Transcriptome Analysis Provides Insights into the Mechanism of Astaxanthin Enrichment in a Mutant of the Ridgetail White Prawn Exopalaemon carinicauda
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Isolation, Sample Pooling, and cDNA Synthesis
2.3. Library Construction and Sequencing
2.4. Assembly and Annotation
2.5. Analysis of Differentially Expressed Unigenes
2.6. Multiple Sequence Alignment and Phylogenetic Analysis
2.7. Validation by Quantitative Real-Time PCR
2.8. Statistical Analyses
3. Results
3.1. Basic Information of the Transcriptome
3.2. Identification and Verification of Differentially Expressed Genes (DEGs)
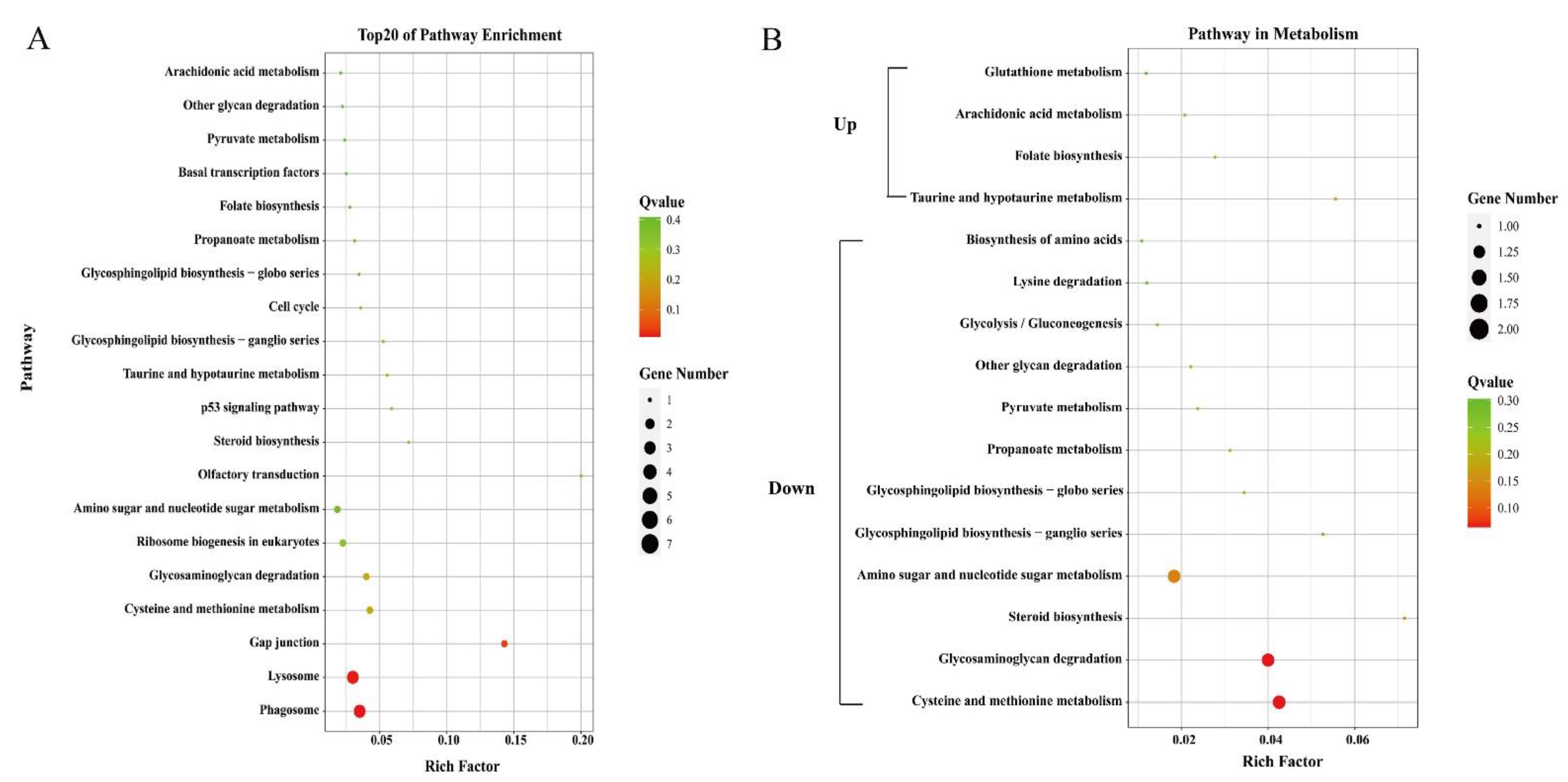
3.3. GO Term and KEGG Pathway Enrichment Analysis of DEGs
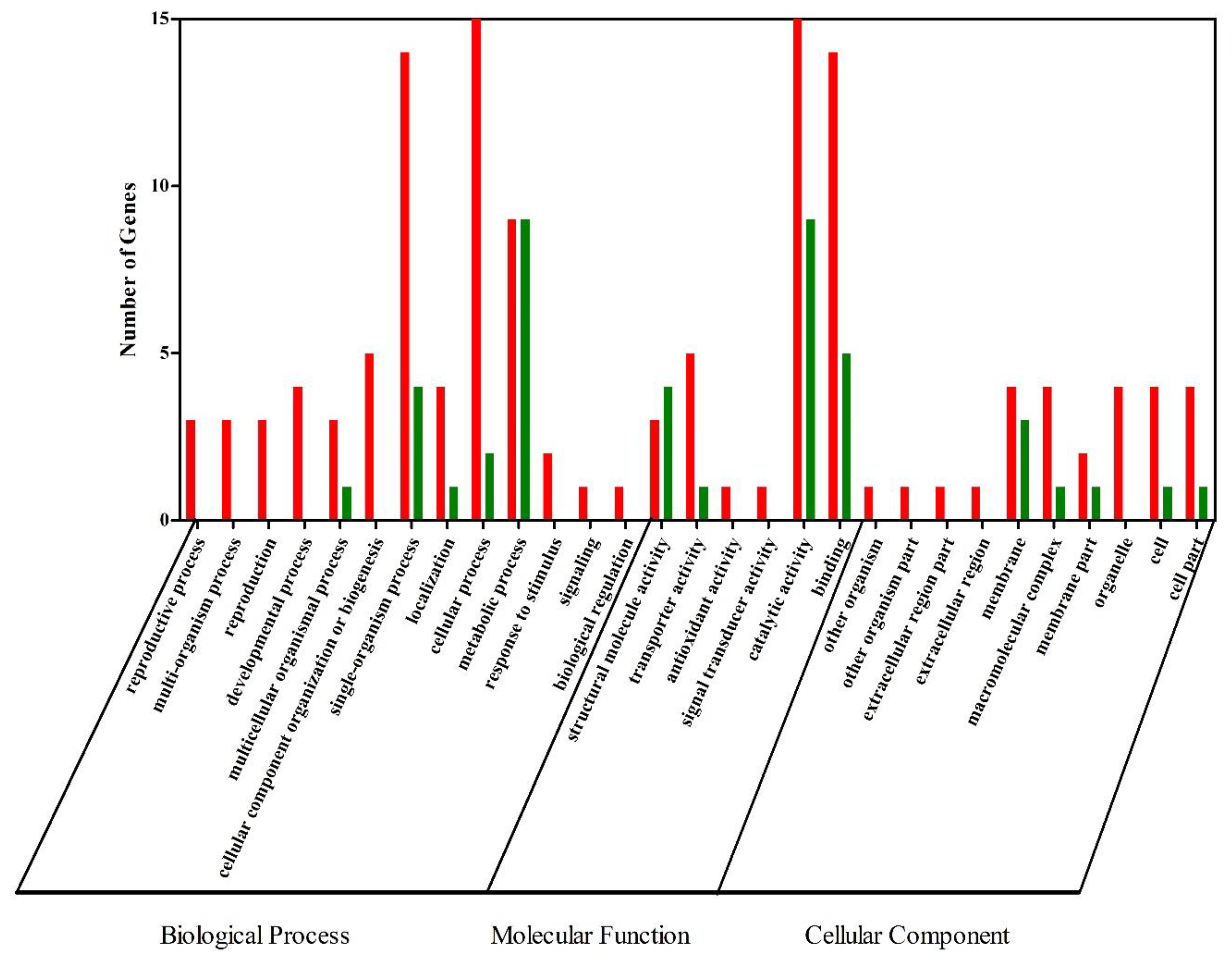
3.4. Functional Classification of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Britton, G.; Liaaen-Jensen, S.; Pfander, H. Carotenoids Handbook; Birkhäuser: Basel, Switzerland, 2004. [Google Scholar]
- Maoka, T. Recent progress in structural studies of carotenoids in animals and plants. Arch. Biochem. Biophys. 2009, 483, 191–195. [Google Scholar] [CrossRef]
- Maoka, T. Carotenoids as natural functional pigments. J. Nat. Med. 2020, 74, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, A.V.; Rao, L.G. Carotenoids and human health. Pharm. Res. 2007, 55, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Dall, W.S.; Smith, D.M.; Moore, L.E. Carotenoids in the tiger prawn Penaeus esculentus during ovarian maturation. Mar. Biol. 1995, 123, 435–441. [Google Scholar] [CrossRef]
- Liñán-Cabello, M.A.; Paniagua-Michel, J. Induction factors derived from carotenoids and vitamin A during the ovarian maturation of Litopenaeus vannamei. Aquac. Int. 2004, 12, 583–592. [Google Scholar] [CrossRef]
- Stradi, R.; Pini, E.; Celentano, G. Carotenoids in bird plumage: The complement of red pigments in the plumage of wild and captive bullfinch (Pyrrhula pyrrhula). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2001, 128, 529–535. [Google Scholar] [CrossRef]
- Lehnert, S.J.; Christensen, K.A.; Vandersteen, W.E.; Sakhrani, D.; Pitcher, T.E.; Heath, J.W.; Koop, B.F.; Heath, D.D.; Devlin, R.H. Carotenoid pigmentation in salmon: Variation in expression at BCO2-l locus controls a key fitness trait affecting red coloration. Proc. Biol. Sci. 2019, 286, 20191588. [Google Scholar] [CrossRef] [Green Version]
- Lopes, R.J.; Johnson, J.D.; Toomey, M.B.; Ferreira, M.S.; Araujo, P.M.; Melo-Ferreira, J.; Andersson, L.; Hill, G.E.; Corbo, J.C.; Carneiro, M. Genetic Basis for Red Coloration in Birds. Curr. Biol. 2016, 26, 1427–1434. [Google Scholar] [CrossRef] [Green Version]
- Mundy, N.I.; Stapley, J.; Bennison, C.; Tucker, R.; Twyman, H.; Kim, K.W.; Burke, T.; Birkhead, T.R.; Andersson, S.; Slate, J. Red Carotenoid Coloration in the Zebra Finch Is Controlled by a Cytochrome P450 Gene Cluster. Curr. Biol. 2016, 26, 1435–1440. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Zhang, Y.Q. Identification and analysis of the pigment composition and sources in the colored cocoon of the silkworm, Bombyx mori, by HPLC-DAD. J. Insect Sci. 2014, 14, 31. [Google Scholar] [CrossRef] [Green Version]
- Wybouw, N.; Kurlovs, A.H.; Greenhalgh, R.; Bryon, A.; Kosterlitz, O.; Manabe, Y.; Osakabe, M.; Vontas, J.; Clark, R.M.; Van Leeuwen, T. Convergent evolution of cytochrome P450s underlies independent origins of keto-carotenoid pigmentation in animals. Proc. Biol. Sci. 2019, 286, 20191039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundvold, H.; Helgeland, H.; Baranski, M.; Omholt, S.W.; Vage, D.I. Characterisation of a novel paralog of scavenger receptor class B member I (SCARB1) in Atlantic salmon (Salmo salar). BMC Genet. 2011, 12, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Hu, J.J.; Wang, S.; Cheng, J.; Hu, X.L.; Lu, Z.Y.; Lin, Z.J.; Zhu, W.M.; Bao, Z.M. Isolation and identification of the main carotenoid pigment from the rare orange muscle of the Yesso scallop. Food Chem. 2010, 118, 616–619. [Google Scholar] [CrossRef]
- Coral-Hinostroza, G.N.; Bjerkeng, B. Astaxanthin from the red crab langostilla (Pleuroncodes planipes): Optical R/S isomers and fatty acid moieties of astaxanthin esters. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2002, 133, 437–444. [Google Scholar] [CrossRef]
- Wade, N.; Goulter, K.C.; Wilson, K.J.; Hall, M.R.; Degnan, B.M. Esterified astaxanthin levels in lobster epithelia correlate with shell colour intensity: Potential role in crustacean shell colour formation. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2005, 141, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Nm, S.; Narayan, B.; Mahendrakar, N. Carotenoids in different body components of Indian shrimps. J. Sci. Food Agric. 2005, 85, 167–172. [Google Scholar]
- Toews, D.P.L.; Hofmeister, N.R.; Taylor, S.A. The evolution and genetics of carotenoid processing in animals. Trends Genet. 2017, 33, 171–182. [Google Scholar] [CrossRef]
- Reboul, E. Absorption of vitamin A and carotenoids by the enterocyte: Focus on transport proteins. Nutrients 2013, 5, 3563–3581. [Google Scholar] [CrossRef] [Green Version]
- Carriere, F.; Barrowman, J.A.; Verger, R.; Laugier, R. Secretion and contribution to lipolysis of gastric and pancreatic lipases during a test meal in humans. Gastroenterology 1993, 105, 876–888. [Google Scholar] [CrossRef]
- van Het Hof, K.H.; West, C.E.; Weststrate, J.A.; Hautvast, J.G. Dietary factors that affect the bioavailability of carotenoids. J. Nutr. 2000, 130, 503–506. [Google Scholar] [CrossRef]
- Parker, R.S. Absorption, metabolism, and transport of carotenoids. FASEB J. 1996, 10, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Pointer, M.A.; Prager, M.; Andersson, S.; Mundy, N.I. A novel method for screening a vertebrate transcriptome for genes involved in carotenoid binding and metabolism. Mol. Ecol. Resour. 2012, 12, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Walsh, N.; Dale, J.; McGraw, K.J.; Pointer, M.A.; Mundy, N.I. Candidate genes for carotenoid coloration in vertebrates and their expression profiles in the carotenoid-containing plumage and bill of a wild bird. Proc. Biol. Sci. 2012, 279, 58–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reboul, E.; Abou, L.; Mikail, C.; Ghiringhelli, O.; Andre, M.; Portugal, H.; Jourdheuil-Rahmani, D.; Amiot, M.J.; Lairon, D.; Borel, P. Lutein transport by Caco-2 TC-7 cells occurs partly by a facilitated process involving the scavenger receptor class B type I (SR-BI). Biochem. J. 2005, 387, 455–461. [Google Scholar] [CrossRef] [PubMed]
- During, A.; Doraiswamy, S.; Harrison, E.H. Xanthophylls are preferentially taken up compared with β-carotene by retinal cells via a SRBI-dependent mechanism. J. Lipid Res. 2008, 49, 1715–1724. [Google Scholar] [CrossRef] [Green Version]
- van Bennekum, A.; Werder, M.; Thuahnai, S.T.; Han, C.H.; Duong, P.; Williams, D.L.; Wettstein, P.; Schulthess, G.; Phillips, M.C.; Hauser, H. Class B scavenger receptor-mediated intestinal absorption of dietary β-carotene and cholesterol. Biochemistry 2005, 44, 4517–4525. [Google Scholar] [CrossRef]
- Toomey, M.B.; Lopes, R.J.; Araujo, P.M.; Johnson, J.D.; Gazda, M.A.; Afonso, S.; Mota, P.G.; Koch, R.E.; Hill, G.E.; Corbo, J.C.; et al. High-density lipoprotein receptor SCARB1 is required for carotenoid coloration in birds. Proc. Natl. Acad. Sci. USA 2017, 114, 5219–5224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiefer, C.; Sumser, E.; Wernet, M.F.; Von Lintig, J. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc. Natl. Acad. Sci. USA 2002, 99, 10581–10586. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Zheng, H.; Zhang, H.; Deng, L.; Liu, W.; Wang, S.; Meng, F.; Wang, Y.; Guo, Z.; Li, S.; et al. A de novo transcriptome of the noble scallop, Chlamys nobilis, focusing on mining transcripts for carotenoid-based coloration. BMC Genom. 2015, 16, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuchida, K.; Arai, M.; Tanaka, Y.; Ishihara, R.; Ryan, R.O.; Maekawa, H. Lipid transfer particle catalyzes transfer of carotenoids between lipophorins of Bombyx mori. Insect Biochem. Mol. Biol. 1998, 28, 927–934. [Google Scholar] [CrossRef]
- During, A.; Dawson, H.D.; Harrison, E.H. Carotenoid transport is decreased and expression of the lipid transporters SR-BI, NPC1L1, and ABCA1 is downregulated in Caco-2 cells treated with ezetimibe. J. Nutr. 2005, 135, 2305–2312. [Google Scholar] [CrossRef]
- Ando, S.; Hatano, M. Bilirubin-binding protein in the serum of spawning-migrating chum salmon, Oncorhynchus keta: Its identity with carotenoid-carrying lipoprotein. Fish. Physiol. Biochem. 1988, 5, 69–78. [Google Scholar] [CrossRef]
- Eriksson, J.; Larson, G.; Gunnarsson, U.; Bed’hom, B.; Tixier-Boichard, M.; Stromstedt, L.; Wright, D.; Jungerius, A.; Vereijken, A.; Randi, E.; et al. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 2008, 4, e1000010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Wang, S.; Xun, X.; Zhang, M.; Wang, S.; Li, H.; Zhao, L.; Fu, Q.; Wang, H.; Li, T.; et al. A carotenoid oxygenase is responsible for muscle coloration in scallop. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 966–975. [Google Scholar] [CrossRef]
- Weesie, R.J.; Askin, D.; Jansen, F.J.; de Groot, H.J.; Lugtenburg, J.; Britton, G. Protein-chromophore interactions in α-crustacyanin, the major blue carotenoprotein from the carapace of the lobster, Homarus gammarus. A study by 13C magic angle spinning NMR. FEBS Lett. 1995, 362, 34–38. [Google Scholar] [CrossRef] [Green Version]
- Zagalsky, P.F. β-Crustacyanin, the blue-purple carotenoprotein of lobster carapace: Consideration of the bathochromic shift of the protein-bound astaxanthin. Acta Cryst. D Biol. Cryst. 2003, 59, 1529–1531. [Google Scholar] [CrossRef] [Green Version]
- Ertl, N.G.; Elizur, A.; Brooks, P.; Kuballa, A.V.; Anderson, T.A.; Knibb, W.R. Molecular characterisation of colour formation in the prawn Fenneropenaeus merguiensis. PLoS ONE 2013, 8, e56920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weaver, R.J.; Gonzalez, B.K.; Santos, S.R.; Havird, J.C. Red Coloration in an Anchialine Shrimp: Carotenoids, Genetic Variation, and Candidate Genes. Biol. Bull. 2020, 238, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Yu, Y.; Zhang, C.; Li, S.; Zhang, X.; Li, F. Characterization and function analysis of the β-carotene oxygenase-like genes in carotenoids metabolism of the ridgetail white prawn Exopalaemon carinicauda. Front. Physiol. 2020, 11, 745. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Su, F.; Li, S.; Yu, Y.; Xiang, J.; Liu, J.; Li, F. Isolation and identification of the main carotenoid pigment from a new variety of the ridgetail white prawn Exopalaemon carinicauda. Food Chem. 2018, 269, 450–454. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, X.; Wei, J.; Sun, X.; Yuan, J.; Li, F.; Xiang, J. Whole Transcriptome Analysis Provides Insights into Molecular Mechanisms for Molting in Litopenaeus vannamei. PLoS ONE 2015, 10, e0144350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef]
- Avarre, J.C.; Lubzens, E.; Babin, P.J. Apolipocrustacein, formerly vitellogenin, is the major egg yolk precursor protein in decapod crustaceans and is homologous to insect apolipophorin II/I and vertebrate apolipoprotein B. BMC Evol. Biol. 2007, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Sun, Z. Random local neighbor joining: A new method for reconstructing phylogenetic trees. Mol. Phylogenet. Evol. 2008, 47, 117–128. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Johnson, G.; Nour, A.A.; Nolan, T.; Huggett, J.; Bustin, S. Minimum information necessary for quantitative real-time PCR experiments. Methods Mol. Biol. 2014, 1160, 5–17. [Google Scholar]
- Livak, K.; Schmittgen, T. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCt Method. Methods 2000, 25, 402–408. [Google Scholar] [CrossRef]
- Chimsung, N.; Lall, S.P.; Tantikitti, C.; Verlhac-Trichet, V.; Milley, J.E. Effects of dietary cholesterol on astaxanthin transport in plasma of Atlantic salmon (Salmo salar). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2013, 165, 73–81. [Google Scholar] [CrossRef]
- Deming, D.M.; Erdman, J.W. Mammalian carotenoid absorption and metabolism. Pure Appl. Chem. 1999, 71, 2213–2223. [Google Scholar] [CrossRef] [Green Version]
- Borel, P. Factors affecting intestinal absorption of highly lipophilic food microconstituents (fat-soluble vitamins, carotenoids and phytosterols). Clin. Chem. Lab. Med. 2003, 41, 979–994. [Google Scholar] [CrossRef]
- Breithaupt, D.E.; Bamedi, A.; Wirt, U. Carotenol fatty acid esters: Easy substrates for digestive enzymes? Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2002, 132, 721–728. [Google Scholar] [CrossRef]
- Bhosale, P.; Bernstein, P.S. Vertebrate and invertebrate carotenoid-binding proteins. Arch. Biochem. Biophys. 2007, 458, 121–127. [Google Scholar] [CrossRef] [Green Version]
- Parker, R.S. Carotenoids in human blood and tissues. J. Nutr. 1989, 119, 101–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hata, T.R.; Scholz, T.A.; Ermakov, I.V.; McClane, R.W.; Khachik, F.; Gellermann, W.; Pershing, L.K. Non-invasive raman spectroscopic detection of carotenoids in human skin. J. Investig. Derm. 2000, 115, 441–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rassart, E.; Bedirian, A.; Do Carmo, S.; Guinard, O.; Sirois, J.; Terrisse, L.; Milne, R. Apolipoprotein D. Biochim. Biophys. Acta 2000, 1482, 185–198. [Google Scholar] [CrossRef]
- Yang, F.; Wang, M.R.; Ma, Y.G.; Ma, W.M.; Yang, W.J. Prawn lipocalin: Characterization of a color shift induced by gene knockdown and ligand binding assay. J. Exp. Zool. A Ecol. Genet. Physiol. 2011, 315, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Ando, S.; Takeyama, T.; Hatano, M. Transport Associated with Serum Vitellogenin of Carotenoid in Chum Salmon (Oncorhynchus keta). Agric. Biol. Chem. 1986, 50, 557–563. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Vachali, P.; Frederick, J.M.; Bernstein, P.S. Identification of StARD3 as a lutein-binding protein in the macula of the primate retina. Biochemistry 2011, 50, 2541–2549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shandala, T.; Lim, C.; Sorvina, A.; Brooks, D.A. A Drosophila model to image phagosome maturation. Cells 2013, 2, 188–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trimble, W.S.; Coppolino, M.G. Phagosome Maturation. In Molecular Mechanisms of Phagocytosis; Rosales, C., Ed.; Springer: Boston, MA, USA, 2005; pp. 133–150. [Google Scholar]
- Xu, H.; Ren, D. Lysosomal physiology. Annu. Rev. Physiol. 2015, 77, 57–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheesman, D.; Lee, W.; Zagalsky, P. Carotenoproteins in Invertebrates. Biol. Rev. 2008, 42, 131–160. [Google Scholar] [CrossRef]
- Chayen, N.E.; Cianci, M.; Grossmann, J.G.; Habash, J.; Helliwell, J.R.; Nneji, G.A.; Raftery, J.; Rizkallah, P.J.; Zagalsky, P.F. Unravelling the structural chemistry of the colouration mechanism in lobster shell (vol D59, pg 2072, 2003). Acta Cryst. D 2005, 61, 1312. [Google Scholar] [CrossRef] [Green Version]
- Cianci, M.; Rizkallah, P.J.; Olczak, A.; Raftery, J.; Chayen, N.E.; Zagalsky, P.F.; Helliwell, J.R. The molecular basis of the coloration mechanism in lobster shell: β-crustacyanin at 3.2-angstrom resolution. Proc. Natl. Acad. Sci. USA 2002, 99, 9795–9800. [Google Scholar] [CrossRef] [Green Version]
- Kuballa, A.V.; Merritt, D.J.; Elizur, A. Gene expression profiling of cuticular proteins across the moult cycle of the crab Portunus pelagicus. BMC Biol. 2007, 5, 45. [Google Scholar] [CrossRef] [Green Version]
- Xiong, G.; Tong, X.; Yan, Z.; Hu, H.; Duan, X.; Li, C.; Han, M.; Lu, C.; Dai, F. Cuticular protein defective Bamboo mutant of Bombyx mori is sensitive to environmental stresses. Pestic. Biochem. Physiol. 2018, 148, 111–115. [Google Scholar] [CrossRef]
- Asano, T.; Taoka, M.; Shinkawa, T.; Yamauchi, Y.; Isobe, T.; Sato, D. Identification of a cuticle protein with unique repeated motifs in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2013, 43, 344–351. [Google Scholar] [CrossRef]
- Xiong, G.; Tong, X.; Gai, T.; Li, C.; Qiao, L.; Monteiro, A.; Hu, H.; Han, M.; Ding, X.; Wu, S.; et al. Body Shape and Coloration of Silkworm Larvae Are Influenced by a Novel Cuticular Protein. Genetics 2017, 207, 1053–1066. [Google Scholar] [CrossRef] [Green Version]



| Gene ID | Description | Sequence (5′ to 3′) | Tm (°C) |
|---|---|---|---|
| Unigene0059050 | Esterase FE4 | F: GTTGGACCAAATTTCGGCTCTT R: TGGAGAGAGCGCATGAAAGTTA | 59 |
| Unigene0054180 | Crustacyanin subunit A | F: TCCTGAGGGAACGCACGAGA R: TGGAGAAGACGAAGGCGAACT | 58 |
| Unigene0036561 | Apolipoprotein D | F: CTGGGCACAGATTACGAGAACT R: GGTGGGTACGATGGAAGAGG | 64 |
| Unigene0037069 | Cuticular protein 34 | F: CAGAAGCCACGAGCCAGAAG R: GGGTCCTACGAGGATGATGTTAT | 57 |
| Unigene0038920 | Cuticle proprotein | F: CGTAACTGGCACCTGGATGA R: AGAGGATAGCGTGGGCTGGA | 59 |
| Unigene0043447 | Cuticular protein 34 | F: TCGTCAGAGCCGATGGAAAC R: TGGGAGTGCCATCATCAGTG | 58 |
| Unigene0030347 | None | F: CTATGCGAATGAATAAGATGAGGAG R: CCAGCGTACCAAGTAATACTGAAA | 57 |
| Unigene0033842 | Retrovirus-related Pol polyprotein | F: CGATTCACTGTCCCCACTACTC R: GTCTATTTCCTTGATGCTCTTACCA | 56 |
| Unigene0047842 | Lathosterol oxidase | F: AGTGTAAGAATCCACCAATGCC R: GCCAAGTTTCCAATGAGACAGC | 58 |
| 18S rRNA | 18S rRNA | F: TATACGCTAGTGGAGCTGGAA R: GGGGAGGTAGTGACGAAAAAT | 55 |
| Sample | Raw Reads | Clean Reads | Mapping Ratio | Unigene Number | Ratio |
|---|---|---|---|---|---|
| EcW-1 | 76558224 | 73335584 | 64071854 (87.71%) | 73,576 | 94.06% |
| EcW-2 | 75407822 | 72788448 | 63707819 (87.67%) | 74,174 | 94.82% |
| EcW-3 | 78269646 | 75141044 | 66104983 (88.14%) | 74,006 | 94.61% |
| EcR-1 | 84068372 | 80832778 | 71088041 (88.17%) | 73,697 | 94.21% |
| EcR-2 | 72076762 | 69700338 | 60751470 (87.70%) | 73,497 | 93.96% |
| EcR-3 | 70924248 | 68597552 | 60474700 (88.34%) | 72,320 | 92.45% |
| EcW | 230235692 | 221265076 | 193884656 (87.84%) | 76,247 | 97.47% |
| EcR | 227069382 | 219130668 | 192314211 (88.07%) | 75,947 | 97.09% |
| All | 457305074 | 440395744 | 387591019 (88.01%) | 77,079 | 98.54% |
| Gene | log2 Ratio (EcR/EcW) | Description | Species |
|---|---|---|---|
| Unigene0039421 | −2.98 | Calcification-associated peptide−2 | Procambarus clarkii |
| Unigene0037661 | −2.56 | Calcification-associated peptide−2 | P. clarkii |
| Unigene0019577 | −6.61 | Calcified cuticle protein CP14.1 | Callinectes sapidus |
| Unigene0010114 | −3.86 | Calcified cuticle protein CP19.0 isoform A | C. sapidus |
| Unigene0072453 | −6.58 | Calcified cuticle protein CP19.0 isoform B | C. sapidus |
| Unigene0042544 | −5.85 | Calcified cuticle protein CP19.0 isoform B | C. sapidus |
| Unigene0038920 | −3.89 | Cuticle proprotein, partial | Palaemon varians |
| Unigene0032773 | −5.13 | Cuticle protein 1 | Cherax quadricarinatus |
| Unigene0056168 | −4.36 | Cuticle protein 18.6, isoform B | Lepeophtheirus salmonis |
| Unigene0016948 | −4.23 | Cuticle protein BD2 | Portunus pelagicus |
| Unigene0035595 | −2.25 | Cuticle protein, partial | Daphnia magna |
| Unigene0003529 | −1.74 | Cuticular protein | Tenebrio molitor |
| Unigene0073146 | −9.77 | Cuticular protein 34 | Eriocheir sinensis |
| Unigene0035142 | −5.96 | Cuticular protein 34 | E. sinensis |
| Unigene0037069 | −5.30 | Cuticular protein 34 | E. sinensis |
| Unigene0017103 | −3.76 | Cuticular protein 34 | E. sinensis |
| Unigene0043447 | −2.56 | Cuticular protein 34 | E. sinensis |
| Unigene0061823 | −1.05 | Cuticular protein analogous to peritrophins 1-F, partial | D. magna |
| Unigene0019752 | −1.54 | Cuticular protein RR-2 motif 78 precursor | Bombyx mori |
| Unigene0029471 | −2.05 | DD5 | Marsupenaeus japonicus |
| Unigene0019787 | −1.97 | DD5 | M. japonicus |
| Unigene0029470 | −1.69 | DD5 | M. japonicus |
| Unigene0044640 | −2.05 | DD9A, partial | M. japonicus |
| Unigene0001656 | −1.55 | Obstructor B | Locusta migratoria |
| Unigene0050714 | −1.99 | Post-molt protein 1 | C. quadricarinatus |
| Unigene0022558 | −1.80 | Strongly chitin-binding protein-1 | P. clarkii |
| Unigene0046800 | −1.19 | Pupal cuticle protein | Cephus cinctus |
| Unigene0008712 | −1.12 | Pupal cuticle protein | C. cinctus |
| Unigene0023218 | −1.42 | Cuticle protein 6 | Blaberus craniifer |
| Unigene0064526 | −1.29 | Cuticle protein 6 | B. craniifer |
| Unigene0025406 | −3.35 | Cuticle protein CP1876 | Cancer pagurus |
| Functional Categories | GeneID | log2 Ratio (EcR/EcW) | Description | Species |
|---|---|---|---|---|
| Transport | Unigene0065550 | 1.57 | Vitellogenin 2 | Pandalopsis japonica |
| Unigene0063752 | 3.65 | Vitellogenin | Pandalus hypsinotus | |
| Unigene0016779 | 1.04 | Vitellogenin | E. carinicauda | |
| Unigene0002542 | 1.52 | StAR-related lipid transfer protein 3 | Penaeus monodon | |
| Unigene0028810 | 3.54 | Apolipoprotein D | Zootermopsis nevadensis | |
| Unigene0036561 | −2.46 | Apolipoprotein D | D. magna | |
| Unigene0044620 | −1.26 | Apolipoprotein D | D. magna | |
| Binding | Unigene0029918 | 2.56 | Tubulin | Tetrahymena thermophila |
| Unigene0073421 | 2.13 | Tubulin/FtsZ family | T. thermophila | |
| Unigene0068154 | 1.79 | Tubulin α-1 chain | Trichinella murrelli | |
| Unigene0013764 | 1.07 | Tubulin α-1 chain-like | Halyomorpha halys | |
| Unigene0026012 | 1.92 | Fibronectin-like | Ciona intestinalis | |
| Unigene0022024 | 1.53 | Fibrillin-1 | Exaiptasia pallida | |
| Unigene0036597 | 1.06 | Hemocyanin | E. carinicauda | |
| Unigene0008214 | 1.15 | Hemocyanin | E. carinicauda | |
| Unigene0027904 | −1.80 | Hemocyanin | Macrobrachium nipponense | |
| Unigene0054180 | −1.85 | Crustacyanin subunit A | F. merguiensis |
| GeneID | log2 Ratio (EcR/EcW) | Description | Species |
|---|---|---|---|
| Unigene0033404 | 1.02 | Cathepsin B | Pandalus borealis |
| Unigene0049424 | −1.25 | Crustapain | Pandalus borealis |
| Unigene0018100 | −2.52 | Cathepsin L2 | Litopenaeus vannamei |
| Unigene0024515 | 2.31 | Cathepsin L | D. magna |
| Unigene0066106 | −3.44 | Heparan-α-glucosaminide N-acetyltransferase | D. magna |
| Unigene0022872 | −1.11 | N-acetyl-β-d-glucosaminidase | M. nipponense |
| Unigene0017001 | −2.85 | Ecdysteroid regulated-like protein | Litopenaeus vannamei |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, Y.; Li, S.; Yu, Y.; Zhang, C.; Zhang, X.; Li, F. Transcriptome Analysis Provides Insights into the Mechanism of Astaxanthin Enrichment in a Mutant of the Ridgetail White Prawn Exopalaemon carinicauda. Genes 2021, 12, 618. https://doi.org/10.3390/genes12050618
Jin Y, Li S, Yu Y, Zhang C, Zhang X, Li F. Transcriptome Analysis Provides Insights into the Mechanism of Astaxanthin Enrichment in a Mutant of the Ridgetail White Prawn Exopalaemon carinicauda. Genes. 2021; 12(5):618. https://doi.org/10.3390/genes12050618
Chicago/Turabian StyleJin, Yue, Shihao Li, Yang Yu, Chengsong Zhang, Xiaojun Zhang, and Fuhua Li. 2021. "Transcriptome Analysis Provides Insights into the Mechanism of Astaxanthin Enrichment in a Mutant of the Ridgetail White Prawn Exopalaemon carinicauda" Genes 12, no. 5: 618. https://doi.org/10.3390/genes12050618
APA StyleJin, Y., Li, S., Yu, Y., Zhang, C., Zhang, X., & Li, F. (2021). Transcriptome Analysis Provides Insights into the Mechanism of Astaxanthin Enrichment in a Mutant of the Ridgetail White Prawn Exopalaemon carinicauda. Genes, 12(5), 618. https://doi.org/10.3390/genes12050618






