Diverse Regulation but Conserved Function: SOX9 in Vertebrate Sex Determination
Abstract
:1. Introduction
2. Unique Cellular and Morphological Changes during Male Gonad Development
3. SRY and DMRT1: Key Switches in Sex Determination
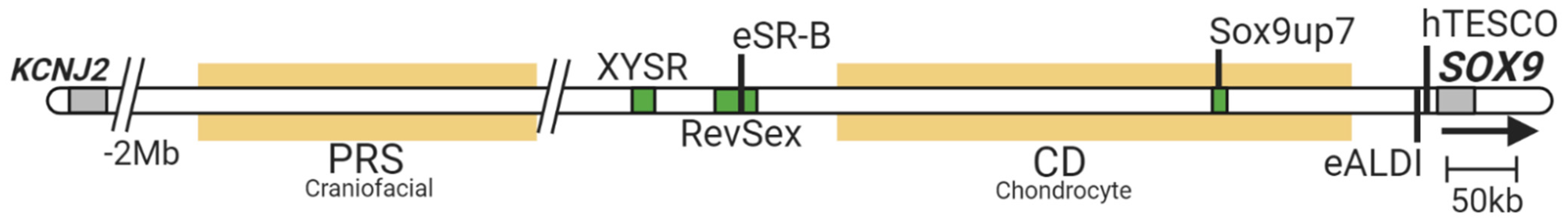
4. Identification of Testis-Specific Enhancers of SOX9/Sox9
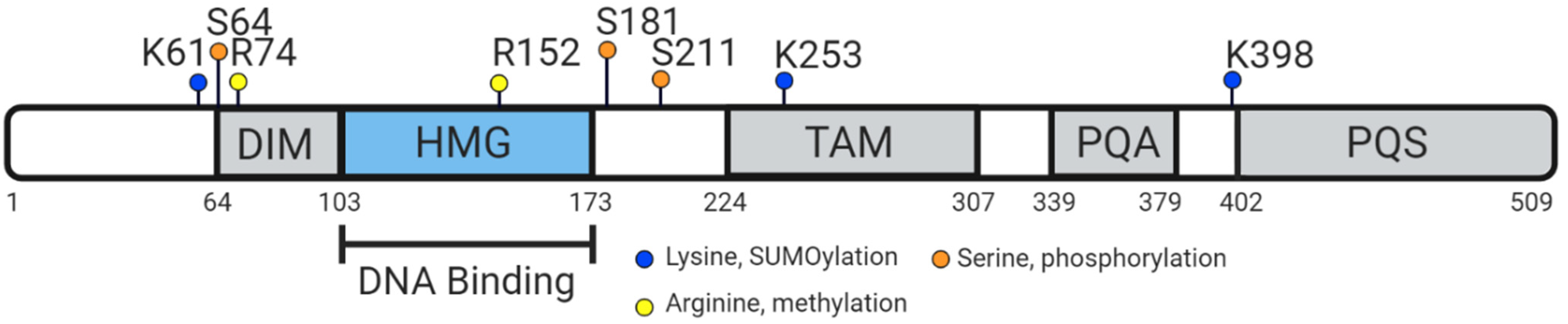
5. Vertebrate SOX9 Proteins
6. Conserved Function of Vertebrate SOX9 Protein

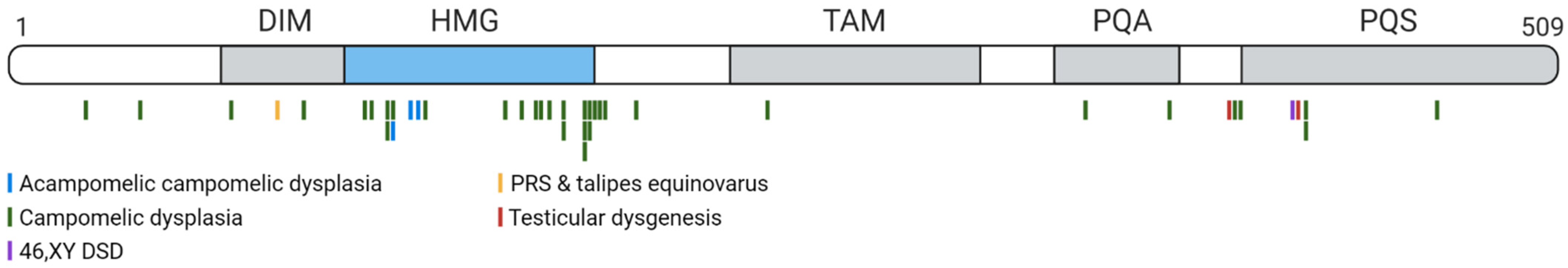
7. Disorders Arising from SOX9 Mutations in Humans
8. Gonad Plasticity: The Role of SOX9 in Transdifferentiation
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stevens, N.M. A study of the germ cells of Aphis rosae and Aphis œnotherae. J. Exp. Zool. 1904, 2, 313–337. [Google Scholar] [CrossRef] [Green Version]
- Bridges, C.B. Non-disjunction as proof of the chromosome theory of heredity. Genetics 1916, 1, 1–52. [Google Scholar] [CrossRef] [PubMed]
- Jost, A. The age factor in the castration of male rabbit fetuses. Proc. Soc. Exp. Biol. Med. 1947, 66, 302. [Google Scholar] [CrossRef]
- Hodgkin, J. Sex determination in the nematode C. Elegans: Analysis of tra-3 suppressors and characterization of fem genes. Genetics 1986, 114, 15–52. [Google Scholar] [CrossRef] [PubMed]
- Western, P.S.; Harry, J.L.; Graves, J.A.M.; Sinclair, A.H. Temperature-dependent sex determination: Upregulation of SOX9 expression after commitment to male development. Dev. Dyn. 1999, 214, 171–177. [Google Scholar] [CrossRef]
- Shoemaker, C.; Ramsey, M.; Queen, J.; Crews, D. Expression of Sox9, Mis, and Dmrt1 in the gonad of a species with temperature-dependent sex determination. Dev. Dyn. 2007, 236, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, C.M.; Queen, J.; Crews, D. Response of candidate sex-determining genes to changes in temperature reveals their involvement in the molecular network underlying temperature-dependent sex determination. Mol. Endocrinol. 2007, 21, 2750–2763. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.A.; Roeszler, K.N.; Ohnesorg, T.; Cummins, D.M.; Farlie, P.G.; Doran, T.J.; Sinclair, A.H. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 2009, 461. [Google Scholar] [CrossRef] [PubMed]
- da Silva, S.M.; Hacker, A.; Harley, V.; Goodfellow, P.; Swain, A.; Lovell-Badge, R. Sox9 expression during gonadal development implies a conserved role for the gene in testis differentiation in mammals and birds. Nat. Genet. 1996, 14, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Grützner, F.; Rens, W.; Tsend-Ayush, E.; El-Mogharbel, N.; O’Brien, P.C.M.; Jones, R.C.; Ferguson-Smith, M.A.; Marshall Graves, J.A. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature 2004, 432, 913–917. [Google Scholar] [CrossRef]
- Rens, W.; O’Brien, P.C.M.; Grützner, F.; Clarke, O.; Graphodatskaya, D.; Tsend-Ayush, E.; Trifonov, V.A.; Skelton, H.; Wallis, M.C.; Johnston, S.; et al. The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z. Genome Biol. 2007, 8, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuyama, H.; Yamada, M.; Kamei, Y.; Fujiwara-Ishikawa, T.; Todo, T.; Nagahama, Y.; Matsuda, M. Dmrt1 mutation causes a male-to-female sex reversal after the sex determination by Dmy in the medaka. Chromosome Res. 2012, 20, 163–176. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, M.; Nagahama, Y.; Kobayashi, T.; Matsuda, C.; Hamaguchi, S.; Sakaizumi, M. The sex determining gene of medaka: A Y-specific DM domain gene (DMY) is required for male development. Fish Physiol. Biochem. 2003, 28, 135–139. [Google Scholar] [CrossRef]
- Uhlenhaut, N.H.; Jakob, S.; Anlag, K.; Eisenberger, T.; Sekido, R.; Kress, J.; Treier, A.C.; Klugmann, C.; Klasen, C.; Holter, N.I.; et al. Somatic Sex Reprogramming of Adult Ovaries to Testes by FOXL2 Ablation. Cell 2009, 139, 1130–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matson, C.K.; Murphy, M.W.; Sarver, A.L.; Griswold, M.D.; Bardwell, V.J.; Zarkower, D. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 2011, 476, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Furman, B.L.S.; Metzger, D.C.H.; Darolti, I.; Wright, A.E.; Sandkam, B.A.; Almeida, P.; Shu, J.J.; Mank, J.E. Sex Chromosome Evolution: So Many Exceptions to the Rules. Genome Biol. Evol. 2020, 12, 750–763. [Google Scholar] [CrossRef] [PubMed]
- Conover, D.O.; Kynard, B.E. Environmental Sex Determination: Interaction of Temperature and Genotype in a Fish. Science 1981, 213, 577–579. [Google Scholar] [CrossRef] [PubMed]
- Manolakou, P.; Lavranos, G.; Angelopoulou, R. Molecular patterns of sex determination in the animal kingdom: A comparative study of the biology of reproduction. Reprod. Biol. Endocrinol. 2006, 4, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Koopman, P.; Münsterberg, A.; Capel, B.; Vivian, N.; Lovell-Badge, R. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature 1990, 348, 450–452. [Google Scholar] [CrossRef]
- Sinclair, A.H.; Berta, P.; Palmer, M.; Hawkins, J.R.; Griffiths, B.L.; Smith, M.J.; Foster, J.W.; Frischauf, A.; Lovell-Badge, R.; Goodfellow, P.N. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 1990, 346, 240–244. [Google Scholar] [CrossRef] [Green Version]
- Waters, P.D.; Wallis, M.C.; Marshall Graves, J.A. Mammalian sex - Origin and evolution of the Y chromosome and SRY. Semin. Cell Dev. Biol. 2007, 18, 389–400. [Google Scholar] [CrossRef]
- Sarre, S.D.; Ezaz, T.; Georges, A. Transitions between sex-determining systems in reptiles and amphibians. Annu. Rev. Genomics Hum. Genet. 2011, 12, 391–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brykov, V.A. Mechanisms of sex determination in fish: Evolutionary and practical aspects. Russ. J. Mar. Biol. 2014, 40, 407–417. [Google Scholar] [CrossRef]
- Wallis, M.C.; Delbridge, M.L.; Pask, A.J.; Alsop, A.E.; Grützner, F.; O’Brien, P.C.M.; Rens, W.; Ferguson-Smith, M.A.; Graves, J.A.M. Mapping platypus SOX genes; autosomal location of SOX9 excludes it from sex determining role. Cytogenet. Genome Res. 2007, 116, 232–234. [Google Scholar] [CrossRef] [PubMed]
- Berta, P.; Hawkins, J.R.; Sinclair, A.H.; Taylor, A.; Griffiths, B.L.; Goodfellow, P.N.; Fellous, M. Genetic evidence equating SRY and the testis-determining factor. Nature 1990, 348, 448–450. [Google Scholar] [CrossRef]
- Ferguson-Smith, M. The evolution of sex chromosomes and sex determination in vertebrates and the key role of DMRT1. Sex. Dev. 2006, 1, 2–11. [Google Scholar] [CrossRef]
- Hu, Y.C.; Okumura, L.M.; Page, D.C. Gata4 Is Required for Formation of the Genital Ridge in Mice. PLoS Genet. 2013, 9, e1003629. [Google Scholar] [CrossRef] [Green Version]
- Yildirim, E.; Aksoy, S.; Onel, T.; Yaba, A. Gonadal development and sex determination in mouse. Reprod. Biol. 2020, 20, 115–126. [Google Scholar] [CrossRef]
- Nef, S.; Stévant, I.; Greenfield, A. Characterizing the bipotential mammalian gonad. In Current Topics in Developmental Biology; Capel, B., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 134, pp. 167–194. ISBN 9780128115442. [Google Scholar]
- Molyneaux, K.; Wylie, C. Primordial Germ Cell Migration. Int. J. Dev. Biol. 2004, 48, 537–544. [Google Scholar] [CrossRef]
- Richardson, B.E.; Lehmann, R. Mechanisms guiding primordial germ cell migration: Strategies from different organisms. Nat. Rev. Mol. Cell Biol. 2010, 11, 37–49. [Google Scholar] [CrossRef] [Green Version]
- Ungewitter, E.K.; Yao, H.H.-C. How to make a gonad: Cellular mechanisms governing formation of the testes and ovaries. Sex. Dev. 2013, 7, 7–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginsburg, M.; Snow, M.H.; McLaren, A. Primordial germ cells in the mouse embryo during gastrulation. Development 1990, 110, 521–528. [Google Scholar] [PubMed]
- Schmahl, J.; Capel, B. Cell proliferation is necessary for the determination of male fate in the gonad. Dev. Biol. 2003, 258, 264–276. [Google Scholar] [CrossRef] [Green Version]
- Goodfellow, P.N.; Darling, S.M. Genetics of sex determination in man and mouse. Development 1988, 102, 251–258. [Google Scholar]
- She, Z.Y.; Yang, W.X. Sry and SoxE genes: How they participate in mammalian sex determination and gonadal development? Semin. Cell Dev. Biol. 2017, 63, 13–22. [Google Scholar] [CrossRef]
- Wilhelm, D.; Palmer, S.; Koopman, P. Sex determination and gonadal development in mammals. Physiol. Rev. 2007, 87, 1–28. [Google Scholar] [CrossRef] [Green Version]
- Kanai, Y.; Hiramatsu, R.; Matoba, S.; Kidokoro, T. From SRY to SOX9: Mammalian testis differentiation. J. Biochem. 2005, 138, 13–19. [Google Scholar] [CrossRef]
- Clarkson, M.J.; Harley, V.R. Sex with two SOX on: SRY and SOX9 in testis development. Trends Endocrinol. Metab. 2002, 13, 106–111. [Google Scholar] [CrossRef]
- Sekido, R.; Lovell-Badge, R. Sex determination and SRY: Down to a wink and a nudge? Trends Genet. 2009, 25, 19–29. [Google Scholar] [CrossRef]
- Albrecht, K.H.; Eicher, E.M. Evidence that Sry is expressed in pre-Sertoli cells and Sertoli and granulosa cells have a common precursor. Dev. Biol. 2001, 240, 92–107. [Google Scholar] [CrossRef] [Green Version]
- Mamsen, L.S.; Ernst, E.H.; Borup, R.; Larsen, A.; Olesen, R.H.; Ernst, E.; Anderson, R.A.; Kristensen, S.G.; Andersen, C.Y. Temporal expression pattern of genes during the period of sex differentiation in human embryonic gonads. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hacker, A.; Capel, B.; Goodfellow, P.; Lovell-Badge, R. Expression of Sry, the mouse sex determining gene. Development 1995, 121, 1603–1614. [Google Scholar]
- Wilhelm, D.; Martinson, F.; Bradford, S.; Wilson, M.J.; Combes, A.N.; Beverdam, A.; Bowles, J.; Mizusaki, H.; Koopman, P. Sertoli cell differentiation is induced both cell-autonomously and through prostaglandin signaling during mammalian sex determination. Dev. Biol. 2005, 287, 111–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zheng, M.; Lau, Y.F.C. The sex-determining factors SRY and SOX9 regulate similar target genes and promote testis cord formation during testicular differentiation. Cell Rep. 2014, 8, 723–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuji-Hosokawa, A.; Kashimada, K.; Kato, T.; Ogawa, Y.; Nomura, R.; Takasawa, K.; Lavery, R.; Coschiera, A.; Schlessinger, D.; Harley, V.R.; et al. Peptidyl arginine deiminase 2 (Padi2) is expressed in Sertoli cells in a specific manner and regulated by SOX9 during testicular development. Sci. Rep. 2018, 8, 13263. [Google Scholar] [CrossRef]
- Gonen, N.; Quinn, A.; O’Neill, H.C.; Koopman, P.; Lovell-Badge, R. Normal Levels of Sox9 Expression in the Developing Mouse Testis Depend on the TES/TESCO Enhancer, but This Does Not Act Alone. PLoS Genet. 2017, 13, e1006520. [Google Scholar] [CrossRef] [Green Version]
- Croft, B.; Ohnesorg, T.; Hewitt, J.; Bowles, J.; Quinn, A.; Tan, J.; Corbin, V.; Pelosi, E.; van den Bergen, J.; Sreenivasan, R.; et al. Human sex reversal is caused by duplication or deletion of core enhancers upstream of SOX9. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larney, C.; Bailey, T.L.; Koopman, P. Switching on sex: Transcriptional regulation of the testis-determining gene Sry. Development 2014, 141, 2195–2205. [Google Scholar] [CrossRef] [Green Version]
- Lei, N.; Hornbaker, K.I.; Rice, D.A.; Karpova, T.; Agbor, V.A.; Heckert, L.L. Sex-specific differences in mouse DMRT1 expression are both cell type- and stage-dependent during gonad development. Biol. Reprod. 2007, 77, 466–475. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, T.; Matsuda, M.; Kajiura-Kobayashi, H.; Suzuki, A.; Saito, N.; Nakamoto, M.; Shibata, N.; Nagahama, Y. Two DM domain genes, DMY and DMRT1, involved in testicular differentiation and development in the Medaka, Oryzias latipes. Dev. Dyn. 2004, 231, 518–526. [Google Scholar] [CrossRef]
- Zhang, J. Evolution of DMY, a Newly Emergent Male Sex-Determination Gene of Medaka Fish. Genetics 2004, 166, 1887–1895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matson, C.; Zarkower, D. Sex and the singular DM domain: Insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 2013, 13, 163–174. [Google Scholar] [CrossRef] [Green Version]
- Long, H.K.; Osterwalder, M.; Welsh, I.C.; Hansen, K.; Davies, J.O.J.; Liu, Y.E.; Koska, M.; Adams, A.T.; Aho, R.; Arora, N.; et al. Loss of Extreme Long-Range Enhancers in Human Neural Crest Drives a Craniofacial Disorder. Cell Stem Cell 2020, 27, 765–783.e14. [Google Scholar] [CrossRef] [PubMed]
- Smyk, M.; Akdemir, K.C.; Stankiewicz, P. SOX9 chromatin folding domains correlate with its real and putative distant cis-regulatory elements. Nucleus 2017, 8, 182–187. [Google Scholar] [CrossRef] [Green Version]
- Bagheri-Fam, S.; Barrionuevo, F.; Dohrmann, U.; Günther, T.; Schüle, R.; Kemler, R.; Mallo, M.; Kanzler, B.; Scherer, G. Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev. Biol. 2006, 291, 382–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagheri-Fam, S.; Sreenivasan, R.; Bernard, P.; Knower, K.C.; Sekido, R.; Lovell-Badge, R.; Just, W.; Harley, V.R. Sox9 gene regulation and the loss of the XY/XX sex-determining mechanism in the mole vole Ellobius lutescens. Chromosom. Res. 2012, 20, 191–199. [Google Scholar] [CrossRef] [Green Version]
- Sekido, R.; Lovell-Badge, R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 2008, 453, 930–934. [Google Scholar] [CrossRef]
- Bagheri-Fam, S.; Sim, H.; Bernard, P.; Jayakody, I.; Taketo, M.M.; Scherer, G.; Harley, V.R. Loss of Fgfr2 leads to partial XY sex reversal. Dev. Biol. 2008, 314, 71–83. [Google Scholar] [CrossRef] [Green Version]
- Symon, A.; Harley, V. SOX9: A genomic view of tissue specific expression and action. Int. J. Biochem. Cell Biol. 2017, 87, 18–22. [Google Scholar] [CrossRef]
- Georg, I.; Bagheri-Fam, S.; Knower, K.C.; Wieacker, P.; Scherer, G.; Harley, V.R. Mutations of the SRY-responsive enhancer of SOX9 are uncommon in XY gonadal dysgenesis. Sex. Dev. 2010, 4, 321–325. [Google Scholar] [CrossRef]
- Bagheri-Fam, S.; Sinclair, A.H.; Koopman, P.; Harley, V.R. Conserved regulatory modules in the Sox9 testis-specific enhancer predict roles for SOX, TCF/LEF, Forkhead, DMRT, and GATA proteins in vertebrate sex determination. Int. J. Biochem. Cell Biol. 2010, 42, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Ohnesorg, T.; van den Bergen, J.A.; Belluoccio, D.; Shankara-Narayana, N.; Kean, A.M.; Vasilaras, A.; Ewans, L.; Ayers, K.L.; Sinclair, A.H. A duplication in a patient with 46,XX ovo-testicular disorder of sex development refines the SOX9 testis-specific regulatory region to 24 kb. Clin. Genet. 2017, 92, 347–349. [Google Scholar] [CrossRef]
- Benko, S.; Gordon, C.T.; Mallet, D.; Sreenivasan, R.; Thauvin-Robinet, C.; Brendehaug, A.; Thomas, S.; Bruland, O.; David, M.; Nicolino, M.; et al. Disruption of a long distance regulatory region upstream of SOX9 in isolated disorders of sex development. J. Med. Genet. 2011, 48, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.J.; Sock, E.; Buchberger, A.; Just, W.; Denzer, F.; Hoepffner, W.; German, J.; Cole, T.; Mann, J.; Seguin, J.H.; et al. Copy number variation of two separate regulatory regions upstream of SOX9 causes isolated 46,XY or 46,XX disorder of sex development. J. Med. Genet. 2015, 52, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Ohnesorg, T.; Croft, B.; Tan, J.; Sinclair, A.H. Using ROADMAP Data to Identify Enhancers Associated with Disorders of Sex Development. Sex. Dev. 2016, 10, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Gonen, N.; Futtner, C.R.; Wood, S.; Garcia-Moreno, A.; Salamone, I.M.; Samson, S.C.; Sekido, R.; Poulat, F.; Maatouk, D.M.; Lovell-Badge, R. Sex reversal following deletion of a single far upstream enhancer of Sox9. Science 2018, 360, 1469–1473. [Google Scholar] [CrossRef] [Green Version]
- Akiyama, H.; Stadler, H.S.; Martin, J.F.; Ishii, T.M.; Beachy, P.A.; Nakamura, T.; De Crombrugghe, B. Misexpression of Sox9 in mouse limb bud mesenchyme induces polydactyly and rescues hypodactyly mice. Matrix Biol. 2007, 26, 224–233. [Google Scholar] [CrossRef]
- Montero, J.A.; Lorda-Diez, C.I.; Francisco-Morcillo, J.; Chimal-Monroy, J.; Garcia-Porrero, J.A.; Hurle, J.M. Sox9 expression in amniotes: Species-specific differences in the formation of digits. Front. Cell Dev. Biol. 2017, 5, 5–10. [Google Scholar] [CrossRef] [Green Version]
- Bi, W.; Deng, J.M.; Zhang, Z.; Behringer, R.R.; De Crombrugghe, B. Sox9 is required for cartilage formation. Nat. Genet. 1999, 22, 85–89. [Google Scholar] [CrossRef]
- Song, H.; Park, K.H. Regulation and function of SOX9 during cartilage development and regeneration. Semin. Cancer Biol. 2020, 67, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, M.K.; Francis, J.C.; Swain, A. The role of Sox9 in prostate development. Differentiation 2008, 76, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Bulanenkova, S.S.; Snezhkov, E.V.; Akopov, S.B. SOX9 as One of the Central Units of Regulation Axis of Pancreas Embryogenesis and Cancer Progression. Mol. Genet. Microbiol. Virol. 2019, 34, 159–169. [Google Scholar] [CrossRef]
- Harley, V.R.; Lovell-Badge, R.; Goodfellow, P.N. Definition of a consensus DNA binding site for SRY. Nucleic Acids Res. 1994, 22, 1500–1501. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Harley, V.R.; Pontiggia, A.; Goodfellow, P.N.; Lovell-Badge, R.; Bianchi, M.E. SRY, like HMG1, recognizes sharp angles in DNA. EMBO J. 1992, 11, 4497–4506. [Google Scholar] [CrossRef]
- Hou, L.; Srivastava, Y.; Jauch, R. Molecular basis for the genome engagement by Sox proteins. Semin. Cell Dev. Biol. 2017, 63, 2–12. [Google Scholar] [CrossRef] [Green Version]
- Mertin, S.; McDowall, S.G.; Harley, V.R. The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res. 1999, 27, 1359–1364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harley, V.R.; Lovell-Badge, R.; Goodfellow, P.N.; Hextall, P.J. The HMG box of SRY is a calmodulin binding domain. FEBS Lett. 1996, 391, 24–28. [Google Scholar] [CrossRef] [Green Version]
- Harley, V.R.; Layfield, S.; Mitchell, C.L.; Forwood, J.K.; John, A.P.; Briggs, L.J.; McDowall, S.G.; Jans, D.A. Defective importin β recognition and nuclear import of the sex-determining factor SRY are associated with XY sex-reversing mutations. Proc. Natl. Acad. Sci. USA 2003, 100, 7045–7050. [Google Scholar] [CrossRef] [Green Version]
- Sim, H.; Rimmer, K.; Kelly, S.; Ludbrook, L.M.; Clayton, A.H.A.; Harley, V.R. Defective calmodulin-mediated nuclear transport of the sex-determining region of the Y chromosome (SRY) in XY sex reversal. Mol. Endocrinol. 2005, 19, 1884–1892. [Google Scholar] [CrossRef] [Green Version]
- Sim, H.; Argentaro, A.; Czech, D.P.; Bagheri-Fam, S.; Sinclair, A.H.; Koopman, P.; Boizet-Bonhoure, B.; Poulat, F.; Harley, V.R. Inhibition of SRY-calmodulin complex formation induces ectopic expression of ovarian cell markers in developing XY gonads. Endocrinology 2011, 152, 2883–2893. [Google Scholar] [CrossRef] [Green Version]
- McDowall, S.; Argentaro, A.; Ranganathan, S.; Weller, P.; Mertin, S.; Mansour, S.; Tolmie, J.; Harley, V. Functional and structural studies of wild SOX9 and mutations causing campomelic dysplasia. J. Biol. Chem. 1999, 274, 24023–24030. [Google Scholar] [CrossRef] [Green Version]
- Haseeb, A.; Lefebvre, V. The SOXE transcription factors-SOX8, SOX9 and SOX10-share a bi-partite transactivation mechanism. Nucleic Acids Res. 2019, 47, 6917–6931. [Google Scholar] [CrossRef] [PubMed]
- Südbeck, P.; Schmitz, M.L.; Baeuerle, P.A.; Scherer, G. Sex reversal by loss of the C-terminal transactivation domain of human SOX9. Nat. Genet. 1996, 13, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Jankowski, A.; Cheah, K.S.E.; Prabhakar, S.; Jauch, R. SOXE transcription factors form selective dimers on non-compact DNA motifs through multifaceted interactions between dimerization and high-mobility group domains. Sci. Rep. 2015, 5, 10398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.F.; Lefebvre, V. The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 2015, 43, 8183–8203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adam, R.C.; Yang, H.; Rockowitz, S.; Larsen, S.B.; Nikolova, M.; Oristian, D.S.; Polak, L.; Kadaja, M.; Asare, A.; Zheng, D.; et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 2015, 521, 366–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sim, H.; Argentaro, A.; Harley, V.R. Boys, girls and shuttling of SRY and SOX9. Trends Endocrinol. Metab. 2008, 19, 213–222. [Google Scholar] [CrossRef]
- Williams, C.A.C.; Soufi, A.; Pollard, S.M. Post-translational modification of SOX family proteins: Key biochemical targets in cancer? Semin. Cancer Biol. 2020, 67, 30–38. [Google Scholar] [CrossRef]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef] [Green Version]
- Voldoire, E.; Brunet, F.; Naville, M.; Volff, J.N.; Galiana, D. Expansion by whole genome duplication and evolution of the sox gene family in teleost fish. PLoS ONE 2017, 12, e180936. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Marí, A.; Yan, Y.L.; BreMiller, R.A.; Wilson, C.; Cañestro, C.; Postlethwait, J.H. Characterization and expression pattern of zebrafish anti-Müllerian hormone (amh) relative to sox9a, sox9b, and cyp19a1a, during gonad development. Gene Expr. Patterns 2005, 5, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, M.; Suzuki, A.; Matsuda, M.; Nagahama, Y.; Shibata, N. Testicular type Sox9 is not involved in sex determination but might be in the development of testicular structures in the medaka, Oryzias latipes. Biochem. Biophys. Res. Commun. 2005, 333, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Aoki, Y.; Saito, D.; Kuroki, Y.; Fujiyama, A.; Naruse, K.; Tanaka, M. Sox9b/sox9a2-EGFP transgenic medaka reveals the morphological reorganization of the gonads and a common precursor of both the female and male supporting cells. Mol. Reprod. Dev. 2008, 75, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Wallis, M.C.; Waters, P.D.; Delbridge, M.L.; Kirby, P.J.; Pask, A.J.; Grützner, F.; Rens, W.; Ferguson-Smith, M.A.; Graves, J.A.M. Sex determination in platypus and echidna: Autosomal location of SOX3 confirms the absence of SRY from monotremes. Chromosom. Res. 2007, 15, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Capel, B. Vertebrate sex determination: Evolutionary plasticity of a fundamental switch. Nat. Rev. Genet. 2017, 18, 675–689. [Google Scholar] [CrossRef]
- Kent, J.; Wheatley, S.C.; Andrews, J.E.; Sinclair, A.H.; Koopman, P. A male-specific role for SOX9 in vertebrate sex determination. Development 1996, 122, 2813–2822. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Zuckerkandl, E.; Pauling, L. Evolutionary Divergence and Convergence in Proteins. In Evolving Genes and Proteins; Bryson, V., Vogel, H., Eds.; Academic Press: New York, NY, USA, 1965; pp. 97–166. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Foster, J.W.; Dominguez-Steglich, M.A.; Guioli, S.; Kwok, C.; Weller, P.A.; Stevanović, M.; Weissenbach, J.; Mansour, S.; Young, I.D.; Goodfellow, P.N.; et al. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 1994, 372, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Barrionuevo, F.; Bagheri-Fam, S.; Klattig, J.; Kist, R.; Taketo, M.M.; Englert, C.; Scherer, G. Homozygous inactivation of Sox9 causes complete XY sex reversal in mice. Biol. Reprod. 2006, 74, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Wang, S.; Ning, Y.; Lamb, A.N.; Bartley, J. Autosomal XX sex reversal caused by duplication of SOX9. Am. J. Med. Genet. 1999, 87, 349–353. [Google Scholar] [CrossRef]
- Bishop, C.E.; Whitworth, D.J.; Qin, Y.; Agoulnik, A.I.; Agoulnik, I.U.; Harrison, W.R.; Behringer, R.R.; Overbeek, P.A. A transgenic insertion upstream of Sox9 is associated with dominant XX sex reversal in the mouse. Nat. Genet. 2000, 26, 490–494. [Google Scholar] [CrossRef]
- Sun, D.; Zhang, Y.; Wang, C.; Hua, X.; Zhang, X.A.; Yan, J. Sox9-related signaling controls zebrafish juvenile ovary-testis transformation. Cell Death Dis. 2013, 4, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, T.; Zhou, L.Y.; Chaudhari, A.; Iguchi, T.; Nagahama, Y. Dmy initiates masculinity by altering Gsdf/Sox9a2/Rspo1 expression in medaka (Oryzias latipes). Sci. Rep. 2016, 6, 19480. [Google Scholar] [CrossRef] [PubMed]
- Lambeth, L.; Raymond, C.S.; Roeszler, K.N.; Kuroiwa, A.; Nakata, T.; Zarkower, D.; Smith, C.A. Over-expression of DMRT1 induces the male pathway in embryonic chicken gonads. Dev. Biol. 2014, 389, 160–172. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Mendoza, N.; Harley, V.R.; Merchant-Larios, H. Differential expression of SOX9 in gonads of the sea turtle Lepidochelys olivacea at male- or female-promoting temperatures. J. Exp. Zool. 1999, 284, 705–710. [Google Scholar] [CrossRef]
- Torres-Maldonado, L.; Moreno-Mendoza, N.; Landa, A.; Merchant-Larios, H. Timing of SOX9 downregulation and female sex determination in gonads of the sea turtle Lepidochelys olivacea. J. Exp. Zool. 2001, 290, 498–503. [Google Scholar] [CrossRef]
- Rhen, T.; Metzger, K.; Schroeder, A.; Woodward, R. Expression of putative sex-determining genes during the thermosensitive period of gonad development in the snapping turtle, Chelydra serpentina. Sex. Dev. 2007, 1, 255–270. [Google Scholar] [CrossRef]
- Sun, W.; Cai, H.; Zhang, G.; Zhang, H.; Bao, H.; Wang, L.; Ye, J.; Qian, G.; Ge, C. Dmrt1 is required for primary male sexual differentiation in Chinese soft-shelled turtle Pelodiscus sinensis. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Sun, W.; Cai, H.; Bao, H.; Zhang, Y.; Qian, G.; Ge, C. The role of anti-Müllerian hormone in testis differentiation reveals the significance of the TGF-b pathway in reptilian sex determination. Genetics 2019, 213, 1317–1327. [Google Scholar] [CrossRef]
- Piprek, R.P.; Damulewicz, M.; Kloc, M.; Kubiak, J.Z. Transcriptome analysis identifies genes involved in sex determination and development of Xenopus laevis gonads. Differentiation 2018, 100, 46–56. [Google Scholar] [CrossRef] [PubMed]
- El Jamil, A.; Kanhoush, R.; Magre, S.; Boizet-Bonhoure, B.; Penrad-Mobayed, M. Sex-specific expression of SOX9 during gonadogenesis in the amphibian Xenopus tropicalis. Dev. Dyn. 2008, 237, 2996–3005. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, A.; Smith, C.; Western, P.; McClive, P. A comparative analysis of vertebrate sex determination. Novartis Found. Symp. 2002, 244, 102–114. [Google Scholar] [CrossRef]
- Moreno-Mendoza, N.; Harley, V.R.; Merchant-Larios, H. Temperature regulates SOX9 expression in cultured gonads of Lepidochelys olivacea, a species with temperature sex determination. Dev. Biol. 2001, 229, 319–326. [Google Scholar] [CrossRef] [Green Version]
- Oreal, E.; Pieau, C.; Mattei, M.G.; Josso, N.; Picard, J.Y.; Carré-Eusèbe, D.; Magre, S. Early expression of AMH in chicken embryonic gonads precedes testicular SOX9 expression. Dev. Dyn. 1998, 212, 522–532. [Google Scholar] [CrossRef]
- Yamashita, S.; Kataoka, K.; Yamamoto, H.; Kato, T.; Hara, S.; Yamaguchi, K.; Renard-Guillet, C.; Katou, Y.; Shirahige, K.; Ochi, H.; et al. Comparative analysis demonstrates cell type-specific conservation of SOX9 targets between mouse and chicken. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Estermann, M.A.; Williams, S.; Hirst, C.E.; Roly, Z.Y.; Serralbo, O.; Adhikari, D.; Powell, D.; Major, A.T.; Smith, C.A. Insights into Gonadal Sex Differentiation Provided by Single-Cell Transcriptomics in the Chicken Embryo. Cell Rep. 2020, 31, 107491. [Google Scholar] [CrossRef]
- Parma, P.; Veyrunes, F.; Pailhoux, E. Sex Reversal in Non-Human Placental Mammals. Sex. Dev. 2016, 10, 326–344. [Google Scholar] [CrossRef] [PubMed]
- Kist, R.; Schrewe, H.; Balling, R.; Scherer, G. Conditional inactivation of Sox9: A mouse model for campomelic dysplasia. Genesis 2002, 32, 121–123. [Google Scholar] [CrossRef] [PubMed]
- Seymour, P.A.; Freude, K.K.; Dubois, C.L.; Shih, H.-P.; Patel, N.A.; Sander, M. A Dosage-Dependent Requirement for SOX9 in Pancreatic Endocrine Cell Formation. Dev. Biol. 2008, 323, 19–30. [Google Scholar] [CrossRef] [Green Version]
- Unger, S.; Scherer, G.; Superti-Furga, A. Campomelic Dysplasia. In GeneReviews® [Internet]; Adam, M., Ardinger, H., Pagon, R., Al, E., Eds.; University of Washington: Seattle, WA, USA, 2008. [Google Scholar]
- Piper, K.; Ball, S.G.; Keeling, J.W.; Mansoor, S.; Wilson, D.I.; Hanley, N.A. Novel SOX9 expression during human pancreas development correlates to abnormalities in Campomelic dysplasia. Mech. Dev. 2002, 116, 223–226. [Google Scholar] [CrossRef]
- Hughes, I.A.; Houk, C.; Ahmed, S.F.; Lee, P.A.; LWPES Consensus Group; ESPE Consensus Group. Consensus statement on management of intersex disorders. Arch. Dis. Child 2006, 91, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Nordenvall, A.S.; Frisén, L.; Nordenström, A.; Lichtenstein, P.; Nordenskjöld, A. Population based nationwide study of hypospadias in Sweden, 1973 to 2009: Incidence and risk factors. J. Urol. 2014, 191, 783–789. [Google Scholar] [CrossRef]
- Matzuk, M.M.; Lamb, D.J. The biology of infertility: Research advances and clinical challenges. Nat. Med. 2008, 14, 1197–1213. [Google Scholar] [CrossRef]
- Eggers, S.; Sadedin, S.; van den Bergen, J.A.; Robevska, G.; Ohnesorg, T.; Hewitt, J.; Lambeth, L.; Bouty, A.; Knarston, I.M.; Tan, T.Y.; et al. Disorders of sex development: Insights from targeted gene sequencing of a large international patient cohort. Genome Biol. 2016, 17, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, S.; Guo, S.; Liu, L.; Yang, X.; Liang, H. Functional study of a novel c.630delG (p.Y211Tfs*85) mutation in NR5A1 gene in a Chinese boy with 46,XY disorders of sex development. J. Assist. Reprod. Genet. 2020, 37, 477–486. [Google Scholar] [CrossRef] [PubMed]
- León, N.Y.; Reyes, A.P.; Harley, V.R. A clinical algorithm to diagnose differences of sex development. Lancet Diabetes Endocrinol. 2019, 7, 560–574. [Google Scholar] [CrossRef]
- Wagner, T.; Wirth, J.; Meyer, J.; Zabel, B.; Held, M.; Zimmer, J.; Pasantes, J.; Bricarelli, F.D.; Keutel, J.; Hustert, E.; et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 1994, 79, 1111–1120. [Google Scholar] [CrossRef]
- Mansour, S.; Hall, C.M.; Pembrey, M.E.; Young, I.D. A clinical and genetic study of campomelic dysplasia. J. Med. Genet. 1995, 32, 415–420. [Google Scholar] [CrossRef] [Green Version]
- Lefebvre, V.; Dvir-Ginzberg, M. SOX9 and the many facets of its regulation in the chondrocyte lineage. Connect. Tissue Res. 2017, 58, 2–14. [Google Scholar] [CrossRef]
- Cui, Y.; Zhao, H.; Liu, Z.; Liu, C.; Luan, J.; Zhou, X.; Han, J. A systematic review of genetic skeletal disorders reported in Chinese biomedical journals between 1978 and 2012. Orphanet J. Rare Dis. 2012, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michel-Calemard, L.; Lesca, G.; Morel, Y.; Boggio, D.; Plauchu, H.; Attia-Sobo, J. Campomelic acampomelic dysplasia presenting with increased nuchal translucency in the first trimester. Prenat. Diagn. 2004, 24, 519–523. [Google Scholar] [CrossRef]
- Thong, M.K.; Scherer, G.; Kozlowski, K.; Haan, E.; Morris, L. Brief clinical report: Acampomelic campomelic dysplasia with SOX9 mutation. Am. J. Med. Genet. 2000, 93, 421–425. [Google Scholar] [CrossRef]
- Wada, Y.; Nishimura, G.; Nagai, T.; Sawai, H.; Yoshikata, M.; Miyagawa, S.; Hanita, T.; Sato, S.; Hasegawa, T.; Ishikawa, S.; et al. Mutation analysis of SOX9 and single copy number variant analysis of the upstream region in eight patients with campomelic dysplasia and acampomelic campomelic dysplasia. Am. J. Med. Genet. Part A 2009, 149A, 2882–2885. [Google Scholar] [CrossRef] [PubMed]
- Massardier, J.; Roth, P.; Michel-Calemard, L.; Rudigoz, R.C.; Bouvier, R.; Dijoud, F.; Arnould, P.; Combourieu, D.; Gaucherand, P. Campomelic dysplasia: Echographic suspicion in the first trimester of pregnancy and final diagnosis of two cases. Fetal Diagn. Ther. 2009, 24, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, M.; Kitoh, H.; Kaneko, H.; Mishima, K.; Kadono, I.; Ishiguro, N.; Nishimura, G. A novel SOX9 H169Q mutation in a family with overlapping phenotype of mild campomelic dysplasia and small patella syndrome. Am. J. Med. Genet. A 2013, 161A, 2528–2534. [Google Scholar] [CrossRef]
- Tonni, G.; Ventura, A.; Pattacini, P.; Bonasoni, M.P.; Baffico, A.M. p.His165Pro: A novel SOX9 missense mutation of campomelic dysplasia. J. Obstet. Gynaecol. Res. 2013, 39, 1085–1091. [Google Scholar] [CrossRef]
- Staffler, A.; Hammel, M.; Wahlbuhl, M.; Bidlingmaier, C.; Flemmer, A.W.; Pagel, P.; Nicolai, T.; Wegner, M.; Holzinger, A. Heterozygous SOX9 mutations allowing for residual DNA-binding and transcriptional activation lead to the acampomelic variant of campomelic dysplasia. Hum. Mutat. 2010, 31, 1436–1444. [Google Scholar] [CrossRef]
- Karaer, K.; Yüksel, Z.; Yalinbaş, E.; Scherer, G. A case of campomelic dysplasia in whom a new mutation was found in the SOX9 gene. Turk. Pediatr. Ars. 2014, 49, 154–156. [Google Scholar] [CrossRef]
- Preiss, S.; Argentaro, A.; Clayton, A.; John, A.; Jans, D.A.; Ogata, T.; Nagai, T.; Barroso, I.; Schafer, A.J.; Harley, V.R. Compound Effects of Point Mutations Causing Campomelic Dysplasia/Autosomal Sex Reversal upon SOX9 Structure, Nuclear Transport, DNA Binding, and Transcriptional Activation. J. Biol. Chem. 2001, 276, 27864–27872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shotelersuk, V.; Jaruratanasirikul, S.; Sinthuwiwat, T.; Janjindamai, W. A novel nonsense mutation, E150X, in the SOX9 gene underlying campomelic dysplasia. Genet. Mol. Biol. 2006, 29, 617–620. [Google Scholar] [CrossRef] [Green Version]
- Barone, C.; Bartoloni, G.; Baffico, A.M.; Pappalardo, E.; Mura, I.; Ettore, G.; Bianca, S. Novel c.358C>T mutation of SOX9 gene in prenatal diagnosis of campomelic dysplasia. Congenit. Anom. (Kyoto) 2014, 54, 193–194. [Google Scholar] [CrossRef] [Green Version]
- Kwok, C.; Weller, P.A.; Guioli, S.; Foster, J.W.; Mansour, S.; Zuffardi, O.; Punnett, H.H.; Dominguez-Steglich, M.A.; Brook, J.D.; Young, I.D.; et al. Mutations in SOX9, the gene responsible for campomelic dysplasia and autosomal sex reversal. Am. J. Hum. Genet. 1995, 57, 1028–1036. [Google Scholar] [PubMed]
- Gentilin, B.; Forzano, F.; Bedeschi, M.F.; Rizzuti, T.; Faravelli, F.; Izzi, C.; Lituania, M.; Rodriguez-Perez, C.; Bondioni, M.P.; Savoldi, G.; et al. Phenotype of five cases of prenatally diagnosed campomelic dysplasia harboring novel mutations of the SOX9 gene. Ultrasound Obstet. Gynecol. 2010, 36, 315–323. [Google Scholar] [CrossRef]
- Sock, E.; Pagon, R.A.; Keymolen, K.; Lissens, W.; Wegner, M.; Scherer, G. Loss of DNA-dependent dimerization of the transcription factor SOX9 as a cause for campomelic dysplasia. Hum. Mol. Genet. 2003, 12, 1439–1447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takenouchi, T.; Matsuzaki, Y.; Yamamoto, K.; Kosaki, K.; Torii, C.; Takahashi, T.; Kosaki, K. SOX9 dimerization domain mutation mimicking type 2 collagen disorder phenotype. Eur. J. Med. Genet. 2014, 57, 298–301. [Google Scholar] [CrossRef]
- Gogi, K.; Nishijima, E.; Tsugawa, C.; Nishio, H.; Pokharel, R.K.; Matsuo, M. Novel Missense Mutation in the HMG Box of SOX9 Gene in a Japanese XY Male Resulted in Campomelic Dysplasia and Severe Defect in Masculinization. Hum. Mutat. 1998, 116, 114–116. [Google Scholar]
- Posey, J.E.; Harel, T.; Liu, P.; Rosenfeld, J.A.; James, R.A.; Coban Akdemir, Z.H.; Walkiewicz, M.; Bi, W.; Xiao, R.; Ding, Y.; et al. Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation. N. Engl. J. Med. 2017, 376, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Pop, R.; Zaragoza, M.V.; Gaudette, M.; Dohrmann, U.; Scherer, G. A homozygous nonsense mutation in SOX9 in the dominant disorder campomelic dysplasia: A case of mitotic gene conversion. Hum. Genet. 2005, 117, 43–53. [Google Scholar] [CrossRef]
- Katoh-Fukui, Y.; Igarashi, M.; Nagasaki, K.; Horikawa, R.; Nagai, T.; Tsuchiya, T.; Suzuki, E.; Miyado, M.; Hata, K.; Nakabayashi, K.; et al. Testicular dysgenesis/regression without campomelic dysplasia in patients carrying missense mutations and upstream deletion of SOX9. Mol. Genet. Genomic Med. 2015, 3, 550–557. [Google Scholar] [CrossRef]
- Stoeva, R.; Grozdanova, L.; Scherer, G.; Krasteva, M.; Bausch, E.; Krastev, T.; Linev, A.; Stefanova, M. A novel SOX9 nonsense mutation, Q401X, in a case of campomelic dysplasia with XY sex reversal. Genet. Couns. 2011, 22, 49–53. [Google Scholar]
- Meyer, J.; Südbeck, P.; Held, M.; Wagner, T.; Schmitz, M.L.; Bricarelli, F.D.; Eggermont, E.; Friedrich, U.; Haas, O.A.; Kobelt, A.; et al. Mutational analysis of the SOX9 gene in campomelic dysplasia and autosomal sex reversal: Lack of genotype/phenotype correlations. Hum. Mol. Genet. 1997, 6, 91–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattos, E.P.; Sanseverino, M.T.; Magalhães, J.A.A.; Leite, J.C.L.; Félix, T.M.; Todeschini, L.A.; Cavalcanti, D.P.; Schüler-Faccini, L. Clinical and molecular characterization of a Brazilian cohort of campomelic dysplasia patients, and identification of seven new SOX9 mutations. Genet. Mol. Biol. 2015, 38, 14–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, I.A. Disorders of sex development: A new definition and classification. Best Pract. Res. Clin. Endocrinol. Metab. 2008, 22, 119–134. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.A.; Houk, C.P.; Ahmed, S.F.; Hughes, I.A.; International Consensus Conference on Intersex organized by the Lawson Wilkins Pediatric Endocrine Society and the European Society for Paediatric Endocrinology. Consensus statement on management of intersex disorders. Pediatrics 2006, 118, e488–e500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verkauskas, G.; Jaubert, F.; Lortat-Jacob, S.; Malan, V.; Thibaud, E.; Nihoul-Fékété, C. The long-term followup of 33 cases of true hermaphroditism: A 40-year experience with conservative gonadal surgery. J. Urol. 2007, 177, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.M.; Ko, J.M.; Shin, C.H.; Yang, S.W. A Korean boy with 46,XX testicular disorder of sex development caused by SOX9 duplication. Ann. Pediatr. Endocrinol. Metab. 2014, 19, 108–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Hernández, B.; Méndez, J.P.; Coral-Vázquez, R.M.; Benítez-Granados, J.; Zenteno, J.C.; Villegas-Ruiz, V.; Calzada-León, R.; Soderlund, D.; Canto, P. Duplication of SOX9 associated with 46,XX ovotesticular disorder of sex development. Reprod. Biomed. Online 2018, 37, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.J.; Willatt, L.; Homfray, T.; Woods, C.G. A SOX9 Duplication and Familial 46,XX Developmental Testicular Disorder. N. Engl. J. Med. 2011, 364, 91–93. [Google Scholar] [CrossRef] [Green Version]
- Hyon, C.; Chantot-Bastaraud, S.; Harbuz, R.; Bhouri, R.; Perrot, N.; Peycelon, M.; Sibony, M.; Rojo, S.; Piguel, X.; Bilan, F.; et al. Refining the regulatory region upstream of SOX9 associated with 46,XX testicular Disorders of Sex Development (DSD). Am. J. Med. Genet. Part A 2015, 167A, 1851–1858. [Google Scholar] [CrossRef] [Green Version]
- Vetro, A.; Dehghani, M.R.; Kraoua, L.; Giorda, R.; Beri, S.; Cardarelli, L.; Merico, M.; Manolakos, E.; Parada-Bustamante, A.; Castro, A.; et al. Testis development in the absence of SRY: Chromosomal rearrangements at SOX9 and SOX3. Eur. J. Hum. Genet. 2015, 23, 1025–1032. [Google Scholar] [CrossRef]
- Vetro, A.; Ciccone, R.; Giorda, R.; Patricelli, M.G.; Mina, E.D.; Forlino, A.; Zuffardi, O. XX males SRY negative: A confirmed cause of infertility. J. Med. Genet. 2011, 48, 710–712. [Google Scholar] [CrossRef]
- Lybæk, H.; de Bruijn, D.; den Engelsman-van Dijk, A.H.A.; Vanichkina, D.; Nepal, C.; Brendehaug, A.; Houge, G. RevSex duplication-induced and sex-related differences in the SOX9 regulatory region chromatin landscape in human fibroblasts. Epigenetics 2013, 9, 416–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Refai, O.; Friedman, A.; Terry, L.; Jewett, T.; Pearlman, A.; Perle, M.A.; Ostrer, H. De novo 12;17 translocation upstream of SOX9 resulting in 46,XX testicular disorder of sex development. Am. J. Med. Genet. A 2010, 152A, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Vidal, V.P.I.; Chaboissier, M.C.; de Rooij, D.G.; Schedl, A. Sox9 induces testis development in XX transgenic mice. Nat. Genet. 2001, 28, 216–217. [Google Scholar] [CrossRef]
- Bagheri-Fam, S.; Combes, A.N.; Ling, C.K.; Wilhelm, D. Heterozygous deletion of Sox9 in mouse mimics the gonadal sex reversal phenotype associated with campomelic dysplasia in humans. Hum. Mol. Genet. 2021, 29, 3781–3792. [Google Scholar] [CrossRef] [PubMed]
- Ushijima, K.; Ogawa, Y.; Asakura, Y.; Muroya, K.; Hayashi, M.; Ishii, T.; Hasegawa, T.; Takada, S.; Narumi, S.; Sekido, R.; et al. Identification of the first promoter-specific gain-of-function SOX9 missense variant (p.E50K) in a patient with 46, XX ovotesticular disorder of sex development. Am. J. Hum. Genet. A 2021, 1–9. [Google Scholar] [CrossRef]
- Vidal, V.P.I.; Chaboissier, M.C.; Lützkendorf, S.; Cotsarelis, G.; Mill, P.; Hui, C.C.; Ortonne, N.; Ortonne, J.P.; Schedl, A. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr. Biol. 2005, 15, 1340–1351. [Google Scholar] [CrossRef] [Green Version]
- Fantauzzo, K.A.; Kurban, M.; Levy, B.; Christiano, A.M. Trps1 and Its Target Gene Sox9 Regulate Epithelial Proliferation in the Developing Hair Follicle and Are Associated with Hypertrichosis. PLoS Genet. 2012, 8, e1003002. [Google Scholar] [CrossRef] [Green Version]
- Bashamboo, A.; McElreavey, K. Human sex-determination and disorders of sex-development (DSD). Semin. Cell Dev. Biol. 2015, 45, 77–83. [Google Scholar] [CrossRef]
- Lee, P.A.; Nordenström, A.; Houk, C.P.; Ahmed, S.F.; Auchus, R.; Baratz, A.; Baratz Dalke, K.; Liao, L.M.; Lin-Su, K.; Looijenga, L.H.J.; et al. Global disorders of sex development update since 2006: Perceptions, approach and care. Horm. Res. Paediatr. 2016, 85, 158–180. [Google Scholar] [CrossRef]
- de Vries, A.L.C.; Roehle, R.; Marshall, L.; Frisén, L.; van de Grift, T.C.; Kreukels, B.P.C.; Bouvattier, C.; Köhler, B.; Thyen, U.; Nordenström, A.; et al. Mental Health of a Large Group of Adults with Disorders of Sex Development in Six European Countries. Psychosom. Med. 2019, 81, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Burgoyne, P.S.; Buehr, M.; McLaren, A. XY follicle cells in ovaries of XX ⇆ XY female mouse chimaeras. Development 1988, 104, 683–688. [Google Scholar] [PubMed]
- Georges, A.; L’Hôte, D.; Todeschini, A.L.; Auguste, A.; Legois, B.; Zider, A.; Veitia, R.A. The transcription factor FOXL2 mobilizes estrogen signaling to maintain the identity of ovarian granulosa cells. Elife 2014, 3, e04207. [Google Scholar] [CrossRef] [Green Version]
- Vainio, S.; Heikkilä, M.; Kispert, A.; Chin, N.; McMahon, A.P. Female development in mammals is regulated by Wnt-4 signalling. Nature 1999, 397, 405–409. [Google Scholar] [CrossRef]
- Parma, P.; Radi, O.; Vidal, V.; Chaboissier, M.C.; Dellambra, E.; Valentini, S.; Guerra, L.; Schedl, A.; Camerino, G. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat. Genet. 2006, 38, 1304–1309. [Google Scholar] [CrossRef]
- Ottolenghi, C.; Pelosi, E.; Tran, J.; Colombino, M.; Douglass, E.; Nedorezov, T.; Cao, A.; Forabosco, A.; Schlessinger, D. Loss of Wnt4 and Foxl2 leads to female-to-male sex reversal extending to germ cells. Hum. Mol. Genet. 2007, 16, 2795–2804. [Google Scholar] [CrossRef] [Green Version]
- Bernard, P.; Ryan, J.; Sim, H.; Czech, D.P.; Sinclair, A.H.; Koopman, P.; Harley, V.R. Wnt signaling in ovarian development inhibits Sf1 activation of Sox9 via the Tesco enhancer. Endocrinology 2012, 153, 901–912. [Google Scholar] [CrossRef] [Green Version]
- Barrionuevo, F.J.; Hurtado, A.; Kim, G.J.; Real, F.M.; Bakkali, M.; Kopp, J.L.; Sander, M.; Scherer, G.; Burgos, M.; Jiménez, R. Sox9 and Sox8 protect the adult testis from male-to-female genetic reprogramming and complete degeneration. Elife 2016, 5, e15635. [Google Scholar] [CrossRef] [Green Version]
- Georg, I.; Barrionuevo, F.; Wiech, T.; Scherer, G. Sox9 and Sox8 are required for basal lamina integrity of testis cords and for suppression of FOXL2 during embryonic testis development in mice. Biol. Reprod. 2012, 87, 99. [Google Scholar] [CrossRef] [PubMed]

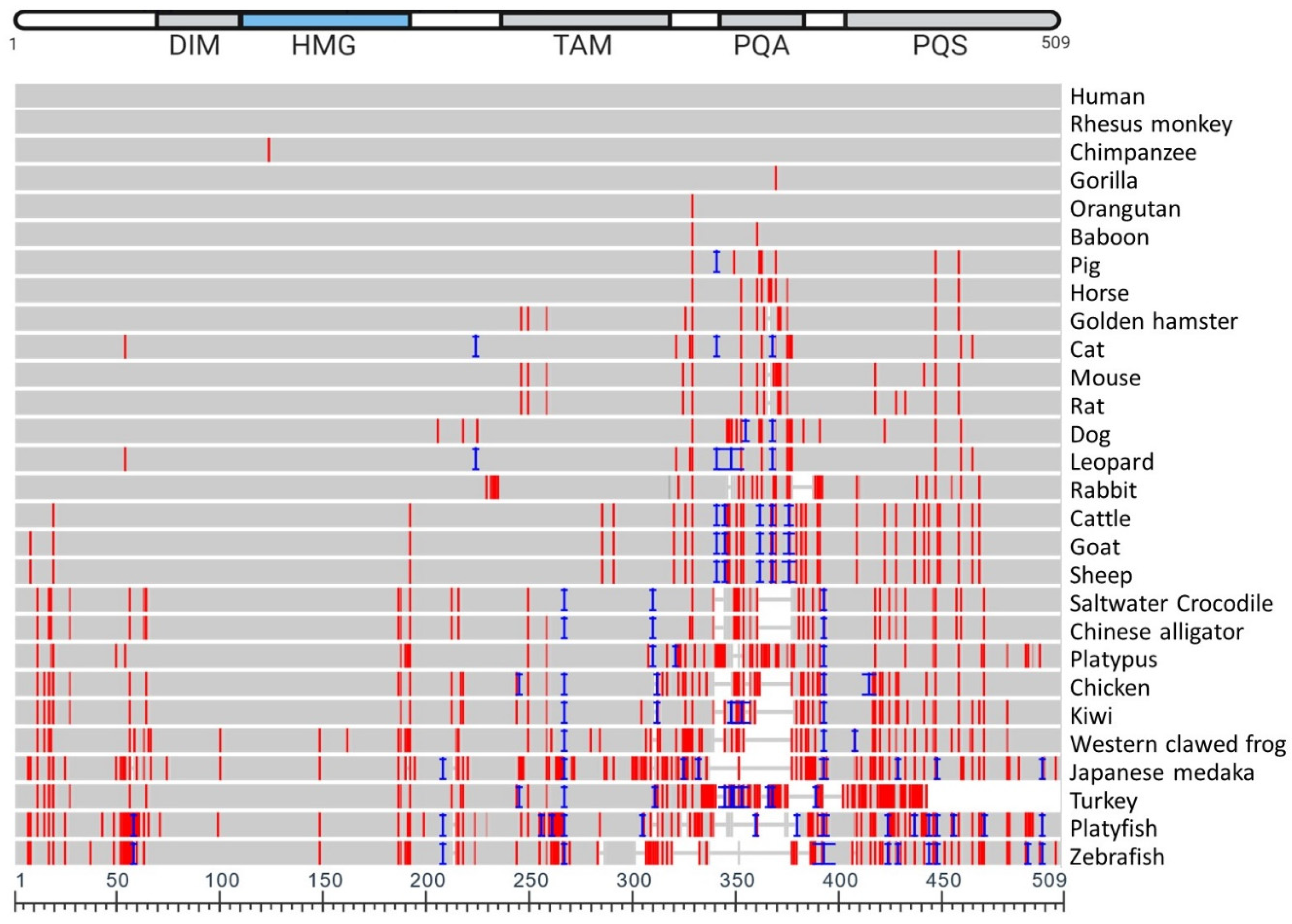

| NCBI Sequence ID | Amino Acid Length | Species Name | Species Common Name | Identity (%) | Coverage (%) | Mismatches |
|---|---|---|---|---|---|---|
| NP_000337.1 | 509 | Homo sapiens | Human | - | - | |
| NP_001028040.1 | 509 | Macaca mulatta | Rhesus monkey | 100 | 100 | 0 |
| XP_009250264.1 | 509 | Pan troglodytes | Chimpanzee | 99.80 | 100 | 1 |
| XP_018883823.1 | 509 | Pongo abelii | Orangutan | 99.80 | 100 | 1 |
| NP_001009029.1 | 509 | Gorilla gorilla gorilla | Gorilla | 99.80 | 100 | 1 |
| XP_003913405.1 | 509 | Papio Anubis | Baboon | 99.61 | 100 | 2 |
| NP_999008.2 | 511 | Sus scrofa | Pig | 98.04 | 100 | 8 |
| XP_023507898.1 | 509 | Equus caballus | Horse | 98.04 | 100 | 10 |
| XP_005070025.1 | 507 | Mesocricetus auratus | Golden hamster | 96.86 | 99.61 | 14 |
| XP_023099583.1 | 511 | Felis catus | Cat | 96.68 | 99.80 | 13 |
| NP_035578.3 | 507 | Mus musculus | Mouse | 96.46 | 99.61 | 16 |
| XP_032769123.1 | 507 | Rattus rattus | Rat | 96.27 | 99.61 | 17 |
| NP_001002978.1 | 513 | Canis lupus familiaris | Dog | 94.75 | 99.80 | 21 |
| XP_019321937.1 | 529 | Panthera pardus | Leopard | 93.40 | 99.80 | 13 |
| XP_008269985.1 | 497 | Oryctolagus cuniculus | Rabbit | 91.75 | 97.64 | 30 |
| XP_024836864.1 | 524 | Bos taurus | Cattle | 90.84 | 100 | 33 |
| XP_017919394.1 | 525 | Capra hircus | Goat | 90.48 | 100 | 34 |
| XP_027829812.1 | 526 | Ovis aries | Sheep | 90.30 | 100 | 34 |
| XP_020858282.1 | 513 | Phascolarctos cinereus | Koala | 89.60 | 98.82 | 38 |
| XP_006029531.1 | 494 | Alligator sinesis | Chinese alligator | 88.13 | 96.07 | 36 |
| XP_019397875.1 | 494 | Crocodylus porosus | Crocodile | 88.52 | 96.07 | 34 |
| XP_001506094.2 | 508 | Ornithorhynchus anatinus | Platypus | 87.55 | 98.82 | 53 |
| NP_989612.1 | 494 | Gallus gallus | Chicken | 83.52 | 94.50 | 45 |
| NP_001016853.1 | 482 | Xenopus tropicalis | Western clawed frog | 81.64 | 94.11 | 61 |
| NP_001098556.1 | 476 | Oryzias latipes | Japanese medaka | 72.74 | 80.94 | 85 |
| XP_025923282.1 | 500 | Apteryx rowi | Kiwi | 82.73 | 94.70 | 46 |
| XP_010719808.1 | 451 | Meleagris gallopavo | Turkey | 71.64 | 80.94 | 66 |
| XP_005807407.1 | 495 | Xiphosphorus maculatus | Platyfish | 70.58 | 91.75 | 88 |
| NP_571718.1 | 462 | Danio rerio | Zebrafish | 69.96 | 85.46 | 60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vining, B.; Ming, Z.; Bagheri-Fam, S.; Harley, V. Diverse Regulation but Conserved Function: SOX9 in Vertebrate Sex Determination. Genes 2021, 12, 486. https://doi.org/10.3390/genes12040486
Vining B, Ming Z, Bagheri-Fam S, Harley V. Diverse Regulation but Conserved Function: SOX9 in Vertebrate Sex Determination. Genes. 2021; 12(4):486. https://doi.org/10.3390/genes12040486
Chicago/Turabian StyleVining, Brittany, Zhenhua Ming, Stefan Bagheri-Fam, and Vincent Harley. 2021. "Diverse Regulation but Conserved Function: SOX9 in Vertebrate Sex Determination" Genes 12, no. 4: 486. https://doi.org/10.3390/genes12040486
APA StyleVining, B., Ming, Z., Bagheri-Fam, S., & Harley, V. (2021). Diverse Regulation but Conserved Function: SOX9 in Vertebrate Sex Determination. Genes, 12(4), 486. https://doi.org/10.3390/genes12040486





