of narB and nirA indicate capacity for assimilatory nitrate reduction, but the MAG does not contain genes for dissimilatory nitrogen metabolism or nitrogen fixation [90]. Although 19 genes associated with methane metabolism were found in the MAG, most of them are involved with various biosynthesis pathways, and there is not a full pathway for methanogenesis or methanotrophy. Notably, hdrA2, hdrB2, and hdrC2, are present, which code for heterodisulfide reductase, an enzyme typically found in methanogens. Previous work has found these genes at 9.8 m depth in Lake Fryxell, and consistent with the content of the MAG, the capacity for methanogenesis (hdrD) was absent [91]. The presence of hdrB may indicate capacity for flavin-based electron bifurcation [92].
3.6. Genes Implicated in the Adaption to Environmental Stress
P. pseudopriestleyi encodes some genes related to osmotic stress. Specifically, the MAG contains several genes related to sodium and potassium antiporters (nhaS2, nhaS3, mrpA, mrpC, trk, and ktr). Additionally, the MAG contains treZ and treY, which support a trehalose biosynthesis pathway, a compatible solute that has been found to have membrane protective features, particularly in filamentous, mat-forming cyanobacteria strains [93]. Trehalose has also been found in cyanobacteria tolerant of desiccation [94,95,96]. The MAG does not contain genes related to glucosylglycercol or glycine betaine, which are compatible solutes that have been identified in halotolerant and halophilic cyanobacteria strains [97]. Sucrose can also be used as an osmolyte in cyanobacteria, and the MAG contains spsA, encoding sucrose phosphate synthase which supports the production of sucrose 6-phosphate, but not spp, encoding sucrose phosphate phosphatase, which is necessary to produce sucrose [98].
Some cyanobacteria synthesize the compounds scytonemin and mycosporine to overcome the harmful effects of long-term UV radiation exposure [99]. The MAG contains no genes related to the biosynthesis of these compounds, though the pathways are not fully understood [100,101]. The photoprotective proteins such as orange carotenoid protein (ocp) and fluorescence recovery protein (frp) protect against high light stress by converting excess excitation energy to heat [87], but the MAG does not contain either of these genes. Another method of dealing with high light stress is to divert electrons away from the photosynthetic electron transport chain using electron valves to prevent over-reduction of photosynthetic machinery. The MAG contains genetic capacity for cyanobacterial electron valves flavodiiron proteins Flv1-4 and another cyanobacterial bidirectional hydrogenase. Additionally, the MAG contains genes for terminal oxidases cytochrome bd-1 and cytochrome c oxidase, which can act as electron valves [87]. Carotenoids are another important molecule for photooxidative stress. The MAG contains crtE, crtB, crtP, crtH, crtQ, cruF, cruG, crtR, and crtO, allowing for carotenoid biosynthesis of myxol-2′-dimehtylfucoside, β-carotene, zeaxanthin, echinenone, and 3’-hydroxyechinenone. It is missing cruE and cruH, and thus does not demonstrate a capacity to produce the carotenoid synechoxanthin [102,103,104]. Additional carotenoid biosynthesis genes crtI_1, crtI_2, and crtI_3 for lycopene and neurosporene phytoene desaturase are present. Although chlorophyll F has been shown to support near-infrared OP, the gene for chlorophyll F synthase (chlf) is not present in the MAG [105].
The MAG contains genes relating to replication, transcription, and translation associated with cold-tolerant organisms. It has the gene for DNA gyrase (gyrA) that helps uncoil DNA that is tightly wound at cold temperatures [106]. The MAG contains genes associated with cold-adapted ribosomal function, including ribosome-binding factor A (rbfA) as well as genes for translational factors Initiation Factor 1 and 2 (IF1 and IF2) (infA, infB) [106]. Genes for a ribosomal rescue system ssrA and smpB are also present. Genes for delta (12)-fatty-acid desaturase (desA) and NADPH-dependent stearoyl-CoA 9-desaturase (desA3) are present in the MAG. These produce unsaturated and branched fatty acids, which help organisms maintain membrane integrity at lower temperatures. None of the csp cold shock proteins are present in the MAG.
Cyanophycin is a copolymer of aspartic acid and arginine that stores nitrogen for when environmental nitrogen levels become deficient [107]. The MAG contains genes for both cyanophycin synthetase to build the polymer (cphA) and cyanophycinase (cphB) to break it down.
4. Discussion
4.1. Sulfide Resistance in P. pseudopriestleyi
Knowing the concentration of sulfide present in the mat at 9.8 m depth is important for understanding the extent of OP inhibition experienced by P. pseudopriestleyi in Lake Fryxell during the winter to spring transition. The early spring concentration was likely higher than the 0.091 mg L−1 sulfide measured at this depth (Table 1), because water samples were collected in mid-January, several months after initiation of oxygenic photosynthesis, which results in sulfide oxidation. Thus, the sulfide concentrations reported here represent minimums for early spring OP inhibition.
Both prior research [12,13] and our field work demonstrate that P. pseudopriestleyi sustains OP in an environment with at least 0.091 mg L−1 sulfide which suggests the presence of a tolerance mechanism for sulfide. One option for a sulfide tolerance mechanism is that the D1 protein is repaired and replaced while OP occurs (response 3 or 4 in Figure 1). Another possible mechanism is that SQR production supports AP in the presence of sulfide (response 2 or 4 in Figure 1). Additionally, the low irradiance in Lake Fryxell may contribute to P. pseudopriestleyi’s sulfide tolerance by minimizing photodamage to the D1 protein and allowing OP to occur at low rates (response 2 in Figure 1).
Sulfide inhibits water from interacting with the water-splitting Mn4CaO5 cluster of the OEC in the D1 protein of PS II (encoded by psbA), preventing the use of H2O as an electron donor and consequently OP [15]. Expression of psbA is increased in cyanobacteria exposed to light stress or DCMU, which inhibits the quinone binding site of PS II and thus electron transfer to PS I [21,22,23]. Similar increased expression of psbA may also occur in response to sulfide stress, although this effect has not been studied in cyanobacteria. P. pseudopriestleyi has multiple copies of the D1 protein, which may assist with increasing expression of psbA. Even though P. pseudopriestleyi grows in a low O2 environment, the MAG appears to only contain genetic capacity for the standard group 4 D1 protein, and there is no evidence that the organism has a microaerobic D1 protein. Thus, although the MAG is incomplete, P. pseudopriestleyi does not appear to use an alternative D1 as part of its sulfide tolerance strategy. If its tolerance mechanism is related to the D1 protein, P. pseudopriestleyi likely overcomes sulfide inhibition by either increasing psbA expression (response 2 in Figure 1) or activating a mechanism that has not been identified. In this scenario, O2 produced from OP will oxidize sulfide in the abiotic reaction presented in reaction 1 at a quick enough rate to prevent sulfide inhibition. This oxidation is passive and may be kinetically very slow at Lake Fryxell temperatures (2 °C), but includes positive feedback between decreasing sulfide and increasing capacity for O2 production.
Alternatively, P. pseudopriestleyi may employ AP by directly oxidizing sulfide to S0 with the membrane protein SQR (response 3 or 4 in Figure 1). This process would deplete sulfide, allowing OP. Cyanobacteria capable of AP pass electrons from SQR oxidation through the quinone pool to PS I to harvest energy [16]. If P. pseudopriestleyi transfers electrons from SQR oxidation to PS I, it gains energy through AP, while sulfide is being depleted. The MAG contains the sqr gene and may be capable of this response. However, laboratory incubation experiments are necessary to determine if the cyanobacterium performs AP, regardless of sqr in the genome [10,26,108].
4.2. Ecology of P. pseudopriestleyi in Lake Fryxell
The seasonality of Lake Fryxell controls how P. pseudopriestleyi shapes the redox potential of its environment by producing O2 through OP. Complete darkness from mid-April to mid-August allows sulfide to accumulate in the mats, creating a reduced, sulfidic environment [12]. As light becomes available starting in mid-August, OP or AP may initiate, leading to the oxidation of sulfide. Constant summer irradiance starts at the end of October, with irradiance sufficient to allow OP, resulting in the accumulation of O2 the mats [12]. OP slows down as light levels fall from mid-February to mid-April and then ceases in total darkness, and sulfide reaccumulates.
The low irradiance in P. pseudopriestleyi’s spring and summer environment may contribute to its sulfide resistance. Even during peak irradiance in the summer, the daily mean photon flux at 9.8 m depth is 1–2 µmol m−2 s−1. Previous research demonstrates that low irradiance allowed the hot spring cyanobacteria Planktothrix str. FS34 to perform OP uninhibited in up to 230 µM sulfide (or 7.83 mg L−1 sulfide) even though sulfide inhibited photosynthesis at higher light fluxes [18]. Lower light levels may reduce photo-damage on the photosensitive D1 protein, allowing more D1 proteins in the thylakoid membrane to perform sulfide-tolerant OP. If sulfide levels are below the threshold for OP inhibition, the O2 from OP will oxidize sulfide, eventually allowing O2 to accumulate. If this mechanism is happening in Lake Fryxell, the balance of irradiance and sulfide levels in Lake Fryxell plays an important role in P. pseudopriestleyi’s ability to perform OP in an extreme environment.
The effect of low irradiance on P. pseudopriestleyi is amplified by a partial mismatch between the wavelengths of light available and the pigments it can produce. The most penetrating waveband at 9 m in Lake Fryxell is 520–580 nm because the ice cover transmits blue light and absorbs wavelengths longer than 600 nm [38,109]. The MAG contains genes for phycoerythrocyanin, phycocyanin, and allophycocyanin, which have peak absorptions at 575, 620, and 650 nm, respectively. Without the genes for phycoerythrin, absorption of wavelengths between 495–560 nm is limited. The combination of pigments and available light suggests that phycoerythrocyanin harvests most of the light for photosynthesis in the Lake Fryxell environment. Although there is a mismatch between available light and pigments for photosynthesis, the low irradiance of the environment is consistent with the absence of UV exposure genes in the MAG.
Continuous illumination during summer, even at low levels, requires a persistent nutrient source. The MAG has genes to create and break down cyanophycin, a nitrogen storage molecule synthesized by cyanobacteria. This may aid P. pseudopriestleyi in meeting peak nitrogen demands during the summer in Lake Fryxell, which is nitrogen limited [110].
4.3. P. pseudopriestleyi in Other Environments
In addition to dominating a narrow sulfide-rich photic benthic zone of Lake Fryxell, P. pseudopriestleyi has been reported from several ponds and lakes across Antarctica, based on 16S rRNA gene analyses [80,81]. P. pseudopriestleyi’s 16S gene sequence reported in Jungblut et al. [13] has a 99.45%, 91.28%, and 99.9% identity to 16S rRNA gene sequences from Antarctic Salt Pond, Fresh Pond, and Ace Lake, respectively (accession numbers AY541534, AY541575, and DQ181670) according to a blastn search. Salt and Fresh Ponds are meltwater ponds on the McMurdo Ice Shelf that experience high UV radiation [111]. Salt Pond is hypersaline with a conductivity of up to 52.9 mS cm−1, with high salinity originating from diluted seawater or sulfate salts from chemical weathering of sedimentary material [80,112,113]. Ace Lake is considered hyposaline with conductivity of 25.4–26.4 mS cm−1 and is permanently stratified [81,114]. P. pseudopriestleyi is present in both locations, suggesting it is adapted to varying levels of salinity, UV, and high light stress in addition to sulfide. Future DNA sequencing beyond 16S amplicons in these environments may reveal whether or not P. pseudopriestleyi possesses additional stress tolerance genes related to these conditions that are absent in the Lake Fryxell MAG.
5. Conclusions
P. pseudopriestleyi is the first example of a cyanobacterium capable of sulfide-tolerant OP in a cold, Antarctic environment. The MAG reconstructed for P. pseudopriestleyi revealed a genome that is consistent with its ability to produce O2 in a sulfidic, cold, low-light environment of the perennially ice-covered Lake Fryxell, Antarctica. The MAG has a genomic capacity to deal with sulfide with multiple copies of a psbA, the D1 protein that is the site of water splitting, or by using SQR to deplete sulfide through AP or through sulfide oxidation. The low light levels at 9.8 m in Lake Fryxell may also contribute to its sulfide tolerance. Thus, there are likely several methods for dealing with sulfide stress on OP in a low light environment. Sulfide tolerance varies widely among cyanobacteria, and the consideration of light level may have implications on a response to sulfide and should be studied further. Specifically, microelectrode measurements combined with gene expression data are likely to uncover the molecular mechanism P. pseudopriestleyi uses to perform sulfide-tolerant OP.
Besides Lake Fryxell, P. pseudopriestleyi has been found in shallow freshwater and hypersaline ice shelf melt water ponds and lakes, indicating a widespread distribution in Antarctica, and ability to thrive in a range of environmental conditions. If genomic data from P. pseudopriestleyi living in these environments can be obtained, a comparison with the MAG from Lake Fryxell can provide insight about the effects of various Antarctic conditions, such as UV exposure, high light levels, or salinity, on a genome. Additionally, the isolation and sequencing of other Antarctic cyanobacteria will allow for more in-depth genomic comparisons between Antarctic cyanobacteria genomes beyond the three genomes currently published.
A deeper understanding of the ecology of cold cyanobacteria ecosystems will provide insights into the production of O2 through Earth history. Specifically, primary productivity during “Snowball Earth” glaciations was required to sustain the biosphere through these climatic crises. In some cases, sulfide appears to have been abundant [3,4,5,115], suggesting that P. pseudopriestleyi may provide a model for how OP persisted in some “Snowball Earth” ecosystems.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/12/3/426/s1, Table S1: Full list of KEGG annotated genes in P. pseudopriestleyi MAG. Alignment S1: Alignment between D1 sequences in P. pseudopriestleyi MAG and D1 and D2 sequences from Cardona et al. [25].
Author Contributions
Author Contributions: Conceptualization, J.E.L., D.Y.S., A.D.J. and M.L.D.; methodology, J.E.L., D.Y.S., A.D.J., and C.L.G.; software, J.E.L.; validation, J.E.L.; formal analysis, J.E.L., M.L.D., and C.L.G.; investigation, J.E.L., D.Y.S., T.J.M., I.H., P.T.D., and M.L.D.; resources, D.Y.S., P.T.D., I.H., T.J.M., G.J.D., and A.D.J.; data curation, J.E.L., M.L.D., G.J.D., and A.D.J.; writing—original draft preparation, J.E.L., D.Y.S., A.D.J., and C.L.G.; writing—review and editing, J.E.L., A.D.J., M.L.D., I.H., P.T.D., T.J.M., G.J.D., C.L.G., and D.Y.S.; visualization, J.E.L., D.Y.S., A.D.J., and C.L.G.; supervision, D.Y.S.; project administration, D.Y.S. and J.E.L.; funding acquisition, D.Y.S., P.T.D., G.J.D., and J.E.L. All authors have read and agreed to the published version of the manuscript.
Funding
Support for this project was provided by NASA Astrobiology (Grant NN13AI60G), the National Science Foundation (Grants OPP-1745341 and EAR-1637066), and a NSF Graduate Research Fellowship to J.E.L. Funding for sample collection and field logistics was provided by the McMurdo Long Term Ecological Research project (NSF Grants OPP-115245 and OPP-1637708) and Antarctic New Zealand Program.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The high-throughput sequencing data are available via NCBI’s Sequence Read Archive. Metagenomic reads from the Lake Fryxell microbial mats are available at the accessions SAMN09937182-SAMN09937191. The P. pseudopriestleyi laboratory culture reads are available at the accession SAMN17478355. The P. pseudopriestleyi MAG has been deposited at GenBank under the accession JAFLQW000000000. The version described in this paper is version JAFLQW010000000.
Acknowledgments
We would like to thank Tanai Cardona for D1 and D2 sequences from Cardona et al. [25] and Colin Hillmann for field assistance.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Kasting, J.F. What Caused the Rise of Atmospheric O2? Chem. Geol. 2013, 362, 13–25. [Google Scholar] [CrossRef]
- Canfield, D.E. A New Model for Proterozoic Ocean Chemistry. Nature 1998, 396, 450–453. [Google Scholar] [CrossRef]
- Parnell, J.; Adrian, J.B. Microbial Sulphate Reduction during Neoproterozoic Glaciation, Port Askaig Formation, UK. J. Geol. Soc. 2017, 174, 850–854. [Google Scholar] [CrossRef]
- Dahl, T.W.; Canfield, D.E.; Rosing, M.T.; Frei, R.E.; Gordon, G.W.; Knoll, A.H.; Anbar, A.D. Molybdenum Evidence for Expansive Sulfidic Water Masses in ~750Ma Oceans. Earth Planet. Sci. Lett. 2011, 311, 264–274. [Google Scholar] [CrossRef]
- Wang, P.; Algeo, T.J.; Zhou, Q.; Yu, W.; Du, Y.; Qin, Y.; Xu, Y.; Yuan, L.; Pan, W. Large Accumulations of 34S-Enriched Pyrite in a Low-Sulfate Marine Basin: The Sturtian Nanhua Basin, South China. Precambrian Res. 2019, 335. [Google Scholar] [CrossRef]
- Voorhies, A.A.; Biddanda, B.A.; Kendall, S.T.; Jain, S.; Marcus, D.N.; Nold, S.C.; Sheldon, N.D.; Dick, G.J. Cyanobacterial Life at Low O2: Community Genomics and Function Reveal Metabolic Versatility and Extremely Low Diversity in a Great Lakes Sinkhole Mat. Geobiology 2012, 10, 250–267. [Google Scholar] [CrossRef] [PubMed]
- Quesada, A.; Vincent, W.F. Cyanobacteria in the Cryosphere: Snow, Ice and Extreme Cold. In Ecology of Cyanobacteria II: Their Diversity in Space and Time; Whitton, B.A., Ed.; Springer: Dordrecht, The Netherlands, 2012; pp. 387–399. [Google Scholar] [CrossRef]
- Klatt, J.M.; Meyer, S.; Häusler, S.; Macalady, J.L.; de Beer, D.; Polerecky, L. Structure and Function of Natural Sulphide-Oxidizing Microbial Mats under Dynamic Input of Light and Chemical Energy. ISME J. 2016, 10, 921–933. [Google Scholar] [CrossRef] [PubMed]
- Macalady, J.L.; Lyon, E.H.; Koffman, B.; Albertson, L.K.; Meyer, K.; Galdenzi, S.; Mariani, S. Dominant Microbial Populations in Limestone-Corroding Stream Biofilms, Frasassi Cave System, Italy. Appl. Environ. Microbiol. 2006, 72, 5596–5609. [Google Scholar] [CrossRef]
- De Beer, D.; Weber, M.; Chennu, A.; Hamilton, T.; Lott, C.; Macalady, J.; Klatt, J.M. Oxygenic and Anoxygenic Photosynthesis in a Microbial Mat from an Anoxic and Sulfidic Spring. Environ. Microbiol. 2017, 19, 1251–1265. [Google Scholar] [CrossRef]
- Hamilton, T.L.; Klatt, J.M.; de Beer, D.; Macalady, J.L. Cyanobacterial Photosynthesis under Sulfidic Conditions: Insights from the Isolate Leptolyngbya Sp. Strain Hensonii. ISME J. 2018, 12, 568–584. [Google Scholar] [CrossRef]
- Sumner, D.Y.; Hawes, I.; Mackey, T.J.; Jungblut, A.D.; Doran, P.T. Antarctic Microbial Mats: A Modern Analog for Archean Lacustrine Oxygen Oases. Geology 2015, 43, 887–890. [Google Scholar] [CrossRef]
- Jungblut, A.D.; Hawes, I.; Mackey, T.J.; Krusor, M.; Doran, P.T.; Sumner, D.Y.; Eisen, J.A.; Hillman, C.; Goroncy, A.K. Microbial Mat Communities along an Oxygen Gradient in a Perennially Ice-Covered Antarctic Lake. Appl. Environ. Microbiol. 2016, 82, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Oren, A.; Padan, E.; Malkin, S. Sulfide Inhibition of Photosystem II in Cyanobacteria (Blue-Green Algae) and Tobacco Chloroplasts. Biochim. Biophys. Acta 1979, 546, 270–279. [Google Scholar] [CrossRef]
- Miller, S.R.; Bebout, B.M. Variation in Sulfide Tolerance of Photosystem II in Phylogenetically Diverse Cyanobacteria from Sulfidic Habitats. Appl. Environ. Microbiol. 2004, 70, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Cohen, Y.; Jørgensen, B.B.; Revsbech, N.P.; Poplawski, R. Adaptation to Hydrogen Sulfide of Oxygenic and Anoxygenic Photosynthesis among Cyanobacteria. Appl. Environ. Microbiol. 1986, 51, 398–407. [Google Scholar] [CrossRef]
- Dick, G.J.; Grim, S.L.; Klatt, J.M. Controls on O2 Production in Cyanobacterial Mats and Implications for Earth’s Oxygenation. Annu. Rev. Earth Planet. Sci. 2018, 46. [Google Scholar] [CrossRef]
- Klatt, J.M.; Haas, S.; Yilmaz, P.; de Beer, D.; Polerecky, L. Hydrogen Sulfide Can Inhibit and Enhance Oxygenic Photosynthesis in a Cyanobacterium from Sulfidic Springs. Environ. Microbiol. 2015, 17, 3301–3313. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. Toward Understanding the Origin and Evolution of Cellular Organisms. Protein Sci. Publ. Protein Soc. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Bishop, N.I. The Influence of the Herbicide, DCMU, on the Oxygen-Evolving System of Photosynthesis. Biochim. Biophys. Acta 1958, 27, 205–206. [Google Scholar] [CrossRef]
- Mulo, P.; Sicora, C.; Aro, E. Cyanobacterial PsbA Gene Family: Optimization of Oxygenic Photosynthesis. Cell. Mol. Life Sci. 2009, 66, 3697. [Google Scholar] [CrossRef]
- Mulo, P.; Sakurai, I.; Aro, E. Strategies for PsbA Gene Expression in Cyanobacteria, Green Algae and Higher Plants: From Transcription to PSII Repair. Biochim. Biophys. Acta 2012, 1817, 247–257. [Google Scholar] [CrossRef]
- Summerfield, T.C.; Toepel, J.; Sherman, L.A. Low-Oxygen Induction of Normally Cryptic PsbA Genes in Cyanobacteria. Biochemistry 2008, 47, 12939–12941. [Google Scholar] [CrossRef] [PubMed]
- Cardona, T.; Murray, J.W.; Rutherford, A.W. Origin and Evolution of Water Oxidation before the Last Common Ancestor of the Cyanobacteria. Mol. Biol. Evol. 2015, 32, 1310–1328. [Google Scholar] [CrossRef]
- Bronstein, M.; Schütz, M.; Hauska, G.; Padan, E.; Shahak, Y. Cyanobacterial Sulfide-Quinone Reductase: Cloning and Heterologous Expression. J. Bacteriol. 2000, 182, 3336–3344. [Google Scholar] [CrossRef] [PubMed]
- Garlick, S.; Oren, A.; Padan, E. Occurrence of Facultative Anoxygenic Photosynthesis among Filamentous and Unicellular Cyanobacteria. J. Bacteriol. 1997, 129, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Cohen, Y.; Padan, E.; Shilo, M. Facultative Anoxygenic Photosynthesis in the Cyanobacterium Oscillatoria Limnetica. J. Bacteriol. 1975, 123, 855–861. [Google Scholar] [CrossRef]
- Cohen, Y.; Jørgensen, B.B.; Padan, E.; Shilo, M. Sulphide-Dependent Anoxygenic Photosynthesis in the Cyanobacterium Oscillatoria Limnetica. Nature 1975, 257, 489. [Google Scholar] [CrossRef]
- Green, W.J.; Lyons, W.B. The Saline Lakes of the McMurdo Dry Valleys, Antarctica. Aquat. Geochem. 2009, 15, 321–348. [Google Scholar] [CrossRef]
- McKnight, D.M.; Niyogi, D.K.; Alger, A.S.; Bomblies, A.; Conovitz, P.A.; Tate, C.M. Dry Valley Streams in Antarctica: Ecosystems Waiting for Water. BioScience 1999, 49, 985–995. [Google Scholar] [CrossRef]
- Lawrence, M.J.F.; Hendy, C.H. Water Column and Sediment Characteristics of Lake Fryxell, Taylor Valley, Antarctica. N. Z. J. Geol. Geophys. 1985, 28, 543–552. [Google Scholar] [CrossRef]
- Vincent, W.F. Production Strategies in Antarctic Inland Waters: Phytoplankton Eco-Physiology in a Permanently Ice-Covered Lake. Ecology 1981, 62, 1215–1224. [Google Scholar] [CrossRef]
- Lyons, W.B.; Frape, S.K.; Welch, K.A. History of McMurdo Dry Valley Lakes, Antarctica, from Stable Chlorine Isotope Data. Geology 1999, 27, 527–530. [Google Scholar] [CrossRef]
- Roberts, E.C.; Laybourn-Parry, J.; McKnight, D.M.; Novarino, G. Stratification and Dynamics of Microbial Loop Communities in Lake Fryxell, Antarctica. Freshw. Biol. 2000, 44, 649–661. [Google Scholar] [CrossRef]
- Laybourn-Parry, J.; James, M.R.; McKnight, D.M.; Priscu, J.; Spaulding, S.A.; Shiel, R. The Microbial Plankton of Lake Fryxell, Southern Victoria Land, Antarctica during the Summers of 1992 and 1994. Polar Biol. 1997, 17, 54–61. [Google Scholar] [CrossRef]
- Spigel, R.H.; Priscu, J.C. Physical Limnology of the McMurdo Dry Valleys Lakes. In Ecosystem Dynamics in a Polar Desert: The McMurdo Dry Valleys, Antarctica; American Geophysical Union (AGU): Washington, DC, USA, 2013; pp. 153–187. [Google Scholar] [CrossRef]
- Howard-Williams, C.; Schwarz, A.; Hawes, I.; Priscu, J.C. Optical Properties of the McMurdo Dry Valley Lakes, Antarctica. In Ecosystem Dynamics in a Polar Desert: The McMurdo Dry Valleys, Antarctica; American Geophysical Union (AGU): Washington, DC, USA, 2013; pp. 189–203. [Google Scholar] [CrossRef]
- Lizotte, M.P.; Priscu, J.C. Photosynthesis-Irradiance Relationships in Phytoplankton from the Physically Stable Water Column of a Perennially Ice-Covered Lake (Lake Bonney, Antarctica). J. Phycol. 1992, 28, 179–185. [Google Scholar] [CrossRef]
- Miner, G. Standard Methods for the Examination of Water and Wastewater, 21st Edition. Am. Water Works Assoc. J. 2006, 98, 130. [Google Scholar] [CrossRef]
- Hillman, C. Structure of Benthic Microbial Mat Assemblages in Lake Fryxell, Antarctica. 2013. Available online: https://ir.canterbury.ac.nz/handle/10092/8737 (accessed on 28 November 2018).
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria. Microbiology 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A Fast and Accurate Illumina Paired-End ReAd MergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Crusoe, M.R.; Alameldin, H.F.; Awad, S.; Boucher, E.; Caldwell, A.; Cartwright, R.; Charbonneau, A.; Constantinides, B.; Edvenson, G.; Fay, S.; et al. The Khmer Software Package: Enabling Efficient Nucleotide Sequence Analysis. F1000Research 2015, 4. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.; Luo, R.; Sadakane, K.; Lam, T. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. Available online: http://arxiv.org/abs/1303.3997 (accessed on 20 May 2019).
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Esen, Ö.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An Advanced Analysis and Visualization Platform for ‘omics Data. PeerJ 2015, 3. [Google Scholar] [CrossRef]
- Delmont, T.O.; Eren, A.M. Linking Pangenomes and Metagenomes: The Prochlorococcus Metapangenome. PeerJ 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Chaumeil, P.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A Toolkit to Classify Genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7. [Google Scholar] [CrossRef]
- Matsen, F.A.; Kodner, R.B.; Armbrust, E.V. Pplacer: Linear Time Maximum-Likelihood and Bayesian Phylogenetic Placement of Sequences onto a Fixed Reference Tree. BMC Bioinform. 2010, 11, 538. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast Genome and Metagenome Distance Estimation Using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Sukumaran, J.; Holder, M.T. DendroPy: A Python Library for Phylogenetic Computing. Bioinformatics 2010, 26, 1569–1571. [Google Scholar] [CrossRef] [PubMed]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array Programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. Comput. Resour. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.W.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.T.; Moritz, D.; O’Brien, M.P.; Reidl, F.; Reiter, T.; Sullivan, B.D. Exploring Neighborhoods in Large Metagenome Assembly Graphs Using Spacegraphcats Reveals Hidden Sequence Diversity. Genome Biol. 2020, 21, 164. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Shahak, Y.; Hauska, H. Advances in Photosynthesis and Respiration. In Sulfur Metabolism in Phototrophic Organisms, Sulfide Oxidation from Cyanobacteria to Humans: Sulfide–Quinone Oxidoreductase (SQR); Springer: Dordrecht, The Netherlands, 2008; pp. 319–335. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldónm, T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Chrismas, N.A.M.; Williamson, C.J.; Yallop, M.L.; Anesio, A.M.; Sánchez-Baracaldo, P. Photoecology of the Antarctic Cyanobacterium Leptolyngbya sp. BC1307 Brought to Light through Community Analysis, Comparative Genomics and in Vitro Photosphysiology. Mol. Ecol. 2018, 27, 5279–5293. [Google Scholar] [CrossRef]
- Grettenberger, C.L.; Sumner, D.Y.; Wall, K.C.; Brown, C.T.; Eisen, J.A.; Mackey, T.J.; Hawes, I.; Jospin, G.; Jungblut, A.D. A Phylogenetically Novel Cyanobacterium Most Closely Related to Gloeobacter. ISME J. 2020, 14, 2142–2152. [Google Scholar] [CrossRef]
- Tang, J.; Du, L.-M.; Lian, Y.-M.; Daroch, M. Complete Genome Sequence and Comparative Analysis of Synechococcus sp. CS-601 (SynAce01), a Cold-Adapted Cyanobacterium from an Oligotrophic Antarctic Habitat. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Lara, Y.; Durieu, B.; Cornet, L.; Verlaine, O.; Rippka, R.; Pessi, I.S.; Misztak, A.; Joris, B.; Javaux, E.J.; Baurain, D.; et al. Draft Genome Sequence of the Axenic Strain Phormidesmis priestleyi ULC007, a Cyanobacterium Isolated from Lake Bruehwiler (Larsemann Hills). Genome Announc. 2017, 5. [Google Scholar] [CrossRef]
- Chrismas, N.A.M.; Barker, G.; Anesio, A.M.; Sánchez-Baracaldo, P. Genomic Mechanisms for Cold Tolerance and Production of Exopolysaccharides in the Arctic Cyanobacterium Phormidesmis Priestleyi BC1401. BMC Genom. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Shih, P.M.; Wu, D.; Latifi, A.; Axen, S.D.; Fewer, D.P.; Talla, E.; Calteau, A.; Cai, F.; de Marsac, N.T.; Rippka, R.; et al. Improving the Coverage of the Cyanobacterial Phylum Using Diversity-Driven Genome Sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 1053–1058. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The Enveomics Collection: A Toolbox for Specialized Analyses of Microbial Genomes and Metagenomes. PeerJ Inc. 2016. [Google Scholar] [CrossRef]
- Jungblut, A.D.; Hawes, I.; Mountfort, D.; Hitzfeld, B.; Dietrich, D.R.; Burns, B.P.; Neilan, B.A. Diversity within Cyanobacterial Mat Communities in Variable Salinity Meltwater Ponds of McMurdo Ice Shelf, Antarctica. Environ. Microbiol. 2005, 7, 519–529. [Google Scholar] [CrossRef]
- Taton, A.; Grubisic, S.; Balthasart, P.; Hodgson, D.A.; Laybourn-Parry, J.; Wilmotte, A. Biogeographical Distribution and Ecological Ranges of Benthic Cyanobacteria in East Antarctic Lakes. FEMS Microbiol. Ecol. 2006, 57, 272–289. [Google Scholar] [CrossRef] [PubMed]
- Barco, R.A.; Garrity, G.M.; Scott, J.J.; Amend, J.P.; Nealson, K.H.; Emerson, D. A Genus Definition for Bacteria and Archaea Based on a Standard Genome Relatedness Index. MBio 2020, 11. [Google Scholar] [CrossRef]
- Bogorad, L. Phycobiliproteins and Complementary Chromatic Adaptation. Annu. Rev. Plant Physiol. 1975, 26, 369–401. [Google Scholar] [CrossRef]
- Durán, R.V.; Hervás, M.; de la Rosa, M.; Navarro, J.A. The Efficient Functioning of Photosynthesis and Respiration in Synechocystis sp. PCC 6803 Strictly Requires the Presence of Either Cytochrome c6 or Plastocyanin. J. Biol. Chem. 2004, 279, 7229–7233. [Google Scholar] [CrossRef] [PubMed]
- Nagy, C.I.; Vass, I.; Rákhely, G.; Vass, I.Z.; Tóth, A.; Duzs, A.; Peca, L.; Kruk, J.; Kós, P.B. Coregulated Genes Link Sulfide:Quinone Oxidoreductase and Arsenic Metabolism in Synechocystis Sp. Strain PCC6803. J. Bacteriol. 2014, 196, 3430–3440. [Google Scholar] [CrossRef]
- Lea-Smith, D.J.; Bombelli, P.; Vasudevan, R.; Howe, C.J. Photosynthetic, Respiratory and Extracellular Electron Transport Pathways in Cyanobacteria. Biochim. Biophys. Acta 2016, 1857, 247–255. [Google Scholar] [CrossRef]
- Mullineaux, C.W. Electron Transport and Light-Harvesting Switches in Cyanobacteria. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.W. Sequence Variation at the Oxygen-Evolving Centre of Photosystem II: A New Class of ‘Rogue’ Cyanobacterial D1 Proteins. Photosynth. Res. 2012, 110, 177–184. [Google Scholar] [CrossRef]
- Vermaas, W.; Charité, J.; Shen, G.Z. Glu-69 of the D2 Protein in Photosystem II Is a Potential Ligand to Mn Involved in Photosynthetic Oxygen Evolution. Biochemistry 1990, 29, 5325–5332. [Google Scholar] [CrossRef]
- Varin, T.; Lovejoy, C.; Jungblut, A.D.; Vincent, W.F.; Corbeil, J. Metagenomic Profiling of Arctic Microbial Mat Communities as Nutrient Scavenging and Recycling Systems. Limnol. Oceanogr. 2010, 55, 1901–1911. [Google Scholar] [CrossRef]
- Dillon, M.L.; Hawes, I.; Jungblut, A.D.; Mackey, T.J.; Eisen, J.A.; Doran, P.T.; Sumner, D.Y. Energetic and Environmental Constraints on the Community Structure of Benthic Microbial Mats in Lake Fryxell, Antarctica. FEMS Microbiol. Ecol. 2020, 96. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.R.; Grein, F.; Oliveira, G.P.; Venceslau, S.S.; Keller, K.L.; Wall, J.D.; Pereira, I.A.C. The FlxABCD-HdrABC Proteins Correspond to a Novel NADH Dehydrogenase/Heterodisulfide Reductase Widespread in Anaerobic Bacteria and Involved in Ethanol Metabolism in Desulfovibrio Vulgaris Hildenborough. Environ. Microbiol. 2015, 17, 2288–2305. [Google Scholar] [CrossRef] [PubMed]
- Hincha, D.K.; Hagemann, M. Stabilization of Model Membranes during Drying by Compatible Solutes Involved in the Stress Tolerance of Plants and Microorganisms. Biochem. J. 2004, 383, 277–283. [Google Scholar] [CrossRef]
- Hershkovitz, N.; Oren, A.; Cohen, Y. Accumulation of Trehalose and Sucrose in Cyanobacteria Exposed to Matric Water Stress. Appl. Environ. Microbiol. 1991, 57, 645–648. [Google Scholar] [CrossRef]
- Page-Sharp, M.; Behm, C.A.; Smith, G.D. Involvement of the Compatible Solutes Trehalose and Sucrose in the Response to Salt Stress of a Cyanobacterial Scytonema Species Isolated from Desert Soils. Biochim. Biophys. Acta 1999, 1472, 519–528. [Google Scholar] [CrossRef]
- Potts, M.; Slaughter, S.M.; Hunneke, F.U.; Garst, J.F.; Helm, R.F. Desiccation Tolerance of Prokaryotes: Application of Principles to Human Cells. Integr. Comp. Biol. 2005, 45, 800–809. [Google Scholar] [CrossRef]
- Hagemann, M. Molecular Biology of Cyanobacterial Salt Acclimation. FEMS Microbiol. Rev. 2011, 35, 87–123. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, F.; Klähn, S.; Hagemann, M. Salt-Regulated Accumulation of the Compatible Solutes Sucrose and Glucosylglycerol in Cyanobacteria and Its Biotechnological Potential. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Ehling-Schulz, M.; Scherer, S. UV Protection in Cyanobacteria. Eur. J. Phycol. 1999, 34, 329–338. [Google Scholar] [CrossRef]
- Wada, N.; Sakamoto, T.; Matsugo, S. Multiple Roles of Photosynthetic and Sunscreen Pigments in Cyanobacteria Focusing on the Oxidative Stress. Metabolites 2013, 3, 463–483. [Google Scholar] [CrossRef]
- Sorrels, C.M.; Proteau, P.J.; Gerwick, W.H. Organization, Evolution, and Expression Analysis of the Biosynthetic Gene Cluster for Scytonemin, a Cyanobacterial UV-Absorbing Pigment. Appl. Environ. Microbiol. 2009, 75, 4861–4869. [Google Scholar] [CrossRef]
- Mills, L.A.; McCormick, A.J.; Lea-Smith, D.J. Current Knowledge and Recent Advances in Understanding Metabolism of the Model Cyanobacterium Synechocystis sp. PCC 6803. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Raven, J.A. Aquatic Photosynthesis, 2nd ed.; Princeton University Press: Princeton, NJ, USA, 2013. [Google Scholar]
- Zhu, Y.; Graham, J.E.; Ludwig, M.; Xiong, W.; Alvey, R.M.; Shen, G.; Bryant, D.A. Roles of Xanthophyll Carotenoids in Protection against Photoinhibition and Oxidative Stress in the Cyanobacterium Synechococcus Sp. Strain PCC 7002. Arch. Biochem. Biophys. 2010, 504, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Behrendt, L.; Brejnrod, A.; Schliep, M.; Sørensen, S.J.; Larkum, A.W.D.; Kühl, M. Chlorophyll f -Driven Photosynthesis in a Cavernous Cyanobacterium. ISME J. 2015, 9, 2108–2111. [Google Scholar] [CrossRef]
- Rodrigues, D.F.; Tiedje, J.M. Coping with Our Cold Planet. Appl. Environ. Microbiol. 2008, 74, 1677–1686. [Google Scholar] [CrossRef] [PubMed]
- Watzer, B.; Forchhammer, K. Cyanophycin Synthesis Optimizes Nitrogen Utilization in the Unicellular Cyanobacterium Synechocystis Sp. Strain PCC 6803. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef]
- Klatt, J.M.; Al-Najjar, M.A.A.; Yilmaz, P.; Lavik, G.; de Beer, D.; Polerecky, L. Anoxygenic Photosynthesis Controls Oxygenic Photosynthesis in a Cyanobacterium from a Sulfidic Spring. Appl. Environ. Microbiol. 2015, 81, 2025–2031. [Google Scholar] [CrossRef] [PubMed]
- Howard-Williams, C.; Pridmore, R.; Downes, M.T.; Vincent, W.F. Microbial Biomass, Photosynthesis and Chlorophyll a Related Pigments in the Ponds of the McMurdo Ice Shelf, Antarctica. Antarct. Sci. 1989, 1, 125–131. [Google Scholar] [CrossRef]
- Priscu, J.C. Phytoplankton Nutrient Deficiency in Lakes of the McMurdo Dry Valleys, Antarctica. Freshw. Biol. 1995, 34, 215–227. [Google Scholar] [CrossRef]
- Roos, J.C.; Vincent, W.F. Temperature Dependence of UV Radiation Effects on Antarctic Cyanobacteria. J. Phycol. 1998, 34, 118–125. [Google Scholar] [CrossRef]
- Keys, J.R.; Williams, K. Origin of Crystalline, Cold Desert Salts in the McMurdo Region, Antarctica. Geochim. Cosmochim. Acta 1981, 2299–2309. [Google Scholar] [CrossRef]
- De Mora, S.J.; Whitehead, R.F.; Gregory, M. The Chemical Composition of Glacial Melt Water Ponds and Streams on the McMurdo Ice Shelf, Antarctica. Antarct. Sci. 1994, 6, 17–27. [Google Scholar] [CrossRef]
- Hodgson, D.A.; Verleyen, E.; Sabbe, K.; Squier, A.H.; Keely, B.J.; Leng, M.J.; Saunders, K.M.; Vyverman, W. Late Quaternary Climate-Driven Environmental Change in the Larsemann Hills, East Antarctica, Multi-Proxy Evidence from a Lake Sediment Core. Quat. Res. 2005, 64, 83–99. [Google Scholar] [CrossRef]
- Hawes, I.; Jungblut, A.D.; Matys, E.D.; Summons, R.E. The ‘Dirty Ice’ of the McMurdo Ice Shelf: Analogues for Biological Oases during the Cryogenian. Geobiology 2018, 16, 369–377. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).