Regulation of DNA (de)Methylation Positively Impacts Seed Germination during Seed Development under Heat Stress
Abstract
1. Introduction
2. Material and Methods
Biological Material and Heat Treatment
3. Physiological and Phenotypic Characterizations
4. Morpho-Anatomical Measurements
5. Total RNA Isolation and Transcriptomic Sequencing
6. DNA Isolation and Whole-Genome Bisulfite Sequencing
7. Results
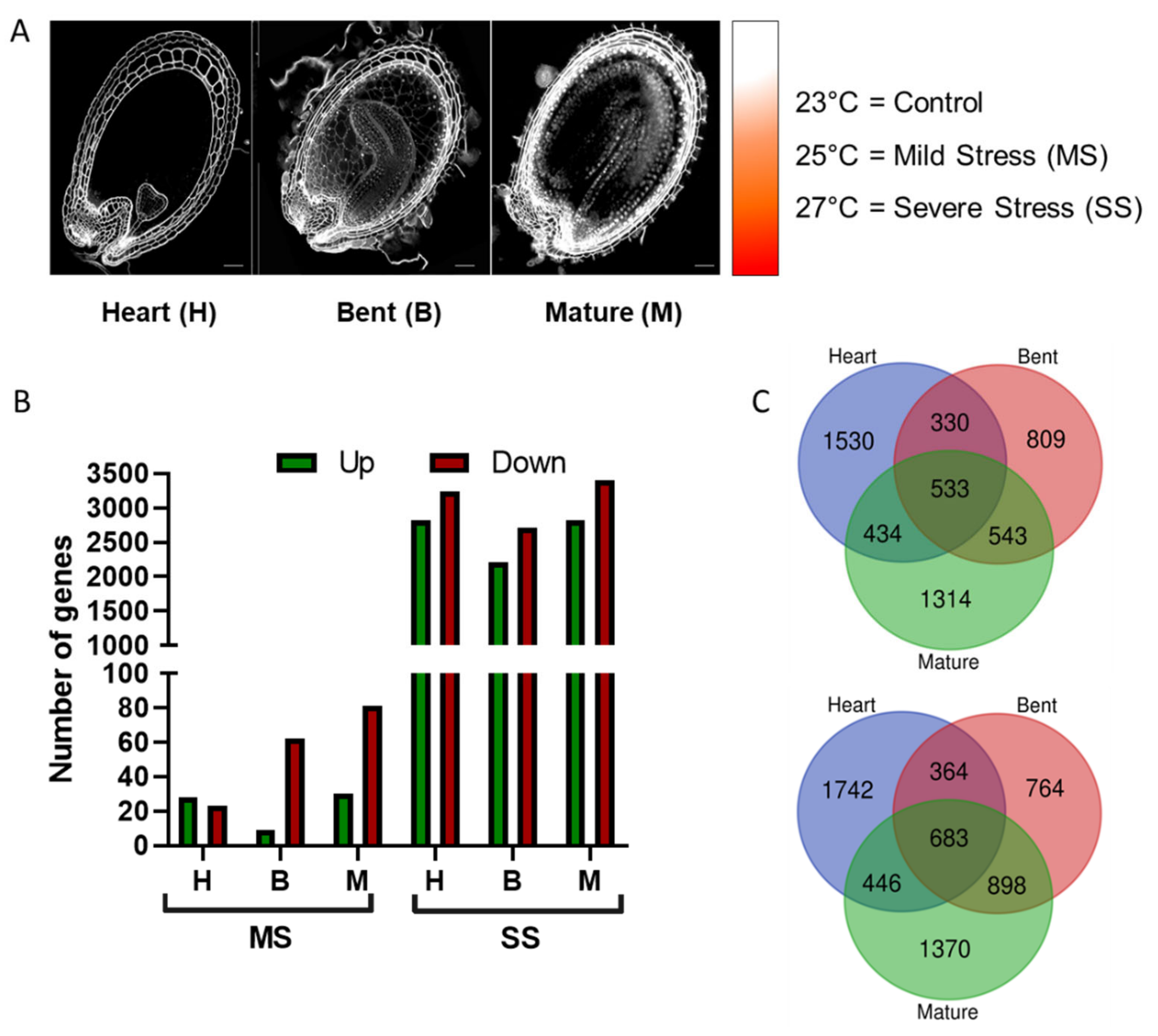
Impact of Constant Heat Stress on A. thaliana Seeds
8. Gene Expression during Seed Development under Heat Stress
9. Severe Heat Stress Effect on Methylation-Related Mutants
10. Whole-Genome DNA Methylation of Seeds under Heat Stress
11. Discussion
Constant Heat Stress Does Not Affect Seed Embryogenesis but Negatively Impacts Seed Germination and Longevity
12. Heat Stress Induces Impaired Cell Development and Metabolic Imbalance
13. DNA Demethylation Is Partially Responsible for Ensuring Seed Germination under Heat Stress Conditions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Venterea, R.T. Climate Change 2014: Mitigation of Climate Change. J. Environ. Qual. 2014, 38, 837. [Google Scholar] [CrossRef]
- Walther, G.; Post, E.; Convey, P.; Menzel, A.; Parmesank, C.; Beebee, T.J.C.; Fromentin, J.; Hoegh-Guldberg, O.; Bairlein, F. Ecological response to recent climate change. Nature 2002, 416, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, E.I.; Fischer, G.; Van Velthuizen, H.; Walter, C.; Ewert, F. Global hot-spots of heat stress on agricultural crops due to climate change. Agric. For. Meteorol. 2013, 170, 206–215. [Google Scholar] [CrossRef]
- Penuelas, J.; Rutishauser, T.; Filella, I. Phenology Feedbacks on Climate Change. Science 2009, 324, 887–888. [Google Scholar] [CrossRef] [PubMed]
- Fitter, A.H.; Fitter, R.S.R. Rapid changes in flowering time in British plants. Science 2002, 296, 1689–1691. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Sureshkumar, S.; Lempe, J.; Weigel, D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2006, 2, 0980–0989. [Google Scholar] [CrossRef]
- Li, Y.; Cheng, R.; Spokas, K.A.; Palmer, A.A.; Borevitz, J.O. Genetic variation for life history sensitivity to seasonal warming in Arabidopsis thaliana. Genetics 2014, 196, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Patel, D.; Franklin, K.A. Temperature-regulation of plant architecture. Plant Signal. Behav. 2009, 4, 577–579. [Google Scholar] [CrossRef]
- Antoun, M.; Ouellet, F. Growth temperature affects inflorescence architecture in Arabidopsis thaliana. Botany 2013, 91, 642–651. [Google Scholar] [CrossRef]
- Long, S.P.; Ort, D.R. More than taking the heat: Crops and global change. Curr. Opin. Plant Biol. 2010, 13, 240–247. [Google Scholar] [CrossRef]
- Donohue, K. Completing the cycle: Maternal effects as the missing link in plant life histories. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 1059–1074. [Google Scholar] [CrossRef]
- Huang, Z.; Footitt, S.; Finch-Savage, W.E. The effect of temperature on reproduction in the summer and winter annual Arabidopsis thaliana ecotypes Bur and Cvi. Ann. Bot. 2014, 113, 921–929. [Google Scholar] [CrossRef]
- Pauli, H.; Gottfried, M.; Dullinger, S.; Abdaladze, O.; Akhalkatsi, M.; Alonso, J.L.B.; Coldea, G.; Dick, J.; Erschbamer, B.; Calzado, R.F.; et al. Recent Plant Diversity Changes on Europe’s Mountain Summits. Science 2012, 336, 353–355. [Google Scholar] [CrossRef] [PubMed]
- Lobell, D.B.; Schlenker, W.; Costa-Roberts, J. Climate Trends and Global Crop Production Since 1980. Science 2011, 333, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Donohue, K.; Rubio De Casas, R.; Burghardt, L.; Kovach, K.; Willis, C.G. Germination, postgermination adaptation, and species ecological ranges. Annu. Rev. Ecol. Evol. Syst. 2010, 41, 293–319. [Google Scholar] [CrossRef]
- Burghardt, L.T.; Edwards, B.R.; Donohue, K. Multiple paths to similar germination behavior in Arabidopsis thaliana. New Phytol. 2016, 209, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- Walck, J.L.; Hidayati, S.N.; Dixon, K.W.; Thompson, K.; Poschlod, P. Climate change and plant regeneration from seed. Glob. Chang. Biol. 2011, 17, 2145–2161. [Google Scholar] [CrossRef]
- Righetti, K.; Vu, J.L.; Pelletier, S.; Vu, B.L.; Glaab, E.; Lalanne, D.; Pasha, A.; Patel, R.V.; Provart, N.J.; Verdier, J.; et al. Inference of Longevity-Related Genes from a Robust Coexpression Network of Seed Maturation Identifies Regulators Linking Seed Storability to Biotic Defense-Related Pathways. Plant Cell 2015, 27, 2692–2708. [Google Scholar] [CrossRef]
- Folsom, J.J.; Begcy, K.; Hao, X.; Wang, D.; Walia, H. Rice Fertilization-Independent Endosperm1 Regulates Seed Size under Heat Stress by Controlling Early Endosperm Development. Plant Physiol. 2014, 165, 238–248. [Google Scholar] [CrossRef] [PubMed]
- McClung, C.R.; Davis, S.J. Ambient thermometers in plants: From physiological outputs towards mechanisms of thermal sensing. Curr. Biol. 2010, 20, R1086–R1092. [Google Scholar] [CrossRef] [PubMed]
- Bokszczanin, K.L.; Fragkostefanakis, S. Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Front. Plant Sci. 2013, 4, 315. [Google Scholar] [CrossRef]
- Popova, O.V.; Dinh, H.Q.; Aufsatz, W.; Jonak, C. The RdDM Pathway Is Required for Basal Heat Tolerance in Arabidopsis. Mol. Plant 2013, 6, 396–410. [Google Scholar] [CrossRef] [PubMed]
- Bonasio, R.; Tu, S.; Reinberg, D. Molecular signals of epigenetic states. Science 2010, 330, 612–616. [Google Scholar] [CrossRef]
- Gibney, E.R.; Nolan, C.M. Epigenetics and gene expression. Heredity 2010, 105, 4–13. [Google Scholar] [CrossRef]
- Lukens, L.N.; Zhan, S. The plant genome’s methylation status and response to stress: Implications for plant improvement. Curr. Opin. Plant Biol. 2007, 10, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.K. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 2009, 12, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.W.L.; Henderson, I.R.; Jacobsen, S.E. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005, 6, 351–360. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.-K. Active DNA Demethylation in Plants and Animals. Cold Spring Harb. Symp. Quant. Biol. 2012, 77, 161–173. [Google Scholar] [CrossRef]
- Tang, K.; Lang, Z.; Zhang, H.; Zhu, J.-K. The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications. Nat. Plants 2016, 2, 16169. [Google Scholar] [CrossRef]
- Zilberman, D.; Gehring, M.; Tran, R.K.; Ballinger, T.; Henikoff, S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007, 39, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Boyko, A.; Blevins, T.; Yao, Y.; Golubov, A.; Bilichak, A.; Ilnytskyy, Y.; Hollander, J.; Meins, F.; Kovalchuk, I. Transgenerational Adaptation of Arabidopsis to Stress Requires DNA Methylation and the Function of Dicer-Like Proteins. PLoS ONE 2010, 5, e9514. [Google Scholar] [CrossRef]
- Naydenov, M.; Baev, V.; Apostolova, E.; Gospodinova, N.; Sablok, G.; Gozmanova, M.; Yahubyan, G. High-temperature effect on genes engaged in DNA methylation and affected by DNA methylation in Arabidopsis. Plant Physiol. Biochem. 2015, 87, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Correia, B.; Valledor, L.; Meijón, M.; Rodriguez, J.L.; Dias, M.C.; Santos, C.; Cañal, M.J.; Rodriguez, R.; Pinto, G. Is the Interplay between Epigenetic Markers Related to the Acclimation of Cork Oak Plants to High Temperatures? PLoS ONE 2013, 8, e53543. [Google Scholar] [CrossRef]
- Gao, G.; Li, J.; Li, H.; Li, F.; Xu, K.; Yan, G.; Chen, B.; Qiao, J.; Wu, X. Comparison of the heat stress induced variations in DNA methylation between heat-tolerant and heat-sensitive rapeseed seedlings. Breed. Sci. 2014, 64, 125–133. [Google Scholar] [CrossRef]
- Min, L.; Li, Y.; Hu, Q.; Zhu, L.; Gao, W.; Wu, Y.; Ding, Y.; Liu, S.; Yang, X.; Zhang, X. Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton. Plant Physiol. 2014, 164, 1293–1308. [Google Scholar] [CrossRef] [PubMed]
- Suter, L.; Widmer, A. Phenotypic effects of salt and heat stress over three generations in Arabidopsis thaliana. PLoS ONE 2013, 8, e80819. [Google Scholar] [CrossRef] [PubMed]
- Migicovsky, Z.; Yao, Y.; Kovalchuk, I. Transgenerational phenotypic and epigenetic changes in response to heat stress in Arabidopsis thaliana. Plant Signal. Behav. 2014, 9, e27971. [Google Scholar] [CrossRef]
- Zhong, S.H.; Liu, J.Z.; Jin, H.; Lin, L.; Li, Q.; Chen, Y.; Yuan, Y.X.; Wang, Z.Y.; Huang, H.; Qi, Y.J.; et al. Warm temperatures induce transgenerational epigenetic release of RNA silencing by inhibiting siRNA biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 9171–9176. [Google Scholar] [CrossRef] [PubMed]
- Whittle, C.A.; Otto, S.P.; Johnston, M.O.; Krochko, J.E. Adaptive epigenetic memory of ancestral temperature regime in Arabidopsis thaliana. Botany 2009, 87, 650–657. [Google Scholar] [CrossRef]
- Liu, J.; Feng, L.; Li, J.; He, Z. Genetic and epigenetic control of plant heat responses. Front. Plant Sci. 2015, 6, 267. [Google Scholar] [CrossRef] [PubMed]
- Bartee, L.; Malagnac, F.; Bender, J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001, 15, 1753–1758. [Google Scholar] [CrossRef]
- López Sánchez, A.; Stassen, J.H.M.; Furci, L.; Smith, L.M.; Ton, J. The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J. 2016, 88, 361–374. [Google Scholar] [CrossRef]
- Jones, D.B. Factors for converting percentages of nitrogen in foods and feeds into percentages of protein. Br. Food J. 1932. [Google Scholar]
- Xu, W.; Fiume, E.; Coen, O.; Pechoux, C.; Lepiniec, L.; Magnani, E. Endosperm and nucellus develop antagonistically in arabidopsis seeds. Plant Cell 2016, 28, 1343–1360. [Google Scholar] [CrossRef] [PubMed]
- Maia, J.; Dekkers, B.J.W.; Provart, N.J.; Ligterink, W.; Hilhorst, H.W.M. The re-establishment of desiccation tolerance in germinated arabidopsis thaliana seeds and its associated transcriptome. PLoS ONE 2011, 6, e29123. [Google Scholar] [CrossRef]
- Kanai, M.; Mano, S.; Nishimura, M. An efficient method for the isolation of highly purified RNA from seeds for use in quantitative transcriptome analysis. J. Vis. Exp. 2017, 2017, 4–9. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Narsai, R.; Gouil, Q.; Secco, D.; Srivastava, A.; Karpievitch, Y.V.; Liew, L.C.; Lister, R.; Lewsey, M.G.; Whelan, J. Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biol. 2017, 18, 172. [Google Scholar] [CrossRef]
- Fischer, D.S.; Theis, F.J.; Yosef, N. Impulse model-based differential expression analysis of time course sequencing data. Nucleic Acids Res. 2018, 46, e119. [Google Scholar] [CrossRef] [PubMed]
- Schultz, M.D.; He, Y.; Whitaker, J.W.; Hariharan, M.; Mukamel, E.A.; Leung, D.; Rajagopal, N.; Nery, J.R.; Urich, M.A.; Chen, H.; et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 2015, 523, 212–216. [Google Scholar] [CrossRef]
- De França-Neto, J.B.; Krzyzanowski, F.C. Tetrazolium: An important test for physiological seed quality evaluation. J. Seed Sci. 2019, 41, 359–366. [Google Scholar] [CrossRef]
- Berr, A.; Xu, L.; Gao, J.; Cognat, V.; Steinmetz, A.; Dong, A.; Shen, W.H. Set Domain Group25 encodes a histone methyltransferase and is involved in FLOWERING LOCUS C activation and repression of flowering. Plant Physiol. 2009, 151, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Geyer, R.; van Zanten, M.; Carles, A.; Li, Y.; Hörold, A.; van Nocker, S.; Soppe, W.J.J. Identification of the Arabidopsis reduced dormancy 2 gene uncovers a role for the polymerase associated factor 1 complex in seed dormancy. PLoS ONE 2011, 6, e22241. [Google Scholar] [CrossRef] [PubMed]
- Sozzani, R.; Maggio, C.; Giordo, R.; Umana, E.; Ascencio-Ibañez, J.T.; Hanley-Bowdoin, L.; Bergounioux, C.; Cella, R.; Albani, D. The E2FD/DEL2 factor is a component of a regulatory network controlling cell proliferation and development in Arabidopsis. Plant Mol. Biol. 2010, 72, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Hanada, A.; Yamauchi, Y.; Kuwahara, A.; Kamiya, Y.; Yamaguchi, S. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 2003, 15, 1591–1604. [Google Scholar] [CrossRef]
- Sliwinska, E.; Bassel, G.W.; Bewley, J.D. Germination of Arabidopsis thaliana seeds is not completed as a result of elongation of the radicle but of the adjacent transition zone and lower hypocotyl. J. Exp. Bot. 2009, 60, 3587–3594. [Google Scholar] [CrossRef]
- Willis, I.; Schmidt, P.; Soll, D. A selection for mutants of the RNA polymerase III transcription apparatus: PCF1 stimulates transcription of tRNA and 5S RNA genes. EMBO J. 1989, 8, 4281–4288. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Liu, X.; Li, X.; Huo, P.; Ge, J.; Hou, Y.; Yang, W.; Zhang, J.; Zhang, L.; Zhao, D.; et al. BRF Negatively Regulates Thermotolerance Defect of fes1a in Arabidopsis. Front. Plant Sci. 2020, 11, 171. [Google Scholar] [CrossRef] [PubMed]
- Pachauri, R.; Meyer, L. Climate change 2014: Synthesis report. Geneva Intergov. Panel Clim. Chang. 2015, 16, 408. [Google Scholar]
- Dolzblasz, A.; Dołzbłasz, S. Arabidopsis high temperature stress research. Acta Soc. Bot. Pol. 2018, 87, 3. [Google Scholar] [CrossRef]
- Ten Hove, C.A.; Lu, K.J.; Weijers, D. Building a plant: Cell fate specification in the early arabidopsis embryo. Development 2015, 142, 420–430. [Google Scholar] [CrossRef]
- Palovaara, J.; De Zeeuw, T.; Weijers, D. Tissue and Organ Initiation in the Plant Embryo: A First Time for Everything. Annu. Rev. Cell Dev. Biol. 2016, 32, 47–75. [Google Scholar] [CrossRef] [PubMed]
- Boscá, S.; Knauer, S.; Laux, T. Embryonic development in arabidopsis thaliana: From the zygote division to the shoot meristem. Front. Plant Sci. 2011, 2, 93. [Google Scholar] [CrossRef]
- Larkindale, J.; Hall, J.D.; Knight, M.R.; Vierling, E. Heat Stress Phenotypes of Arabidopsis Mutants Implicate Multiple Signaling Pathways in the Acquisition of Thermotolerance. Plant Physiol. 2005, 138, 882–897. [Google Scholar] [CrossRef]
- Malabarba, J.; Buffon, V.; Mariath, J.E.A.; Gaeta, M.L.; Dornelas, M.C.; Margis-Pinheiro, M.; Pasquali, G.; Revers, L.F. The MADS-box gene Agamous-like 11 is essential for seed morphogenesis in grapevine. J. Exp. Bot. 2017, 68, 1493–1506. [Google Scholar] [CrossRef]
- Bac-Molenaar, J.A.; Fradin, E.F.; Becker, F.F.M.; Rienstra, J.A.; van der Schoot, J.; Vreugdenhil, D.; Keurentjes, J.J.B. Genome-wide association mapping of fertility reduction upon heat stress reveals developmental stage-specific QTLs in Arabidopsis Thaliana. Plant Cell 2015, 27, 1857–1874. [Google Scholar] [CrossRef]
- Zinn, K.E.; Tunc-Ozdemir, M.; Harper, J.F. Temperature stress and plant sexual reproduction: Uncovering the weakest links. J. Exp. Bot. 2010, 61, 1959–1968. [Google Scholar] [CrossRef]
- Warner, R.M.; Erwin, J.E. Naturally occurring variation in high temperature induced floral bud abortion across Arabidopsis thaliana accessions. Plant Cell Environ. 2005, 28, 1255–1266. [Google Scholar] [CrossRef]
- Kim, S.Y.; Hong, C.B.; Lee, I. Heat shock stress causes stage-specific male sterility in Arabidopsis thaliana. J. Plant Res. 2001, 114, 301–307. [Google Scholar] [CrossRef]
- Endo, M.; Tsuchiya, T.; Hamada, K.; Kawamura, S.; Yano, K.; Ohshima, M.; Higashitani, A.; Watanabe, M.; Kawagishi-Kobayashi, M. High temperatures cause male sterility in rice plants with transcriptional alterations during pollen development. Plant Cell Physiol. 2009, 50, 1911–1922. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Kamiyama, M.; Iwata, T.; Makita, N.; Furukawa, H.; Ikeda, H. Moderate increase of mean daily temperature adversely affects fruit set of Lycopersicon esculentum by disrupting specific physiological processes in male reproductive development. Ann. Bot. 2006, 97, 731–738. [Google Scholar] [CrossRef]
- Hedhly, A. Sensitivity of flowering plant gametophytes to temperature fluctuations. Environ. Exp. Bot. 2011, 74, 9–16. [Google Scholar] [CrossRef]
- Saini, H.S.; Aspinall, D. Abnormal sporogenesis in wheat (Triticum aestivum L.) induced by short periods of high temperature. Ann. Bot. 1982, 49, 835–846. [Google Scholar] [CrossRef]
- Schmuths, H.; Bachmann, K.; Weber, W.E.; Horres, R.; Hoffmann, M.H. Effects of preconditioning and temperature during germination of 73 natural accessions of Arabidopsis thaliana. Ann. Bot. 2006, 97, 623–634. [Google Scholar] [CrossRef]
- He, H.; De Souza Vidigal, D.; Basten Snoek, L.; Schnabel, S.; Nijveen, H.; Hilhorst, H.; Bentsink, L. Interaction between parental environment and genotype affects plant and seed performance in Arabidopsis. J. Exp. Bot. 2014, 65, 6603–6615. [Google Scholar] [CrossRef]
- Silva-Correia, J.; Freitas, S.; Tavares, R.M.; Lino-Neto, T.; Azevedo, H. Phenotypic analysis of the Arabidopsis heat stress response during germination and early seedling development. Plant Methods 2014, 10, 7. [Google Scholar] [CrossRef]
- Fenner, M.; Thompson, K. The Ecology of Seeds; Cambridge University Press: Cambridge, UK, 2005; Volume 15, ISBN 9780521653114. [Google Scholar]
- Gaff, D.F.; Oliver, M. The evolution of desiccation tolerance in angiosperm plants: A rare yet common phenomenon. Funct. Plant Biol. 2013, 40, 315–328. [Google Scholar] [CrossRef]
- Fait, A.; Angelovici, R.; Less, H.; Ohad, I.; Urbanczyk-Wochniak, E.; Fernie, A.R.; Galili, G. Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol. 2006, 142, 839–854. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, J.V.; Pearce, D.W.; Poole, A.T.; Pharis, R.P.; Mander, L.N. Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol. Plant 2002, 115, 428–441. [Google Scholar] [CrossRef]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The role and regulation of ABI5 (ABA-insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef]
- Liu, H.; Stone, S.L. Regulation of ABI5 turnover by reversiblepost-translational modifications. Plant Signal. Behav. 2014, 9, e27577. [Google Scholar] [CrossRef]
- Vierling, E. The roles of heat shock proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 579–620. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Brunsting, J.F.; Stege, G.J.J.; Burgman, P.W.J.J.; Konings, A.W.T. Thermal protein denaturation and protein aggregation in cells made thermotolerant by various chemicals: Roleof heat shock proteins. Exp. Cell Res. 1995, 219, 536–546. [Google Scholar] [CrossRef] [PubMed]
- Swidzinski, J.A.; Sweetlove, L.J.; Leaver, C.J. A custom microarray analysis of gene expression during programmed cell death in Arabidopsis thaliana. Plant J. 2002, 30, 431–446. [Google Scholar] [CrossRef] [PubMed]
- Vacca, R.A.; De Pinto, M.C.; Valenti, D.; Passarella, S.; Marra, E.; De Gara, L. Production of reactive oxygen species, alteration of cytosolic ascorbate peroxidase, and impairment of mitochondrial metabolism are early events in heat shock-induced programmed cell death in tobacco bright-yellow 2 cells. Plant Physiol. 2004, 134, 1100–1112. [Google Scholar] [CrossRef]
- Alfonso, M.; Yruela, I.; Almárcegui, S.; Torrado, E.; Pérez, M.A.; Picorel, R. Unusual tolerance to high temperatures in a new herbicide-resistant D1 mutant from Glycine max (L.) Merr. cell cultures deficient in fatty acid desaturation. Planta 2001, 212, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Sangwan, V.; Örvar, B.L.; Beyerly, J.; Hirt, H.; Dhindsa Rajinder, S. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 2002, 31, 629–638. [Google Scholar] [CrossRef]
- Hall, A.E. Crop Responses to Environment; CRC Press: Boca Raton, FL, USA, 2001; ISBN 9780429126543. [Google Scholar]
- Dat, J.F.; Foyer, C.H.; Scott, I.M. Changes in salicylic acid and antioxidants during induced thermotolerance in mustard seedlings. Plant Physiol. 1998, 118, 1455–1461. [Google Scholar] [CrossRef] [PubMed]
- Larkindale, J.; Knight, M.R. Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol. 2002, 128, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Gong, M.; Li, Y.J.; Chen, S.Z. Abscisic acid-induced thermotolerance in maize seedlings is mediated by calcium and associated with antioxidant systems. J. Plant Physiol. 1998, 153, 488–496. [Google Scholar] [CrossRef]
- Singh, P.; Yekondi, S.; Chen, P.W.; Tsai, C.H.; Yu, C.W.; Wu, K.; Zimmerli, L. Environmental history modulates Arabidopsis pattern-triggered immunity in a histone acetyltransferase1-dependent manner. Plant Cell 2014, 26, 2676–2688. [Google Scholar] [CrossRef]
- Lang-Mladek, C.; Popova, O.; Kiok, K.; Berlinger, M.; Rakic, B.; Aufsatz, W.; Jonak, C.; Hauser, M.T.; Luschnig, C. Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in arabidopsis. Mol. Plant 2010, 3, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Lucyshyn, D.; Jaeger, K.E.; Alós, E.; Alvey, E.; Harberd, N.P.; Wigge, P.A. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 2012, 484, 242–245. [Google Scholar] [CrossRef]
- Kumar, S.V.; Wigge, P.A. H2A.Z-Containing Nucleosomes Mediate the Thermosensory Response in Arabidopsis. Cell 2010, 140, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ren, Y.; Zhang, Y.; Xu, J.; Sun, F.; Zhang, Z.; Wang, Y. Genome-wide identification and expression analysis of heat-responsive and novel microRNAs in Populus tomentosa. Gene 2012, 504, 160–165. [Google Scholar] [CrossRef]
- Chellappan, P.; Vanitharani, R.; Ogbe, F.; Fauquet, C.M. Effect of temperature on geminivirus-induced RNA silencing in plants. Plant Physiol. 2005, 138, 1828–1841. [Google Scholar] [CrossRef]
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Kawakatsu, T.; Nery, J.R.; Castanon, R.; Ecker, J.R. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 2017, 18, 171. [Google Scholar] [CrossRef]
- Hsieh, T.-F.; Ibarra, C.A.; Silva, P.; Zemach, A.; Eshed-Williams, L.; Fischer, R.L.; Zilberman, D. Genome-Wide Demethylation of Arabidopsis Endosperm. Science 2009, 324, 1451–1454. [Google Scholar] [CrossRef]
- Penterman, J.; Uzawa, R.; Fischer, R.L. Genetic interactions between DNA demethylation and methylation in arabidopsis. Plant Physiol. 2007, 145, 1549–1557. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Córdoba-Cañero, D.; Qian, W.; Zhu, X.; Tang, K.; Zhang, H.; Ariza, R.R.; Roldán-Arjona, T.; Zhu, J.K. An AP Endonuclease Functions in Active DNA Dimethylation and Gene Imprinting in Arabidopsis. PLoS Genet. 2015, 11, e1004905. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Macías, M.I.; Qian, W.; Miki, D.; Pontes, O.; Liu, Y.; Tang, K.; Liu, R.; Morales-Ruiz, T.; Ariza, R.R.; Roldán-Arjona, T.; et al. A DNA 3’ Phosphatase Functions in Active DNA Demethylation in Arabidopsis. Mol. Cell 2012, 45, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Miki, D.; Zhang, H.; Liu, Y.; Zhang, X.; Tang, K.; Kan, Y.; La, H.; Li, X.; Li, S.; et al. A Histone Acetyltransferase Regulates Active DNA Demethylation in Arabidopsis. Science 2012, 336, 1445–1448. [Google Scholar] [CrossRef]
- Lang, Z.; Lei, M.; Wang, X.; Tang, K.; Miki, D.; Zhang, H.; Mangrauthia, S.K.; Liu, W.; Nie, W.; Ma, G.; et al. The Methyl-CpG-Binding Protein MBD7 Facilitates Active DNA Demethylation to Limit DNA Hyper-Methylation and Transcriptional Gene Silencing. Mol. Cell 2015, 57, 971–983. [Google Scholar] [CrossRef]
- Brocard-Gifford, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol. 2003, 131, 78–92. [Google Scholar] [CrossRef]
- Katavic, V.; Reed, D.W.; Taylor, D.C.; Giblin, E.M.; Barton, D.L.; Zou, J.; MacKenzie, S.L.; Covello, P.S.; Kunst, L. Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol. 1995, 108, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Focks, N.; Benning, C. Wrinkled 1: A novel, low-seed-oil mutant of arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998, 118, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Hills, M.J. Arabidopsis mutants deficient in diacylglycerol acyltransferase display increased sensitivity to abscisic acid, sugars, and osmotic stress during germination and seedling development. Plant Physiol. 2002, 129, 1352–1358. [Google Scholar] [CrossRef]
- Penfield, S.; Graham, S.; Graham, I.A. Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochem. Soc. Trans. 2005, 33, 380–383. [Google Scholar] [CrossRef]
- Pritchard Sarah, L.; Charlton Wayne, L.; Baker, A.; Graham Ian, A. Germination and storage reserve mobilization are regulated independently in Arabidopsis. Plant J. 2002, 31, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J.D. Seed germination and dormancy. Plant Cell 1997, 9, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J.D.; Bradford, K.J.; Hilhorst, H.W.M.; Nonogaki, H. Seeds: Physiology of Development, Germination and Dormancy, 3rd ed.; Springer: New York, NY, USA, 2013; ISBN 9781461446934. [Google Scholar]
- Borisjuk, L.; Rolletschek, H.; Radchuk, R.; Weschke, W.; Wobus, U.; Weber, H. Seed Development and Differentiation: A Role for Metabolic Regulation. Plant Biol. 2004, 6, 375–386. [Google Scholar] [CrossRef]
- Rodríguez, J.L.; De Diego, J.G.; Rodríguez, F.D.; Cervantes, E. Mitochondrial structures during seed germination and early seedling development in Arabidopsis thaliana. Biologia 2015, 70, 1019–1025. [Google Scholar] [CrossRef]
- Law, S.R.; Narsai, R.; Whelan, J. Mitochondrial biogenesis in plants during seed germination. Mitochondrion 2014, 19, 214–221. [Google Scholar] [CrossRef]
- Paszkiewicz, G.; Gualberto, J.M.; Benamar, A.; Macherel, D.; Logan, D.C. Arabidopsis seed mitochondria are bioenergetically active immediately upon imbibition and specialize via biogenesis in preparation for autotrophic growth. Plant Cell 2017, 29, 109–128. [Google Scholar] [CrossRef]
- Ratajczak, E.; Małecka, A.; Ciereszko, I.; Staszak, A.M. Mitochondria are important determinants of the aging of seeds. Int. J. Mol. Sci. 2019, 20, 1568. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malabarba, J.; Windels, D.; Xu, W.; Verdier, J. Regulation of DNA (de)Methylation Positively Impacts Seed Germination during Seed Development under Heat Stress. Genes 2021, 12, 457. https://doi.org/10.3390/genes12030457
Malabarba J, Windels D, Xu W, Verdier J. Regulation of DNA (de)Methylation Positively Impacts Seed Germination during Seed Development under Heat Stress. Genes. 2021; 12(3):457. https://doi.org/10.3390/genes12030457
Chicago/Turabian StyleMalabarba, Jaiana, David Windels, Wenjia Xu, and Jerome Verdier. 2021. "Regulation of DNA (de)Methylation Positively Impacts Seed Germination during Seed Development under Heat Stress" Genes 12, no. 3: 457. https://doi.org/10.3390/genes12030457
APA StyleMalabarba, J., Windels, D., Xu, W., & Verdier, J. (2021). Regulation of DNA (de)Methylation Positively Impacts Seed Germination during Seed Development under Heat Stress. Genes, 12(3), 457. https://doi.org/10.3390/genes12030457