High SGO2 Expression Predicts Poor Overall Survival: A Potential Therapeutic Target for Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Resource and Description
2.2. SGO2 Expression and Clinicopathological and Survival Analyses
2.3. Univariate and Multivariate Cox Regression Analysis
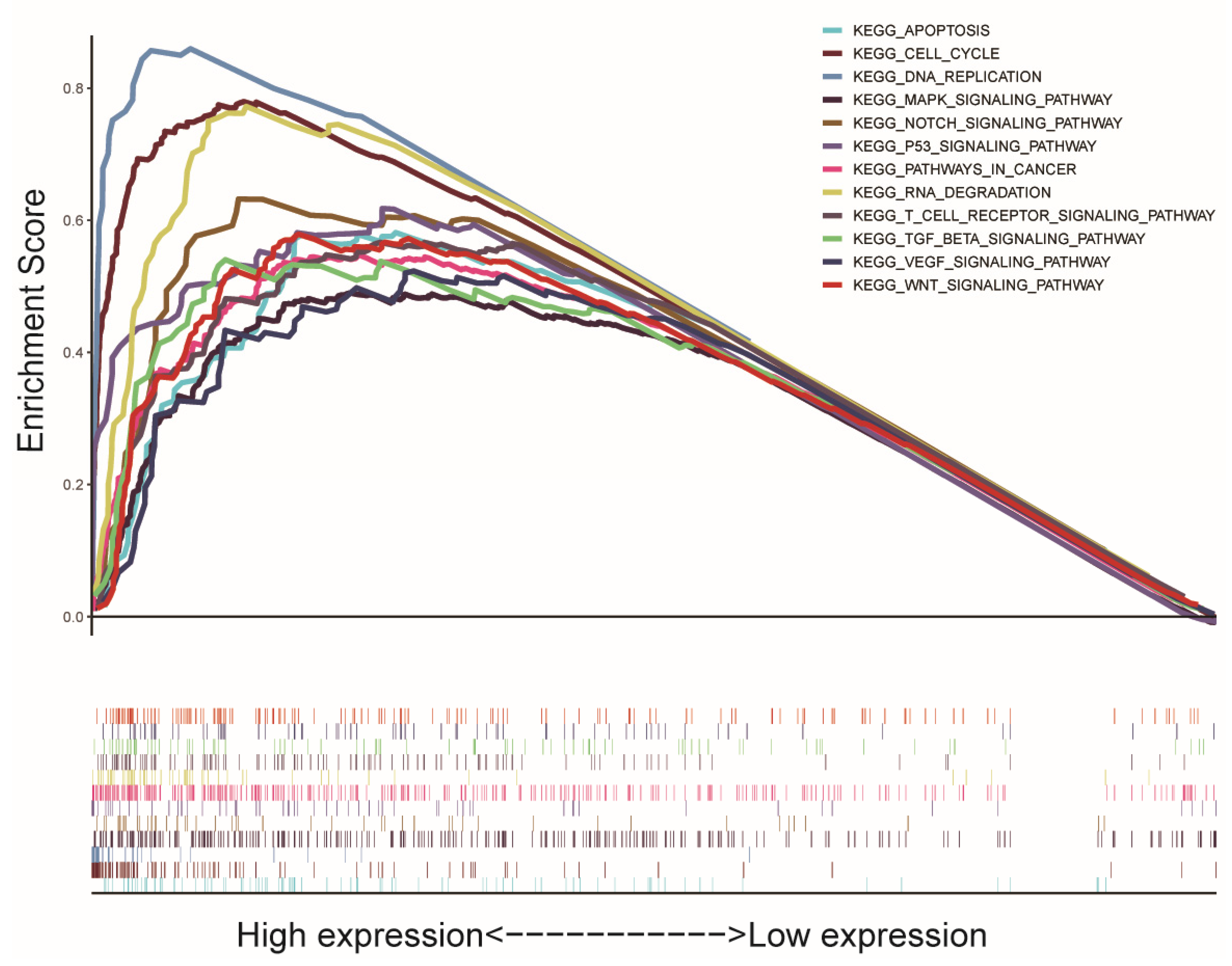
2.4. Gene Set Enrichment Analysis (GSEA)
2.5. Protein–Protein Interaction (PPI) Network and Gene Co-Expression Network Analysis
2.6. Statistical Analysis
3. Results
3.1. SGO2 Expression Comparison
3.2. Associations between SGO2 and Survival in HCC Patients
3.3. Association between SGO2 Expression and Clinicopathological Features in HCC
3.4. Univariate and Multivariate Analysis
3.5. GSEA Biological Process Enrichment
3.6. Protein–Protein Interaction (PPI) Network Construction and Gene Co-Expression Network Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Llovet, J.M.; Zucman-Rossi, J.; Pikarsky, E.; Sangro, B.; Schwartz, M.; Sherman, M.; Gores, G. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2016, 2, 16018. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marrero, J.A.; Kulik, L.M.; Sirlin, C.B.; Zhu, A.X.; Finn, R.S.; Abecassis, M.M.; Roberts, L.R.; Heimbach, J.K. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018, 68, 723–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruix, J.; Gores, G.J.; Mazzaferro, V. Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut 2014, 63, 844–855. [Google Scholar] [CrossRef]
- Orcutt, S.T.; Anaya, D.A. Liver Resection and Surgical Strategies for Management of Primary Liver Cancer. Cancer Control 2018, 25, 1073274817744621. [Google Scholar] [CrossRef]
- Lohitesh, K.; Chowdhury, R.; Mukherjee, S. Resistance a major hindrance to chemotherapy in hepatocellular carcinoma: An insight. Cancer Cell Int. 2018, 18, 44. [Google Scholar] [CrossRef]
- Jindal, A.; Thadi, A.; Shailubhai, K. Hepatocellular Carcinoma: Etiology and Current and Future Drugs. J. Clin. Exp. Hepatol. 2019, 9, 221–232. [Google Scholar] [CrossRef]
- Roderburg, C.; Özdirik, B.; Wree, A.; Demir, M.; Tacke, F. Systemic treatment of hepatocellular carcinoma: From sorafenib to combination therapies. Hepat. Oncol. 2020, 7, HEP20. [Google Scholar] [CrossRef]
- Zongyi, Y.; Xiaowu, L. Immunotherapy for hepatocellular carcinoma. Cancer Lett. 2020, 470, 8–17. [Google Scholar] [CrossRef]
- Shen, Y.; Bu, L.; Li, R.; Chen, Z.; Tian, F.; Lu, N.; Ge, Q.; Bai, Y.; Lu, Z. Screening effective differential expression genes for hepatic carcinoma with metastasis in the peripheral blood mononuclear cells by RNA-seq. Oncotarget 2017, 8, 27976–27989. [Google Scholar] [CrossRef] [Green Version]
- Faridi, R.; Rehman, A.U.; Morell, R.J.; Friedman, P.L.; Demain, L.; Zahra, S.; Khan, A.A.; Tohlob, D.; Assir, M.Z.; Beaman, G.; et al. Mutations of SGO2 and CLDN14 collectively cause coincidental Perrault syndrome. Clin. Genet. 2017, 91, 328–332. [Google Scholar] [CrossRef] [Green Version]
- Qi, J.; Zhou, J.; Tang, X.Q.; Wang, Y. Gene Biomarkers Derived from Clinical Data of Hepatocellular Carcinoma. Interdiscip Sci. 2020, 12, 226–236. [Google Scholar] [CrossRef]
- Wang, L.H.; Yen, C.J.; Li, T.N.; Elowe, S.; Wang, W.C.; Wang, L.H. Sgo1 is a potential therapeutic target for hepatocellular carcinoma. Oncotarget 2015, 6, 2023–2033. [Google Scholar] [CrossRef]
- Yamada, H.Y.; Zhang, Y.; Reddy, A.; Mohammed, A.; Lightfoot, S.; Dai, W.; Rao, C.V. Tumor-promoting/progressing role of additional chromosome instability in hepatic carcinogenesis in Sgo1 (Shugoshin 1) haploinsufficient mice. Carcinogenesis 2015, 36, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Wang, L.; Zhu, B.; Zhang, J.; Guo, S.; Li, Q.; Zhang, T.; Zheng, Z.; Wu, G.; Zhao, Y. Integrated Bioinformatics Analysis of the Clinical Value and Biological Function of ATAD2 in Hepatocellular Carcinoma. BioMed Res. Int. 2020, 2020, 8657468. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; He, B.; Yang, B.; Hu, W.; Cheng, S.; Xiao, H.; Yang, Z.; Wen, X.; Zhou, L.; Xie, H.; et al. Genome-wide CRISPR screen reveals SGOL1 as a druggable target of sorafenib-treated hepatocellular carcinoma. Lab. Investig. 2018, 98, 734–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellmuth, S.; Gómez-H, L.; Pendás, A.M.; Stemmann, O. Securin-independent regulation of separase by checkpoint-induced shugoshin-MAD2. Nature 2020, 580, 536–541. [Google Scholar] [CrossRef]
- Kitajima, T.S.; Sakuno, T.; Ishiguro, K.; Iemura, S.; Natsume, T.; Kawashima, S.A.; Watanabe, Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 2006, 441, 46–52. [Google Scholar] [CrossRef]
- Salic, A.; Waters, J.C.; Mitchison, T.J. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell 2004, 118, 567–578. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Feng, J.; Famulski, J.; Rattner, J.B.; Liu, S.T.; Kao, G.D.; Muschel, R.; Chan, G.K.; Yen, T.J. Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J. Cell Biol. 2007, 177, 413–424. [Google Scholar] [CrossRef]
- Tsukahara, T.; Tanno, Y.; Watanabe, Y. Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature 2010, 467, 719–723. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, D.; Jiang, F.; Shen, Y.; Li, X.; Hu, X.; Wei, P.; Shen, X. Prognostic Prediction Using a Stemness Index-Related Signature in a Cohort of Gastric Cancer. Front. Mol. Biosci. 2020, 7, 570702. [Google Scholar] [CrossRef]
- Li, L.; Huang, K.; Zhao, H.; Chen, B.; Ye, Q.; Yue, J. CDK1-PLK1/SGOL2/ANLN pathway mediating abnormal cell division in cell cycle may be a critical process in hepatocellular carcinoma. Cell Cycle 2020, 19, 1236–1252. [Google Scholar] [CrossRef]
- Jiang, S.S.; Ke, S.J.; Ke, Z.L.; Li, J.; Li, X.; Xie, X.W. Cell Division Cycle Associated Genes as Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma. Front. Mol. Biosci. 2021, 8, 657161. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, P.; Li, M.; Fei, H.; Huang, J.; Zhao, T.; Li, T. Comprehensive Analysis of the Control of Cancer Stem Cell Characteristics in Endometrial Cancer by Network Analysis. Comput. Math. Methods Med. 2021, 2021, 6653295. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Jensen, L.J. Protein-protein interaction databases. Methods Mol. Biol. 2015, 1278, 39–56. [Google Scholar] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cucchetti, A.; Zhong, J.; Berhane, S.; Toyoda, H.; Shi, K.; Tada, T.; Chong, C.; Xiang, B.D.; Li, L.Q.; Lai, P.; et al. The chances of hepatic resection curing hepatocellular carcinoma. J. Hepatol. 2020, 72, 711–717. [Google Scholar] [CrossRef]
- Wang, M.D.; Li, C.; Liang, L.; Xing, H.; Sun, L.Y.; Quan, B.; Wu, H.; Xu, X.F.; Wu, M.C.; Pawlik, T.M.; et al. Early and Late Recurrence of Hepatitis B Virus-Associated Hepatocellular Carcinoma. Oncologist 2020, 25, e1541–e1551. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.M.; Knox, J.; Cosby, R.; Beecroft, J.R.; Chan, K.; Coburn, N.; Feld, J.; Jonker, D.; Mahmud, A.; Ringash, J.; et al. Nonsurgical management of advanced hepatocellular carcinoma: A clinical practice guideline. Curr. Oncol. 2020, 27, e106–e114. [Google Scholar] [PubMed]
- Chen, Z.; Xie, H.; Hu, M.; Huang, T.; Hu, Y.; Sang, N.; Zhao, Y. Recent progress in treatment of hepatocellular carcinoma. Am. J. Cancer Res. 2020, 10, 2993–3036. [Google Scholar]
- Anwanwan, D.; Singh, S.K.; Singh, S.; Saikam, V.; Singh, R. Challenges in liver cancer and possible treatment approaches. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188314. [Google Scholar] [CrossRef]
- Huang, A.; Yang, X.R.; Chung, W.Y.; Dennison, A.R.; Zhou, J. Targeted therapy for hepatocellular carcinoma. Signal Transduct. Target. Ther. 2020, 5, 146. [Google Scholar] [CrossRef]
- Gantchev, J.; Martínez Villarreal, A.; Xie, P.; Lefrançois, P.; Gunn, S.; Netchiporouk, E.; Sasseville, D.; Litvinov, I.V. The Ectopic Expression of Meiosis Regulatory Genes in Cutaneous T-Cell Lymphomas (CTCL). Front. Oncol. 2019, 9, 429. [Google Scholar] [CrossRef]
- Chen, S.; Cao, Q.; Wen, W.; Wang, H. Targeted therapy for hepatocellular carcinoma: Challenges and opportunities. Cancer Lett. 2019, 460, 1–9. [Google Scholar] [CrossRef]
- Liu, X.; Qin, S. Immune Checkpoint Inhibitors in Hepatocellular Carcinoma: Opportunities and Challenges. Oncologist 2019, 24, S3–S10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, E.B.; Qin, X.; Peng, K.; Li, Q.; Tang, C.; Wei, Y.C.; Yu, S.; Gan, L.; Liu, T.S. HnRNPR-CCNB1/CENPF axis contributes to gastric cancer proliferation and metastasis. Aging 2019, 11, 7473–7491. [Google Scholar] [CrossRef]
- Han, X.; Zhang, J.J.; Han, Z.Q.; Zhang, H.B.; Wang, Z.A. Let-7b attenuates cisplatin resistance and tumor growth in gastric cancer by targeting AURKB. Cancer Gene Ther. 2018, 25, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, R.; Calvisi, D.F.; Ladu, S.; Ehemann, V.; Staniscia, T.; Evert, M.; Dombrowski, F.; Schirmacher, P.; Longerich, T. Oncogenic and tumor suppressive roles of polo-like kinases in human hepatocellular carcinoma. Hepatology 2010, 51, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Kwon, C.H.; Park, H.J.; Choi, Y.R.; Kim, A.; Kim, H.W.; Choi, J.H.; Hwang, C.S.; Lee, S.J.; Choi, C.I.; Jeon, T.Y.; et al. PSMB8 and PBK as potential gastric cancer subtype-specific biomarkers associated with prognosis. Oncotarget 2016, 7, 21454–21468. [Google Scholar] [CrossRef] [Green Version]
- Su, T.C.; Chen, C.Y.; Tsai, W.C.; Hsu, H.T.; Yen, H.H.; Sung, W.W.; Chen, C.J. Cytoplasmic, nuclear, and total PBK/TOPK expression is associated with prognosis in colorectal cancer patients: A retrospective analysis based on immunohistochemistry stain of tissue microarrays. PLoS ONE 2018, 13, e0204866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Rajasekaran, M.; Xia, H.; Zhang, X.; Kong, S.N.; Sekar, K.; Seshachalam, V.P.; Deivasigamani, A.; Goh, B.K.; Ooi, L.L.; et al. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the Wnt/β-catenin signalling pathway. Gut 2016, 65, 1522–1534. [Google Scholar] [CrossRef]
- Zhang, L.; Makamure, J.; Zhao, D.; Liu, Y.; Guo, X.; Zheng, C.; Liang, B. Bioinformatics analysis reveals meaningful markers and outcome predictors in HBV-associated hepatocellular carcinoma. Exp. Ther. Med. 2020, 20, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Sents, W.; Meeusen, B.; Kalev, P.; Radaelli, E.; Sagaert, X.; Miermans, E.; Haesen, D.; Lambrecht, C.; Dewerchin, M.; Carmeliet, P.; et al. PP2A Inactivation Mediated by PPP2R4 Haploinsufficiency Promotes Cancer Development. Cancer Res. 2017, 77, 6825–6837. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.; Pain, M.; Wang, H.; Herman, J.A.; Toledo, C.M.; DeLuca, J.G.; Yong, R.L.; Paddison, P.; Zhu, J. Sensitivity to BUB1B Inhibition Defines an Alternative Classification of Glioblastoma. Cancer Res. 2017, 77, 5518–5529. [Google Scholar] [CrossRef] [Green Version]
- Ci, C.; Tang, B.; Lyu, D.; Liu, W.; Qiang, D.; Ji, X.; Qiu, X.; Chen, L.; Ding, W. Overexpression of CDCA8 promotes the malignant progression of cutaneous melanoma and leads to poor prognosis. Int. J. Mol. Med. 2019, 43, 404–412. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Liu, Y.; Shang, L.; Yu, J.; Qu, Q. The FOXM1/BUB1B signaling pathway is essential for the tumorigenicity and radioresistance of glioblastoma. Oncol. Rep. 2017, 38, 3367–3375. [Google Scholar] [CrossRef] [Green Version]
- Katz, L.H.; Likhter, M.; Jogunoori, W.; Belkin, M.; Ohshiro, K.; Mishra, L. TGF-β signaling in liver and gastrointestinal cancers. Cancer Lett. 2016, 379, 166–172. [Google Scholar] [CrossRef] [Green Version]
- Ishimoto, T.; Miyake, K.; Nandi, T.; Yashiro, M.; Onishi, N.; Huang, K.K.; Lin, S.J.; Kalpana, R.; Tay, S.T.; Suzuki, Y.; et al. Activation of Transforming Growth Factor Beta 1 Signaling in Gastric Cancer-associated Fibroblasts Increases Their Motility, via Expression of Rhomboid 5 Homolog 2, and Ability to Induce Invasiveness of Gastric Cancer Cells. Gastroenterology 2017, 153, 191–204.e16. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.J. Notch signalling: A simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006, 7, 678–689. [Google Scholar] [CrossRef]
- Schweisguth, F. Regulation of notch signaling activity. Curr. Biol. 2004, 14, R129–R138. [Google Scholar] [CrossRef] [PubMed]
- Bode, A.M.; Dong, Z. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 2004, 4, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Horn, H.F.; Vousden, K.H. Coping with stress: Multiple ways to activate p53. Oncogene 2007, 26, 1306–1316. [Google Scholar] [CrossRef] [Green Version]
- Qian, Z.; Zhang, Z.; Wang, Y. T cell receptor signaling pathway and cytokine-cytokine receptor interaction affect the rehabilitation process after respiratory syncytial virus infection. PeerJ 2019, 7, e7089. [Google Scholar] [CrossRef] [Green Version]
- Courtney, A.H.; Lo, W.L.; Weiss, A. TCR Signaling: Mechanisms of Initiation and Propagation. Trends Biochem. Sci. 2018, 43, 108–123. [Google Scholar] [CrossRef]
- Matsumoto, T.; Claesson-Welsh, L. VEGF receptor signal transduction. Sci. STKE 2001, 2001, re21. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.D.; Vigne, J.L.; Minchenko, A.; Lebovic, D.I.; Leitman, D.C.; Taylor, R.N. Regulation of vascular endothelial growth factor (VEGF) gene transcription by estrogen receptors alpha and beta. Proc. Natl. Acad. Sci. USA 2000, 97, 10972–10977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wittenberg, C.; Reed, S.I. Cell cycle-dependent transcription in yeast: Promoters, transcription factors, and transcriptomes. Oncogene 2005, 24, 2746–2755. [Google Scholar] [CrossRef] [Green Version]
- Kitao, H.; Iimori, M.; Kataoka, Y.; Wakasa, T.; Tokunaga, E.; Saeki, H.; Oki, E.; Maehara, Y. DNA replication stress and cancer chemotherapy. Cancer Sci. 2018, 109, 264–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houseley, J.; Tollervey, D. The many pathways of RNA degradation. Cell 2009, 136, 763–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Logan, C.Y.; Nusse, R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004, 20, 781–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Characteristics | Variable | Patients (377) | Percentages (%) |
|---|---|---|---|
| Age | <50 years | 72 | 19.10 |
| ≥50 years | 304 | 80.63 | |
| Unknown | 1 | 0.27 | |
| Gender | Male | 255 | 67.64 |
| Female | 122 | 32.36 | |
| Tumor grade | G1 | 55 | 14.59 |
| G2 | 180 | 47.74 | |
| G3 | 124 | 32.89 | |
| G4 | 13 | 3.45 | |
| Unknow | 5 | 1.33 | |
| Pathological stage | I | 175 | 46.42 |
| II | 87 | 23.08 | |
| III | 86 | 22.81 | |
| IV | 5 | 1.33 | |
| Unknown | 24 | 6.36 | |
| T | T1 | 185 | 49.07 |
| T2 | 95 | 25.20 | |
| T3 | 81 | 21.49 | |
| T4 | 13 | 3.45 | |
| TX | 3 | 0.79 | |
| N | N0 | 257 | 68.17 |
| N1 | 4 | 1.06 | |
| NX | 116 | 30.77 | |
| M | M0 | 272 | 72.15 |
| M1 | 4 | 1.06 | |
| MX | 101 | 26.79 | |
| Vascular invasion | None | 210 | 55.70 |
| Micro | 94 | 24.93 | |
| Macro | 17 | 4.51 | |
| Unknow | 56 | 14.86 | |
| ECOG score | 0 | 166 | 44.03 |
| 1 | 86 | 22.81 | |
| 2 | 26 | 6.90 | |
| 3 | 12 | 3.18 | |
| 4 | 3 | 0.80 | |
| Unknow | 84 | 22.28 | |
| Vital status | Alive | 249 | 66.05 |
| Death | 128 | 33.95 |
| Clinicopathological Parameters | SGO2 Expression | Total | p-Value | |
|---|---|---|---|---|
| High (n = 212) | Low (n = 212) | |||
| Age | ||||
| <50 years | 45 (64.3) | 25 (35.7) | 70 | 0.184 |
| ≥50 years | 164 (54.7) | 136 (45.3) | 300 | |
| Gender | ||||
| Male | 55 (45.1) | 67 (54.9) | 122 | 0.034 |
| Female | 35 (63.6) | 20 (36.4) | 55 | |
| Tumor grade | ||||
| G1 and G2 | 112 (48.3) | 120 (51.7) | 232 | <0.001 |
| G3 and G4 | 95 (70.9) | 39 (29.1) | 134 | |
| Pathological stage | ||||
| I and II | 136 (52.9) | 121 (47.1) | 257 | 0.050 |
| III and IV | 59 (65.7) | 31 (34.3) | 90 | |
| T classification | ||||
| T1 and T2 | 148 (53.8) | 127 (46.2) | 275 | 0.063 |
| T3 and T4 | 61 (65.6) | 32 (34.4) | 93 | |
| Vascular invasion | ||||
| None | 103 (50) | 103 (50) | 206 | 0.095 |
| positive | 66 (60.6) | 43 (39.4) | 109 | |
| ECOG score | ||||
| 0 to 1 | 123 (50) | 123 (50) | 246 | 0.004 |
| 2–4 | 31 (75.6) | 10 (24.4) | 41 | |
| Clinicopathological Parameters | Total (N) | Odds Ratio in SGO2 Expression | p-Value |
|---|---|---|---|
| Age | |||
| <50 vs. ≥50 | 370 | 0.868 (0.514–1.462) | 0.596 |
| Gender | |||
| Male vs. Female | 178 | 2.278 (1.192–4.446) | 0.014 |
| Tumor grade | |||
| G1 and G2 vs. G3 and G4 | 366 | 2.622 (1.695–4.096) | <0.001 |
| Pathological stage | |||
| Stage II vs. Stage I | 257 | 2.317 (1.370–3.960) | 0.002 |
| Stage III and IV vs. Stage I | 261 | 2.170 (1.295–3.669) | 0.003 |
| T classification | |||
| T3 and T4 vs. T1 and T2 | 368 | 1.523 (1.206–1.936) | <0.001 |
| Vascular invasion | |||
| positive vs. negative | 315 | 1.572 (0.986–2.517) | 0.058 |
| ECOG score | |||
| 0 to 1 vs. 2–4 | 287 | 3.263 (1.607–7.081) | 0.002 |
| Parameter | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p | HR | 95% CI | p | |
| Age | 1.021 | 0.996–1.047 | 0.098 | 1.033 | 1.005–1.063 | 0.022 |
| Gender | 1.627 | 0.893–2.962 | 0.112 | 1.029 | 0.526–2.015 | 0.933 |
| Grade | 1.469 | 0.966–2.236 | 0.072 | 1.490 | 0.887–2.502 | 0.132 |
| Pathological stage | 1.586 | 1.160–2.169 | 0.004 | 2.115 | 0.385–11.615 | 0.389 |
| T | 1.506 | 1.105–2.052 | 0.010 | 0.600 | 0.115–3.143 | 0.546 |
| N | 1.400 | 0.192–10.215 | 0.740 | 0.195 | 0.007–5.777 | 0.345 |
| M | 5.570 | 1.707–18.179 | 0.004 | 4.324 | 0.866–21.587 | 0.074 |
| Vascular invasion | 1.571 | 1.010–2.443 | 0.045 | 1.506 | 0.944–2.405 | 0.086 |
| ECOG score | 1.531 | 0.982–2.388 | 0.060 | 1.429 | 0.869–2.350 | 0.160 |
| SGO2 | 1.412 | 1.123–1.776 | 0.003 | 1.401 | 1.066–1.841 | 0.016 |
| Gene Set Name | NES | NOM p-Value | FDR q-Value |
|---|---|---|---|
| KEGG_CELL_CYCLE | 2.230 | 0.000 | 0.000 |
| KEGG_RNA_DEGRADATION | 2.097 | 0.000 | 0.003 |
| KEGG_DNA_REPLICATION | 2.001 | 0.000 | 0.005 |
| KEGG_P53_SIGNALING_PATHWAY | 1.952 | 0.000 | 0.007 |
| KEGG_WNT_SIGNALING_PATHWAY | 1.920 | 0.002 | 0.008 |
| KEGG_PATHWAYS_IN_CANCER | 1.878 | 0.000 | 0.010 |
| KEGG_NOTCH_SIGNALING_PATHWAY | 1.811 | 0.000 | 0.015 |
| KEGG_APOPTOSIS | 1.751 | 0.004 | 0.022 |
| KEGG_VEGF_SIGNALING_PATHWAY | 1.704 | 0.004 | 0.031 |
| KEGG_MAPK_SIGNALING_PATHWAY | 1.696 | 0.002 | 0.032 |
| KEGG_TGF_β_SIGNALING_PATHWAY | 1.689 | 0.014 | 0.033 |
| KEGG_T_CELL_RECEPTOR_SIGNALING_PATHWAY | 1.658 | 0.016 | 0.040 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, M.; Li, S.; Mei, J.; Lin, W.; Zou, J.; Wei, W.; Guo, R. High SGO2 Expression Predicts Poor Overall Survival: A Potential Therapeutic Target for Hepatocellular Carcinoma. Genes 2021, 12, 876. https://doi.org/10.3390/genes12060876
Deng M, Li S, Mei J, Lin W, Zou J, Wei W, Guo R. High SGO2 Expression Predicts Poor Overall Survival: A Potential Therapeutic Target for Hepatocellular Carcinoma. Genes. 2021; 12(6):876. https://doi.org/10.3390/genes12060876
Chicago/Turabian StyleDeng, Min, Shaohua Li, Jie Mei, Wenping Lin, Jingwen Zou, Wei Wei, and Rongping Guo. 2021. "High SGO2 Expression Predicts Poor Overall Survival: A Potential Therapeutic Target for Hepatocellular Carcinoma" Genes 12, no. 6: 876. https://doi.org/10.3390/genes12060876
APA StyleDeng, M., Li, S., Mei, J., Lin, W., Zou, J., Wei, W., & Guo, R. (2021). High SGO2 Expression Predicts Poor Overall Survival: A Potential Therapeutic Target for Hepatocellular Carcinoma. Genes, 12(6), 876. https://doi.org/10.3390/genes12060876






