Bioinformatics Analysis of Allele Frequencies and Expression Patterns of ACE2, TMPRSS2 and FURIN in Different Populations and Susceptibility to SARS-CoV-2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Analysis of FURIN Gene Expression Patterns in Human Lungs
2.2. eQTL Variant Retrieval
2.3. Retrieval of Population and Clinical Indicator Data
2.4. Pathway Analysis of ACE2, FURIN and TMPRSS2 Interactome
3. Results
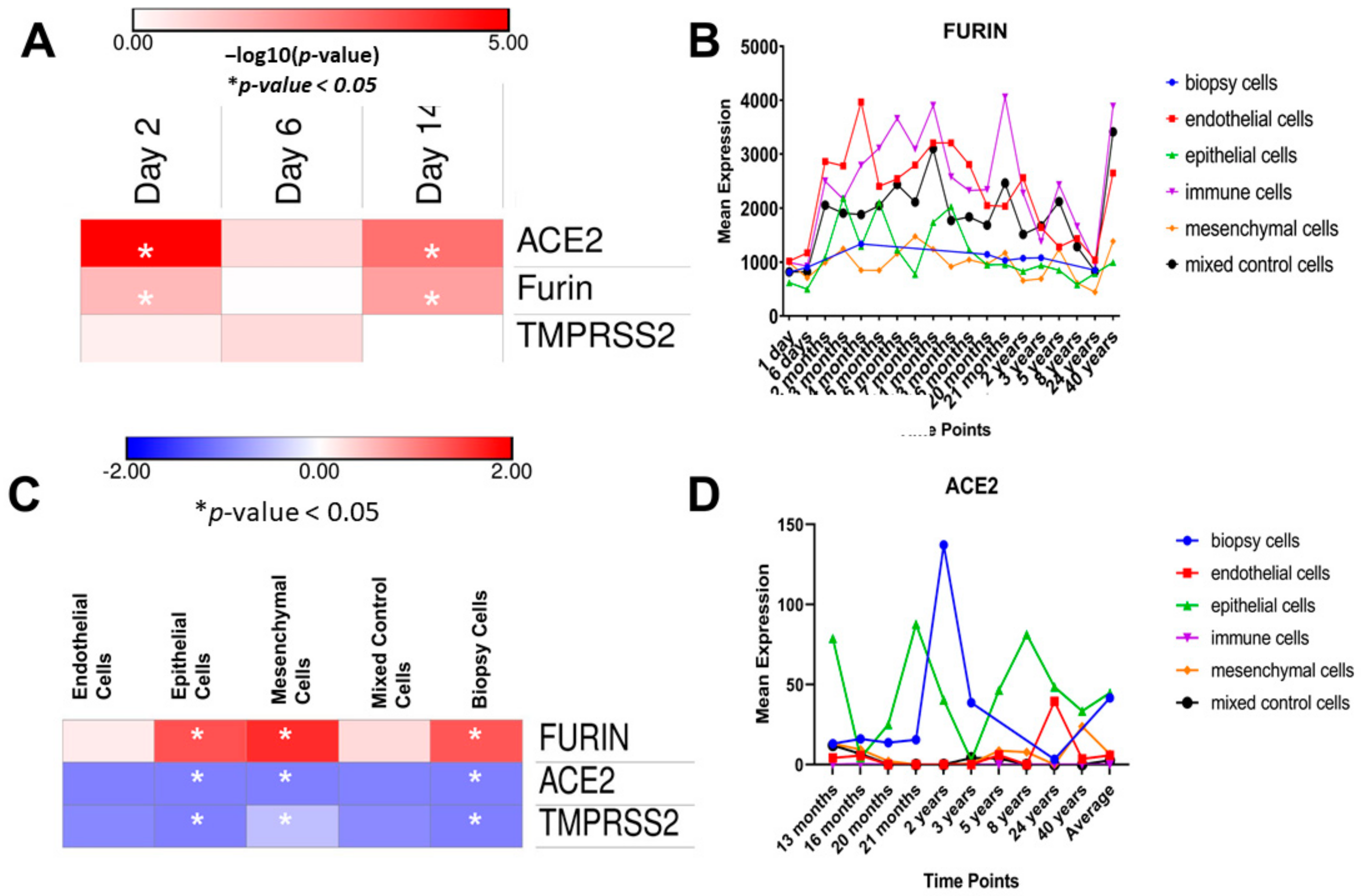
3.1. Gene Expression Analysis in Human Lung Using the LungMAP Database
3.2. Population-Based Frequency Analysis of the Three Studied Genes
3.3. Population-Based Frequencies of FURIN eQTL Variants
3.4. Population-Based Frequencies of TMPRSS2 eQTLs in the Lungs
3.4.1. Frequencies of Regulatory Intronic Variants of TMPRSS2
3.4.2. Frequencies of Common Exonic Variants of TMPRSS2
3.5. Population-Based Frequencies of ACE2 Coding, eQTLs and Intronic Variants
3.6. Pathway Analysis and Interaction Network of ACE2, TMPRSS2 and FURIN
4. Discussion
4.1. Shedding Light on FURIN Expression Patterns and Immune Responses in Relation to SARS-CoV-2 Infection
4.2. Implications of Population-Based Variations of Studied Genes’ eQTL Frequencies
4.2.1. FURIN
4.2.2. TMPRSS2
4.2.3. ACE2
4.3. Possible Contribution of eQTL Frequencies to COVID-19 Patterns of Transmission and Mortality; the Bigger Picture
4.4. Study Limitations and Future Recommendations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Term | Definition |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus-2 |
| MERS | Middle East respiratory syndrome |
| ACE2 | angiotensin-converting enzyme 2 |
| TMPRSS2 | transmembrane protease, serine 2 |
| eQTL | expression quantitative trait loci are genomic loci that explain variations in the expression levels of mRNA transcripts. |
| GEO | Gene Expression Omnibus |
| LoM | lung-only mice |
| GTEx | Genotype-Tissue Expression portal database |
| “GC”, “CC” Genotypes of rs-id of a variant | the nucleotides represent the genotype of a specific eQTL variant, labeled with its unique rs-id. |
| CFR | case fatality ratio |
| TPM | transcripts-per-million |
References
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 18 October 2020).
- Ali, H.; Alshukry, A.; Marafie, S.K.; AlRukhayes, M.; Ali, Y.; Abbas, M.B.; Al-Taweel, A.; Bukhamseen, Y.; Dashti, M.H.; Al-Shammari, A.A.; et al. Outcomes of COVID-19: Disparities by ethnicity. Infect. Genet. Evol. 2021, 87, 104639. [Google Scholar] [CrossRef]
- Medhat, M.A.; El Kassas, M. COVID-19 in Egypt: Uncovered figures or a different situation? J. Glob. Health 2020, 10. [Google Scholar] [CrossRef]
- Shikov, A.E.; Barbitoff, Y.A.; Glotov, A.S.; Danilova, M.M.; Tonyan, Z.N.; Nasykhova, Y.A.; Mikhailova, A.A.; Bespalova, O.N.; Kalinin, R.S.; Mirzorustamova, A.M.; et al. Analysis of the Spectrum of ACE2 Variation Suggests a Possible Influence of Rare and Common Variants on Susceptibility to COVID-19 and Severity of Out-come. Front. Genet. 2020, 11. Available online: https://www.frontiersin.org/articles/10.3389/fgene.2020.551220/full (accessed on 19 February 2021). [CrossRef]
- Nica, A.C.; Dermitzakis, E.T. Expression quantitative trait loci: Present and future. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120362. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. Available online: http://www.sciencedirect.com/science/article/pii/S0092867420302294 (accessed on 10 April 2020). [CrossRef]
- Vaarala, M.H.; Porvari, K.S.; Kellokumpu, S.; Kyllönen, A.P.; Vihko, P.T. Expression of transmembrane serine protease TMPRSS2 in mouse and human tissues. J. Pathol. 2001, 193, 134–140. [Google Scholar] [CrossRef]
- Devaux, C.A.; Rolain, J.-M.; Raoult, D. ACE2 receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020, 53, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Strope, J.D.; Pharm, C.H.C.; Figg, W.D. TMPRSS2: Potential Biomarker for COVID-19 Outcomes. J. Clin. Pharmacol. 2020, 60, 801–807. [Google Scholar] [CrossRef]
- Hou, Y.; Zhao, J.; Martin, W.; Kallianpur, A.; Chung, M.K.; Jehi, L.; Sharifi, N.; Erzurum, S.; Eng, C.; Cheng, F. New insights into genetic susceptibility of COVID-19: An ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020, 18, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Millet, J.K.; Whittaker, G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA 2014, 111, 15214–15219. [Google Scholar] [CrossRef] [Green Version]
- Xia, S.; Lan, Q.; Su, S.; Wang, X.; Xu, W.; Liu, Z.; Zhu, Y.; Wang, Q.; Lu, L.; Jiang, S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct. Target. Ther. 2020, 5, 1–3. [Google Scholar] [CrossRef]
- Chaudhary, M. COVID-19 susceptibility: Potential of ACE2 polymorphisms. Egypt. J. Med. Hum. Genet. 2020, 21, 54. [Google Scholar] [CrossRef]
- Milne, S.; Yang, C.X.; Timens, W.; Bossé, Y.; Sin, D.D. SARS-CoV-2 receptor ACE2 gene expression and RAAS inhibitors. Lancet Respir. Med. 2020, 8, e50–e51. [Google Scholar] [CrossRef]
- Chen, J.; Jiang, Q.; Xia, X.; Liu, K.; Yu, Z.; Tao, W.; Gong, W.; Han, J.J. Individual variation of the SARS-CoV-2 receptor ACE2 gene expression and regulation. Aging Cell 2020, 19, e13168. [Google Scholar] [CrossRef] [PubMed]
- Al-Mulla, F.; Mohammad, A.; Al Madhoun, A.; Haddad, D.; Ali, H.; Eaaswarkhanth, M.; John, S.E.; Nizam, R.; Channanath, A.; Abu-Farha, M.; et al. ACE2 and FURIN variants are potential predictors of SARS-CoV-2 outcome: A time to implement precision medicine against COVID-19. Heliyon 2021, 7, e06133. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Xiang, Z.; Wu, Y.; Gu, Y.; Guo, J.; Geng, F. Analysis of the susceptibility of lung cancer patients to SARS-CoV-2 infection. Mol. Cancer 2020, 19, 80. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7186321/ (accessed on 19 February 2021). [CrossRef]
- LungMAP-Home. Available online: https://lungmap.net/ (accessed on 19 February 2021).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Wahl, A.; Gralinski, L.; Johnson, C.; Yao, W.; Kovarova, M.; Dinnon, K.; Liu, H.; Madden, V.; Krzystek, H.; De, C.; et al. Acute SARS-CoV-2 Infection is Highly Cytopathic, Elicits a Robust Innate Immune Response and is Efficiently Prevented by EIDD-2801. Res. Sq. 2020. [Google Scholar] [CrossRef]
- GTEx Portal. Available online: https://www.gtexportal.org/home/ (accessed on 19 February 2021).
- Ensembl Genome Browser 103. Available online: https://www.ensembl.org/index.html (accessed on 19 February 2021).
- gnomAD. Available online: https://gnomad.broadinstitute.org/ (accessed on 19 February 2021).
- Egyptian Genome. Available online: https://www.egyptian-genome.org/ (accessed on 19 February 2021).
- Bonnet, A.; Lagarrigue, S.; Liaubet, L.; Robert-Granié, C.; Cristobal, M.S.; Tosser-Klopp, G. Pathway results from the chicken data set using GOTM, Pathway Studio and Ingenuity softwares. BMC Proc. 2009, 3, S11. [Google Scholar] [CrossRef] [Green Version]
- Yuryev, A.; Kotelnikova, E.; Daraselia, N. Ariadne’s ChemEffect and Pathway Studio knowledge base. Expert Opin. Drug Discov. 2009, 4, 1307–1318. [Google Scholar] [CrossRef]
- Wohlers, I.; Künstner, A.; Munz, M.; Olbrich, M.; Fähnrich, A.; Calonga-Solís, V.; Ma, C.; Hirose, M.; El-Mosallamy, S.; Salama, M.; et al. An integrated personal and population-based Egyptian genome reference. Nat. Commun. 2020, 11, 4719. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, L.; Feng, Z.; Wan, S.; Huang, P.; Sun, X. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020, 6, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef]
- Saheb Sharif-Askari, N.; Saheb Sharif-Askari, F.; Alabed, M.; Temsah, M.-H.; Al Heialy, S.; Hamid, Q.; Halwani, R. Airways Expression of SARS-CoV-2 Receptor, ACE2, and TMPRSS2 Is Lower in Children Than Adults and Increases with Smoking and COPD. Mol. Ther. Methods Clin. Dev. 2020, 18, 1–6. [Google Scholar] [CrossRef] [PubMed]
- WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19-28 February 2020. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---28-february-2020 (accessed on 19 February 2021).
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Pesu, M.; Watford, W.T.; Wei, L.; Xu, L.; Fuss, I.; Strober, W.; Andersson, J.; Shevach, E.M.; Quezado, M.; Bouladoux, N.; et al. T-cell-expressed proprotein convertase furin is essential for maintenance of peripheral immune tolerance. Nat. Cell Biol. 2008, 455, 246–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oksanen, A.; Aittomäki, S.; Jankovic, D.; Ortutay, Z.; Pulkkinen, K.; Hämäläinen, S.; Rokka, A.; Corthals, G.L.; Watford, W.T.; Junttila, I.; et al. Proprotein convertase FURIN constrains Th2 differentiation and is critical for host resistance against Toxoplasma gondii. J. Immunol. 2014, 193, 5470–5479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izaguirre, G. The Proteolytic Regulation of Virus Cell Entry by Furin and Other Proprotein Convertases. Viruses 2019, 11, 837. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6784293/ (accessed on 19 February 2021). [CrossRef] [Green Version]
- Kieny, M.P.; Lathe, R.; Riviére, Y.; Dott, K.; Schmitt, D.; Girard, M.; Montagnier, L.; Lecocq, J.-P.; Kleny, M. Improved antigenicity of the HIV env protein by cleavage site removal. Protein Eng. Des. Sel. 1988, 2, 219–225. [Google Scholar] [CrossRef] [Green Version]
- Stieneke-Gröber, A.; Vey, M.; Angliker, H.; Shaw, E.; Thomas, G.; Roberts, C.; Klenk, H.; Garten, W. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 1992, 11, 2407–2414. [Google Scholar] [CrossRef]
- Volchkov, V.E.; Volchkova, V.A.; Ströher, U.; Becker, S.; Dolnik, O.; Cieplik, M.; Garten, W.; Klenk, H.D.; Feldmann, H. Proteolytic processing of Marburg virus gly-coprotein. Virology 2000, 268, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Junjhon, J.; Lausumpao, M.; Supasa, S.; Noisakran, S.; Songjaeng, A.; Saraithong, P.; Chaichoun, K.; Utaipat, U.; Keelapang, P.; Kanjanahaluethai, A.; et al. Differential Modulation of prM Cleavage, Extracellular Particle Distribution, and Virus Infectivity by Conserved Residues at Nonfurin Consensus Positions of the Dengue Virus pr-M Junction. J. Virol. 2008, 82, 10776–10791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Day, P.M.; Schiller, J.T. The role of furin in papillomavirus infection. Future Microbiol. 2009, 4, 1255–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, K.; Kim, K.-H.; Lok, A.S.-F.; Tong, S. Characterization of Genotype-Specific Carboxyl-Terminal Cleavage Sites of Hepatitis B Virus e Antigen Precursor and Identification of Furin as the Candidate Enzyme. J. Virol. 2009, 83, 3507–3517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De La Vega, M.-A.; Wong, G.; Kobinger, G.P.; Qiu, X. The Multiple Roles of sGP in Ebola Pathogenesis. Viral. Immunol. 2015, 28, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Vähätupa, M.; Aittomäki, S.; Cordova, Z.M.; May, U.; Prince, S.; Uusitalo-Järvinen, H.; Järvinen, T.A.; Pesu, M. T-cell-expressed proprotein convertase FURIN inhibits DMBA/TPA-induced skin cancer development. Oncoimmunology 2016, 5, e1245266. [Google Scholar] [CrossRef] [Green Version]
- Declercq, J.; Brouwers, B.; Pruniau, V.P.E.G.; Stijnen, P.; Tuand, K.; Meulemans, S.; Prat, A.; Seidah, N.G.; Khatib, A.B.; Creemers, J.W.M. Liver-Specific Inactivation of the Proprotein Convertase FURIN Leads to Increased Hepatocellular Carcinoma Growth. BioMed Res. Int. 2015, 2015, e148651. Available online: https://www.hindawi.com/journals/bmri/2015/148651/ (accessed on 19 February 2021). [CrossRef] [Green Version]
- Huang, Y.-H.; Lin, K.-H.; Liao, C.-H.; Lai, M.-W.; Tseng, Y.-H.; Yeh, C.-T. Furin Overexpression Suppresses Tumor Growth and Predicts a Better Postoperative Disease-Free Survival in Hepatocellular Carcinoma. PLoS ONE 2012, 7, e40738. [Google Scholar] [CrossRef] [PubMed]
- Tomé, M.; Pappalardo, A.; Soulet, F.; López, J.J.; Olaizola, J.; Leger, Y.; Dubreuil, M.; Mouchard, A.; Fessart, D.; Delom, F.; et al. Inactivation of Proprotein Convertases in T Cells Inhibits PD-1 Expression and Creates a Favorable Immune Microenvironment in Colorectal Cancer. Cancer Res. 2019, 79, 5008–5021. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Khatib, A.-M.; Creemers, J.W. Loss of the proprotein convertase Furin in T cells represses mammary tumorigenesis in oncogene-driven triple negative breast cancer. Cancer Lett. 2020, 484, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Van der Veeken, J.; Gonzalez, A.J.; Cho, H.; Arvey, A.; Hemmers, S.; Leslie, C.S.; Rudensky, A.Y. Memory of Inflammation in Regulatory T Cells. Cell 2016, 166, 977–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fathi, N.; Rezaei, N. Lymphopenia in COVID-19: Therapeutic opportunities. Cell Biol. Int. 2020, 44, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Gong, E.; Zhang, B.; Zheng, J.; Gao, Z.; Zhong, Y.; Zou, W.; Zhan, J.; Wang, S.; Xie, Z.; et al. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005, 202, 415–424. [Google Scholar] [CrossRef]
- Wang, H.; Li, N.; Hong, J.; Luo, W.; Yan, Z.; Wang, X.; Zhu, Y.; Shen, Y. Relationship between genetic variation of furin gene and hypercho-lesterolemia and hyper-low-density lipoprotein cholesterolemia in Kazakh general population. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 2014, 36, 168–175. [Google Scholar]
- Cheng, Z.; Zhou, J.; To, K.K.-W.; Chu, H.; Li, C.; Wang, D.; Yang, D.; Zheng, S.; Hao, K.; Bossé, Y.; et al. Identification ofTMPRSS2as a Susceptibility Gene for Severe 2009 Pandemic A(H1N1) Influenza and A(H7N9) Influenza. J. Infect. Dis. 2015, 212, 1214–1221. [Google Scholar] [CrossRef] [Green Version]
- Asselta, R.; Paraboschi, E.M.; Mantovani, A.; Duga, S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging 2020, 12, 10087–10098. [Google Scholar] [CrossRef]
- García-Álvarez, M.; Berenguer, J.; Jiménez-Sousa, M.Á.; Pineda-Tenor, D.; Aldámiz-Echevarria, T.; Tejerina, F.; Diez, C.; Vázquez-Morón, S.; Resino, S. Mx1, OAS1 and OAS2 polymorphisms are associated with the severity of liver disease in HIV/HCV-coinfected patients: A cross-sectional study. Sci. Rep. 2017, 7, 41516. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5282518/ (accessed on 19 February 2021). [CrossRef] [Green Version]
- Fitzgerald, L.M.; Agalliu, I.; Johnson, K.; Miller, M.A.; Kwon, E.M.; Hurtado-Coll, A.; Fazli, L.; Rajput, A.B.; Gleave, M.E.; Cox, M.E.; et al. Association of TMPRSS2-ERG gene fusion with clinical characteristics and outcomes: Results from a population-based study of prostate cancer. BMC Cancer 2008, 8, 230. [Google Scholar] [CrossRef]
- Giri, V.N.; Ruth, K.; Hughes, L.; Uzzo, R.G.; Chen, D.Y.; Boorjian, S.A.; Viterbo, R.; Rebbeck, T.R. Racial differences in prediction of time to prostate cancer diagnosis in a prospective screening cohort of high-risk men: Effect of TMPRSS2 Met160Val. BJU Int. 2010, 107, 466–470. [Google Scholar] [CrossRef] [Green Version]
- Lopera Maya, E.A.; van der Graaf, A.; Lanting, P.; van der Geest, M.; Fu, J.; Swertz, M.; Franke, L.; Wijmenga, C.; Deelen, P.; Zhernakova, A.; et al. Lack of Association Between Genetic Variants at ACE2 and TMPRSS2 Genes Involved in SARS-CoV-2 Infection and Human Quantitative Phenotypes. Front. Genet 2020, 11, 613. Available online: https://www.frontiersin.org/articles/10.3389/fgene.2020.00613/full (accessed on 19 February 2021). [CrossRef]
- Yang, X.; Yang, Q.; Wang, Y.; Wu, Y.; Xu, J.; Yu, Y.; Shang, Y. Thrombocytopenia and its association with mortality in patients with COVID-19. J. Thromb. Haemost. 2020, 18, 1469–1472. [Google Scholar] [CrossRef]
- Chaoxin, J.; Daili, S.; Yanxin, H.; Ruwei, G.; Chenlong, W.; Yaobin, T. The influence of angiotensin-converting enzyme 2 gene polymorphisms on type 2 diabetes mellitus and coronary heart disease. Eur. Rev. Med. Pharmacol. Sci. 2013, 17, 2654–2659. [Google Scholar] [PubMed]
- Yang, M.; Zhao, J.; Xing, L.; Shi, L. The association between angiotensin-converting enzyme 2 polymorphisms and essential hypertension risk: A meta-analysis involving 14,122 patients. J. Renin Angiotensin Aldosterone Syst. 2015, 16, 1240–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.-H.; Li, J.-Y.; Wang, C.; Zhang, L.-M.; Qiao, H. TheACE2G8790A Polymorphism: Involvement in Type 2 Diabetes Mellitus Combined with Cerebral Stroke. J. Clin. Lab. Anal. 2016, 31, e22033. [Google Scholar] [CrossRef] [PubMed]
- Pinto, B.G.G.; Oliveira, A.E.R.; Singh, Y.; Jimenez, L.; Gonçalves, A.N.A.; Ogava, R.L.T.; Creighton, R.; Peron, J.P.S.; Nakaya, H.I. ACE2 Expression is Increased in the Lungs of Patients with Comorbidities Associated with Severe COVID-19. medRxiv 2020. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7276054/ (accessed on 19 February 2021).
- Chiu, R.W.; Tang, N.L.; Hui, D.S.; Chung, G.T.; Chim, S.; Chan, K.A.; Sung, Y.-M.; Chan, L.Y.; Tong, Y.-K.; Lee, W.-S.; et al. ACE2 Gene Polymorphisms Do Not Affect Outcome of Severe Acute Respiratory Syndrome. Clin. Chem. 2004, 50, 1683–1686. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Liu, C.; Guan, T.; Li, Y.; Lai, Y.; Li, F. Association of ACE2 genetic polymorphisms with hypertension-related target organ damages in south Xinjiang. Hypertens. Res. 2019, 42, 681–689. [Google Scholar] [CrossRef] [Green Version]
- Simmons, G.; Zmora, P.; Gierer, S.; Heurich, A.; Pöhlmann, S. Proteolytic activation of the SARS-coronavirus spike protein: Cutting enzymes at the cutting edge of antiviral research. Antivir. Res. 2013, 100, 605–614. [Google Scholar] [CrossRef]
- Glowacka, I.; Bertram, S.; Müller, M.A.; Allen, P.D.; Soilleux, E.J.; Pfefferle, S.; Steffen, I.; Tsegaye, T.S.; He, Y.; Gnirss, K.; et al. Evidence that TMPRSS2 Activates the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Membrane Fusion and Reduces Viral Control by the Humoral Immune Response. J. Virol. 2011, 85, 4122–4134. [Google Scholar] [CrossRef] [Green Version]
- Agoulnik, I.; Bingman, W.E.; Nakka, M.; Li, W.; Wang, Q.; Liu, X.S.; Brown, M.; Weigel, N.L. Target Gene-Specific Regulation of Androgen Receptor Activity by p42/p44 Mitogen-Activated Protein Kinase. Mol. Endocrinol. 2008, 22, 2420–2432. [Google Scholar] [CrossRef] [Green Version]
- Grimes, J.M.; Grimes, K.V. p38 MAPK inhibition: A promising therapeutic approach for COVID-19. J. Mol. Cell. Cardiol. 2020, 144, 63–65. [Google Scholar] [CrossRef]
- Lu, L.; Liu, Q.; Zhu, Y.; Chan, K.-H.; Qin, L.; Li, Y.; Wang, Q.; Chan, J.F.-W.; Du, L.; Yu, F.; et al. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014, 5, 3067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Luo, S.; Libby, P.; Shi, G.-P. Cathepsin L-selective inhibitors: A potentially promising treatment for COVID-19 patients. Pharmacol. Ther. 2020, 213, 107587. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Schroeder, S.; Kleine-Weber, H.; Müller, M.A.; Drosten, C.; Pöhlmann, S. Nafamostat Mesylate Blocks Activation of SARS-CoV-2: New Treatment Option for COVID-19. Antimicrob. Agents Chemother. 2020, 64, e00754-20. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, H.; Pyrc, K.; Van Der Hoek, L.; Geier, M.; Berkhout, B.; Pöhlmann, S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA 2005, 102, 7988–7993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, M.J.; Dorfman, T.; Li, W.; Wong, S.K.; Li, Y.; Kuhn, J.H.; Coderre, J.; Vasilieva, N.; Han, Z.; Greenough, T.C.; et al. Retroviruses Pseudotyped with the Severe Acute Respiratory Syndrome Coronavirus Spike Protein Efficiently Infect Cells Expressing Angiotensin-Converting Enzyme 2. J. Virol. 2004, 78, 10628–10635. [Google Scholar] [CrossRef] [Green Version]
- Purushothaman, K.-R.; Krishnan, P.; Purushothaman, M.; Wiley, J.; Alviar, C.L.; Ruiz, F.J. Expression of angiotensin-converting enzyme 2 and its end product angiotensin 1-7 is increased in diabetic atheroma: Implications for inflammation and neo-vascularization. Cardiovasc. Pathol. 2013, 22, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Turpeinen, H.; Kukkurainen, S.; Pulkkinen, K.; Kauppila, T.; Ojala, K.; Hytönen, V.P.; Pesu, M. Identification of proprotein convertase substrates using genome-wide expression correlation analysis. BMC Genom. 2011, 12, 618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Won, Y.-W.; Lee, M.; Kim, H.A.; Bull, D.A.; Kim, S.W. Post-translational regulated and hypoxia-responsible VEGF plasmid for efficient secretion. J. Control. Release 2012, 160, 525–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.-W.; Chao, T.-L.; Li, C.-L.; Chiu, M.-F.; Kao, H.-C.; Wang, S.-H.; Pang, Y.; Lin, C.; Tsai, Y.; Lee, W.; et al. Furin Inhibitors Block SARS-CoV-2 Spike Protein Cleavage to Suppress Virus Production and Cytopathic Effects. Cell Rep. 2020, 33, 108254. [Google Scholar] [CrossRef] [PubMed]
- Ueland, T.; Holter, J.C.; Holten, A.R.; Müller, K.E.; Lind, A.; Bekken, G.K.; Dudman, S.; Aukrust, P.; Dyrhol-Riise, A.M.; Heggelund, L. Distinct and early increase in circulating MMP-9 in COVID-19 patients with respiratory failure. J. Infect. 2020, 81, e41–e43. [Google Scholar] [CrossRef]
- De Lang, A.; Osterhaus, A.D.M.E.; Haagmans, B.L. Interferon-gamma and interleukin-4 downregulate expression of the SARS coronavirus receptor ACE2 in Vero E6 cells. Virology 2006, 353, 474–481. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.P.; Look, D.C.; Tan, P.; Shi, L.; Hickey, M.; Gakhar, L.; Chappell, M.C.; Wohlford-Lenane, C.; McCray, P.B. Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am. J. Physiol. Cell. Mol. Physiol. 2009, 297, L84–L96. [Google Scholar] [CrossRef] [Green Version]
- Hipp, M.M.; Shepherd, D.; Gileadi, U.; Aichinger, M.C.; Kessler, B.M.; Edelmann, M.J.; Essalmani, R.; Seidah, N.G.; Sousa, C.R.E.; Cerundolo, V. Processing of Human Toll-like Receptor 7 by Furin-like Proprotein Convertases Is Required for Its Accumulation and Activity in Endosomes. Immunity 2013, 39, 711–721. [Google Scholar] [CrossRef] [Green Version]
- Elmorsi, R. The therapeutic potential of targeting ACE2 in COVID-19. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 9744–9747. [Google Scholar]




| RSID | Gene | REF | ALT | AF | AC/AN (Egypt) | Africa | East Asia | European | p-Value | NES | Tissue |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs78164913 | FURIN | T | G | 0.041 | 9 | 50 | 0 | 479 | 1.30 × 10−31 | −1.6 | Lung |
| 220 | 8710 | 1558 | 15,420 | ||||||||
| <0.0001 | <0.0001 | 0.5416 | |||||||||
| rs79742014 | FURIN | C | T | 0.032 | 7 | 48 | 0 | 445 | 3.30 × 10−26 | −1.4 | Lung |
| 220 | 8712 | 1560 | 15,418 | ||||||||
| <0.0001 | <0.0001 | 0.9603 | |||||||||
| rs6226 | FURIN | G | C | 0.682 | 150 | 23,180 | 10,584 | 87,076 | 0.000008 | 0.14 | Esophagus—Mucosa |
| 220 | 24,860 | 19,918 | 128,088 | ||||||||
| 0.0037 | 0.022 | 0.9799 | |||||||||
| rs8039305 | FURIN | T | C | 0.5 | 109 | 7074 | 261 | 7305 | 1.70 × 10−17 | 0.23 | Esophagus—Mucosa |
| 218 | 8688 | 1554 | 15,370 | ||||||||
| <0.0001 | <0.0001 | 0.7117 | |||||||||
| rs464397 | TMPRSS2 | T | C | 0.593 | 128 | 7250 | 1554 | 7458 | 0.00001 | −0.089 | Lung |
| 216 | 8696 | 1558 | 15,392 | ||||||||
| 0.0028 | <0.0001 | 0.0826 | |||||||||
| rs469390 | TMPRSS2 | G | A | 0.537 | 117 | 11,136 | 5216 | 76,812 | 0.0000068 | 0.091 | Lung |
| 218 | 24,930 | 19,938 | 128,862 | ||||||||
| 0.1244 | <0.0001 | 0.3904 | |||||||||
| rs2070788 | TMPRSS2 | G | A | 0.528 | 114 | 6125 | 1034 | 8207 | 8.90 × 10−9 | −0.11 | Lung |
| 216 | 8694 | 1556 | 15,392 | ||||||||
| 0.0155 | 0.0682 | 0.9765 | |||||||||
| rs383510 | TMPRSS2 | T | C | 0.56 | 121 | 5635 | 1051 | 7908 | 1.20 × 10−8 | −0.11 | Lung |
| 216 | 8672 | 1560 | 15,350 | ||||||||
| 0.2161 | 0.14 | 0.5 | |||||||||
| rs112171234 | ACE2 | T | G | 0.043 | 9 | 1159 | 0 | 7 | 0.000041 | −0.85 | Adipose—Visceral (Omentum) |
| 210 | 5882 | 1003 | 10,875 | ||||||||
| <0.0001 | <0.0001 | <0.0001 | |||||||||
| rs12010448 | ACE2 | T | C | 0.023 | 4 | 651 | 0 | 2 | 0.000098 | 0.43 | Muscle—Skeletal |
| 174 | 5757 | 1009 | 10,497 | ||||||||
| 0.0008 | <0.0001 | <0.0001 | |||||||||
| rs200781818 | ACE2 | G | GGGCGCGGTCCTTACGTGT | 0.374 | 74 | 3582 | 785 | 4492 | 2.00 × 10−16 | 0.27 | Nerve—Tibial |
| 198 | 5312 | 836 | 10,036 | ||||||||
| <0.0001 | <0.0001 | 0.2122 | |||||||||
| rs2158082 | ACE2 | G | A | 0.481 | 103 | 5409 | 970 | 5041 | 1.00 × 10−16 | 0.28 | Nerve—Tibial |
| 214 | 5816 | 972 | 10,610 | ||||||||
| <0.0001 | <0.0001 | 0.9632 | |||||||||
| rs4646127 | ACE2 | A | G | 0.563 | 107 | 3834 | 858 | 5756 | 7.50 × 10−9 | 0.2 | Nerve—Tibial |
| 190 | 4926 | 858 | 9249 | ||||||||
| 0.0097 | <0.0001 | 0.4479 | |||||||||
| rs4830974 | ACE2 | G | A | 0.392 | 76 | 4219 | 952 | 5140 | 3.70 × 10−17 | 0.27 | Nerve—Tibial |
| 194 | 5859 | 1004 | 10,670 | ||||||||
| <0.0001 | <0.0001 | 0.1462 | |||||||||
| rs5936011 | ACE2 | C | T | 0.461 | 95 | 4788 | 936 | 4951 | 5.20 × 10−16 | 0.27 | Nerve—Tibial |
| 206 | 5560 | 940 | 10,334 | ||||||||
| <0.0001 | <0.0001 | 0.8084 | |||||||||
| rs5936029 | ACE2 | C | T | 0.441 | 90 | 4761 | 927 | 4921 | 1.40 × 10−14 | 0.26 | Nerve—Tibial |
| 204 | 5504.00 | 931.00 | 10,198.00 | ||||||||
| <0.0001 | <0.0001 | 0.52 | |||||||||
| rs6629110 | ACE2 | T | C | 0.39 | 79.00 | 2459 | 952.00 | 4996.00 | 3.40 × 10−15 | 0.25 | Nerve—Tibial |
| 202.00 | 5772.00 | 989.00 | 10,571.00 | ||||||||
| 0.57 | <0.0001 | 0.18 | |||||||||
| rs6632704 | ACE2 | C | A | 0.35 | 74.00 | 2868 | 944.00 | 5088.00 | 4.60 × 10−16 | 0.26 | Nerve—Tibial |
| 210.00 | 5757.00 | 992.00 | 10,604.00 | ||||||||
| 0.01 | <0.0001 | 0.03 |
| RSID | Gene | REF | ALT | AF | AC/AN (Egypt) | Africa | East Asia | European | Amino Acids | SIFT | POLYPHEN |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs148110342 | FURIN | C | T | 0.014 | 3 | 4 | 0 | 206 | p.Arg81Cys | 0.04 | 0.59 |
| 216 | 24,902 | 19,940 | 128,808 | ||||||||
| <0.0001 | <0.0001 | 0.0003 | |||||||||
| rs150965978 | TMPRSS2 | C | A | 0.004545 | 1 | 83 | 0 | 765 | Intron Variant | ||
| 220 | 8706 | 1560 | 15,408 | ||||||||
| 0.6913 | 0.2541 | 0.0046 | Intron Variant | ||||||||
| rs28401567 | TMPRSS2 | C | T | 0.241 | 53 | 2885 | 1147 | 3161 | - | ||
| 220 | 8696 | 1552 | 15,404 | ||||||||
| 0.0446 | <0.0001 | 0.3371 | |||||||||
| rs2298659 | TMPRSS2 | G | A | 0.125 | 27 | 4592 | 5179 | 28,744 | p.Gly259Gly | synonymous variant | synonymous variant |
| 216 | 23,850 | 19,478 | 122,880 | ||||||||
| 0.0415 | 0.0002 | 0.0024 | |||||||||
| rs17854725 | TMPRSS2 | A | G | 0.427 | 93 | 9066 | 2544 | 67,712 | p.Ile256Ile | synonymous variant | synonymous variant |
| 218 | 23,918 | 19,604 | 122,814 | ||||||||
| 0.3754 | <0.0001 | 0.0438 | |||||||||
| rs12329760 | TMPRSS2 | C | T | 0.188 | 41 | 7265 | 7651 | 29,831 | p.Val160Met | 0 | 0.938 |
| 218 | 24,896 | 19,934 | 128,604 | ||||||||
| 0.0118 | <0.0001 | 0.2485 | |||||||||
| rs3787950 | TMPRSS2 | T | C | 0.185 | 40 | 5000 | 2905 | 9864 | p.Thr75Thr | synonymous variant | synonymous variant |
| 216 | 24,832 | 19,600 | 127,666 | ||||||||
| 0.6889 | 0.2324 | <0.0001 | |||||||||
| rs2285666 | ACE2 | C | T | 0.15600 | 34.00000 | 4057.00000 | 7336.00000 | 17,240.00000 | c.439+4G>A | intron variant | splice region variant |
| 218.00000 | 18,323.00000 | 13,387.00000 | 86,164.00000 | ||||||||
| 0.06890 | <0.0001 | 0.20460 | |||||||||
| rs35803318 | ACE2 | C | T | 0.00935 | 2.00000 | 133.00000 | 0 | 3935.00000 | p.Val749Val | non-coding transcript exon variant | non-coding transcript exon variant |
| 214.00000 | 18,549.00000 | 13,918.00000 | 88,946.00 | ||||||||
| 0.97210 | <0.0001 | 0.02470 | |||||||||
| rs2106809 | ACE2 | A | G | 0.15000 | 32.00000 | 707.00000 | 533.00000 | 1938.00000 | intron variant | ||
| 214.00000 | 5771.00000 | 954.00000 | 10,584.00000 | ||||||||
| 0.35390 | <0.0001 | 0.33090 | |||||||||
| rs4646142 | ACE2 | G | C | 0.15200 | 32.00000 | 1372.00000 | 557.00000 | 2162.00000 | intron variant | ||
| 210.00000 | 5792.00000 | 979.00000 | 10,704.00000 | ||||||||
| 0.0257 | <0.0001 | 0.16410 | |||||||||
| rs714205 | ACE2 | C | G | 0.13200 | 28.00000 | 707 | 544.00000 | 1927.00000 | intron variant | ||
| 212.00000 | 5865 | 969.00000 | 10,714.00000 | ||||||||
| 0.73400 | <0.0001 | 0.15010 | |||||||||
| rs17264937 | ACE2 | T | C | 0.16000 | 32.00000 | 289.00000 | 382.00000 | 3041.00000 | intron variant | ||
| 200.00000 | 5698.00000 | 946.00000 | 10,493.00000 | ||||||||
| <0.0001 | <0.0001 | 0.00220 | |||||||||
| rs5980163 | ACE2 | C | G | 0.02400 | 5.00000 | 13.00000 | 0.00000 | 128.00000 | intron variant | ||
| 212.00000 | 5887.00000 | 997.00000 | 10,850.00000 | ||||||||
| <0.0001 | <0.0001 | 0.22400 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tarek, M.; Abdelzaher, H.; Kobeissy, F.; El-Fawal, H.A.N.; Salama, M.M.; Abdelnaser, A. Bioinformatics Analysis of Allele Frequencies and Expression Patterns of ACE2, TMPRSS2 and FURIN in Different Populations and Susceptibility to SARS-CoV-2. Genes 2021, 12, 1041. https://doi.org/10.3390/genes12071041
Tarek M, Abdelzaher H, Kobeissy F, El-Fawal HAN, Salama MM, Abdelnaser A. Bioinformatics Analysis of Allele Frequencies and Expression Patterns of ACE2, TMPRSS2 and FURIN in Different Populations and Susceptibility to SARS-CoV-2. Genes. 2021; 12(7):1041. https://doi.org/10.3390/genes12071041
Chicago/Turabian StyleTarek, Mohammad, Hana Abdelzaher, Firas Kobeissy, Hassan A. N. El-Fawal, Mohammed M. Salama, and Anwar Abdelnaser. 2021. "Bioinformatics Analysis of Allele Frequencies and Expression Patterns of ACE2, TMPRSS2 and FURIN in Different Populations and Susceptibility to SARS-CoV-2" Genes 12, no. 7: 1041. https://doi.org/10.3390/genes12071041
APA StyleTarek, M., Abdelzaher, H., Kobeissy, F., El-Fawal, H. A. N., Salama, M. M., & Abdelnaser, A. (2021). Bioinformatics Analysis of Allele Frequencies and Expression Patterns of ACE2, TMPRSS2 and FURIN in Different Populations and Susceptibility to SARS-CoV-2. Genes, 12(7), 1041. https://doi.org/10.3390/genes12071041







